Abstract
Carcinogenic metals, such as nickel, arsenic, and chromium, are widespread environmental and occupational pollutants. Chronic exposure to these metals has been connected with increased risks of numerous cancers and as well as non-carcinogenic health outcomes, including cardiovascular disease, neurologic deficits, neuro-developmental deficits in childhood, and hypertension. However, currently the specific molecular targets for metal toxicity and carcinogenicity are not fully understood. Here, we propose that the iron- and 2-oxoglutaratedependent dioxygenase family enzymes, as well as, other histone modifying enzymes are important intracellular targets that mediate the toxicity and carcinogenicity of nickel, and maybe potential targets in chromium and arsenic induced carcinogenesis. Our data demonstrates that all three metals are capable of inducing post-translational histone modifications and affecting the enzymes that modulate them (i.e. the iron- and 2-oxoglutaratedependent dioxygenase family, including HIF-prolyl hydroxylase PHD2, histone demethylase JHDM2A/JMJD1A, and DNA repair enzymes ABH3 and ABH2, and histone methyltransferases, G9a). Given the effects these metals can exert on the epigenome, future studies of their involvement in histone modifying enzymes dynamics would deepen our understanding on their respective toxicities and carcinogenicities.
Introduction
Occupational and/or environmental exposures to metal compounds, such as nickel (Ni), arsenic (As), and hexavalent chromium (CrVI) have been implicated in the development of numerous health outcomes, including various forms of cancer 1–5. However, even though all three are established human carcinogens, the precise mechanism/s by which they exert their carcinogenic effects are not fully understood 6, 7. Since carcinogenic metals typically tend to be fairly weak mutagens, and with the exception of hexavalent chromium, do not directly interact with DNA, numerous studies have alluded to the fact that epigenetic dysregulation may play an important role in metal-induced carcinogenesis 8–10. The epigenetic effect of carcinogenic metals has been one of the focal research interests in my laboratory. This article will review the work conducted in my laboratory on cellular effects, intracellular targets, epigenetic modifications and changes in gene expression brought on by nickel, chromium and arsenic compounds, as well as discuss future directions for studying metal carcinogenesis.
Iron- and 2-oxoglutarate-dependent Dioxygenases
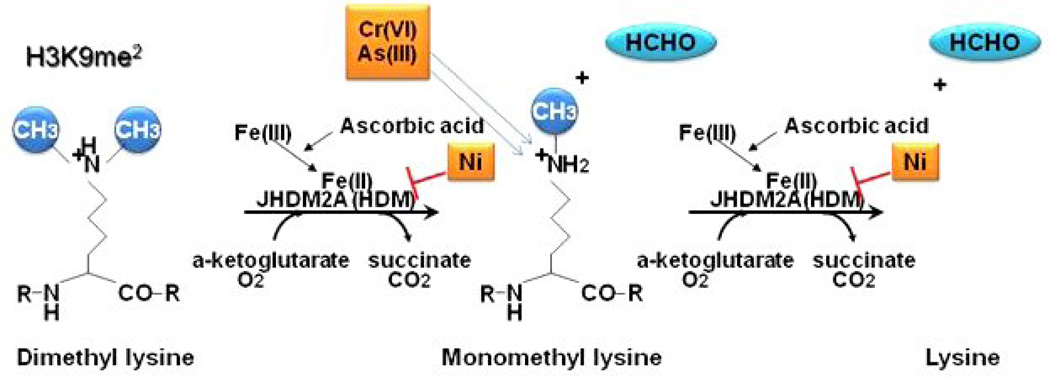
Covalent modifications of core histones play a critical role in chromatin regulation, gene activity and nuclear architecture 11, 12. One such modification, methylation, which can be found on argenine and/or lysine residues, is involved in regulating a wide range of processes, including gene expression, chromatin structure, dosage compensation and epigenetic memory13. JmjC-domain-containing histone demethylases, JHDMs, are a class of enzymes that are capable of dynamically removing histone methylation13. The JmjC domain contains the His/His/carboxylic acid facial triad that binds iron in its catalytic center and is characteristic of all dioxygenases 14. JHDM2A, also known as JMJD1A, belongs to the iron- and 2-oxoglutarate-dependent dioxygenase family and can remove histone H3 lysine 9 (H3K9) mono and dimethylation. The demethylation of H3K9 occurs by catalyzing the generation of highly reactive oxygen species (ROS) in the presence of oxygen, iron (Fe), 2-oxoglutarate, and ascorbic acid. These resultant species attack the methyl groups on histone lysines and produce unstable oxidized intermediates that spontaneously release formaldehyde, resulting in the removal of methyl groups from histone lysines (Figure 1). Given JHDMA2 need for iron and ascorbic acid, it is likely that the replacement of iron from its catalytic center by nickel and depletion of reduced ascorbate by chromate and/or arsenic would attenuate its enzymatic activity in cells.
Figure 1.
Model for iron- and 2-oxoglutarate-dependent dioxygenase dimethylation of lysines.
Nickel
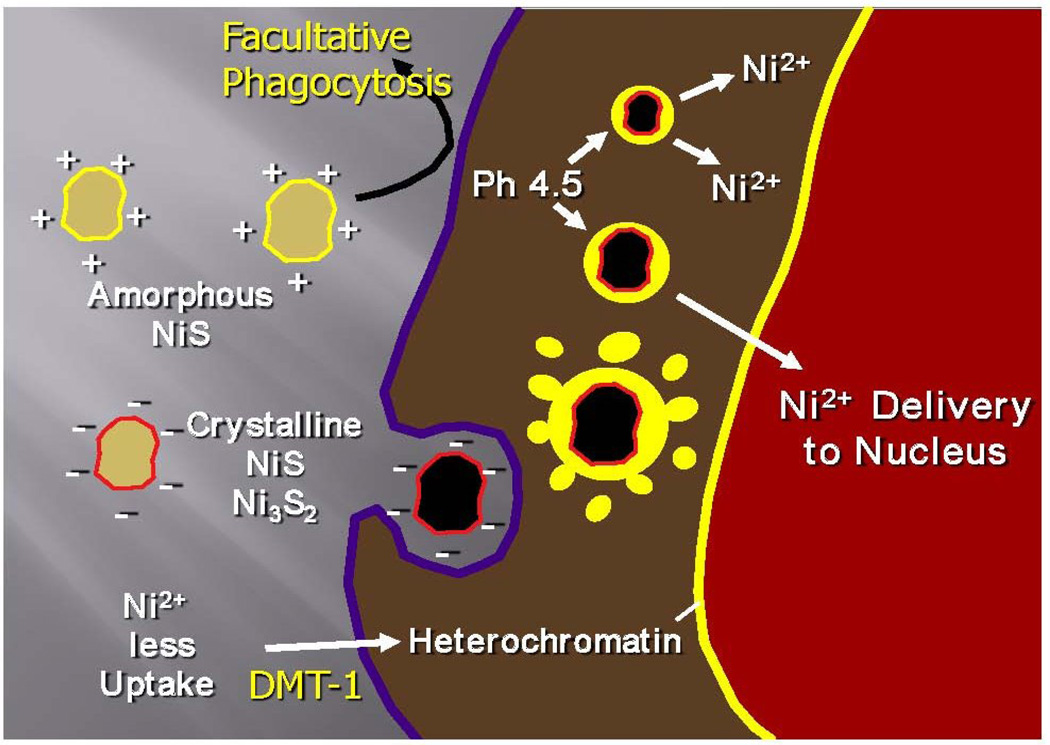
Roughly 150,000 metric tons of nickel, are released into the environment due to volcanic dusts and rock weathering, while approximately 180,000 tons are discharged by anthropogenic sources, including fossil fuel combustion, and industrial use/disposal of nickel compounds and alloys 15, 16. Because of its natural abundance and many uses and applications, the potential for exposure to nickel and its compounds is fairly extensive in both environmental and occupational settings 17. The most potent nickel compounds that cause human cancer via inhalation are those of low water solubility, including crystalline nickel sulfide (NiS), crystalline nickel subsulfide (Ni3S2) and nickel oxides (NOx) 18. Fig. 2 shows the uptake and subcellular distribution of carcinogenic crystalline and non-carciongenic amorphous nickel particles 19. The negatively charged crystalline nickel subsulfide particles, but not the positively charged amorphous nickel sulfide particles, are actively phagocytized by cells. Once phagocytized, these particles form nickel ion (Ni2+) generating intracellular vacuoles that are localized close to the nucleus of the cell. The acidification of these cytoplasmic vacuoles (pH 4.5) accelerates the dissolution of nickel ions from the particle, resulting in the buildup of very high concentrations of the metal inside the cell 20–24. Soluble nickel compounds can also enter cells through the divalent metal transporter DMT1, which is also responsible for transporting iron (Fe) and manganese (Mn) ions 25.
Figure 2.
Model of particular nickel compound uptake and intracellular dissolution.
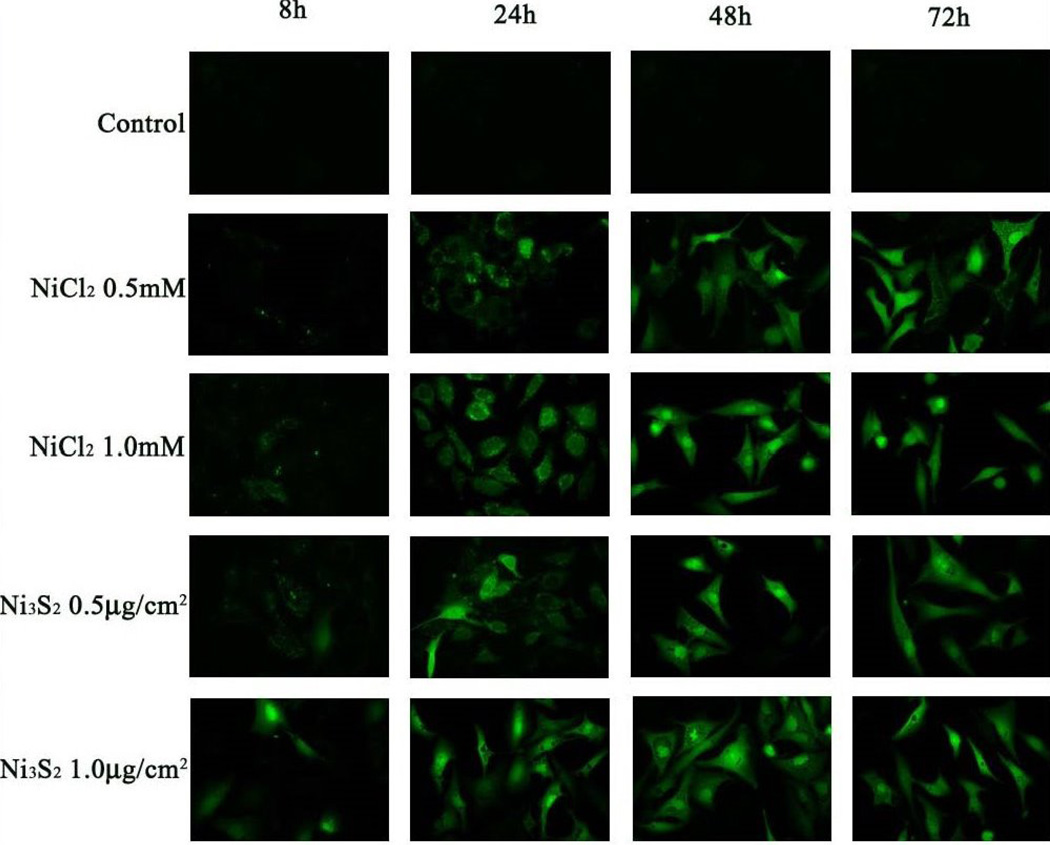
To further investigate the intracellular distribution of ionic nickel in the cell, we utilized a dye that would fluoresce when it binds to ionic nickel ions in the cell. Figure 3 shows the cellular distribution of nickel ions following treatment of human lung cells with either nickel subsulfide (Ni3S2) particles or nickel chloride (NiCl2) using Newport Green Dye. Both water-soluble and insoluble nickel compounds are taken up by cells and accumulate in the cytoplasm and nucleus 26. When the exposure to the nickel was removed, nickel ions from the insoluble form persisted in the nucleus and cytoplasm longer than the ions from the soluble form 26.
Figure 3.
Newport Green fluorescence showing the intracellular distribution of nickel ions following treatment of human lung cells with NiCl2 and Ni3S2. From “Fluorescent tracking of nickel ions in human cultured cells” vol. 219(1) by Ke Q. et al. Copyright 2007 by Toxicology and Applied Pharmacology. Reproduced with permission of Toxicology and Applied Pharmacology via Copyright Clearance Center.
Having worked on nickel for more than three decades, we have now come to understand that one of the major intracellular targets of nickel ions in the cell is the iron- and 2-oxoglutarate-dependent dioxygenase family of enzymes. The first piece of supportive evidence regarding this family of enzymes emerged from our work on nickel-induced hypoxia-mimetic stress and proline hydroxylase domain proteins (PHD), which are responsible for degrading the hypoxia inducible factor-1 alpha (HIF-1 α) 27. These enzymes hydroxylate prolines in the oxygen-dependent domain of (HIF-1 α), recruit the binding a Von-Hippel-Lindau (VHL) E3 ubiquitin ligase complex, and consequently induce the ubiquitin-dependant degradation of (HIF-1 α) 28. Nickel ions are able to stabilize (HIF-1 α) by replacing the iron (Fe) in the HIF-1 alpha proline hydroxylases, resulting in the inhibition of their enzymatic activity and prevention of HIF-1 alpha degradation. We found that the 50% inhibitory concentration (IC50) of nickel was 22 µM for PHD2 in the presence of 100 µM Fe2+, indicating that this enzyme is highly sensitive to nickel 27.
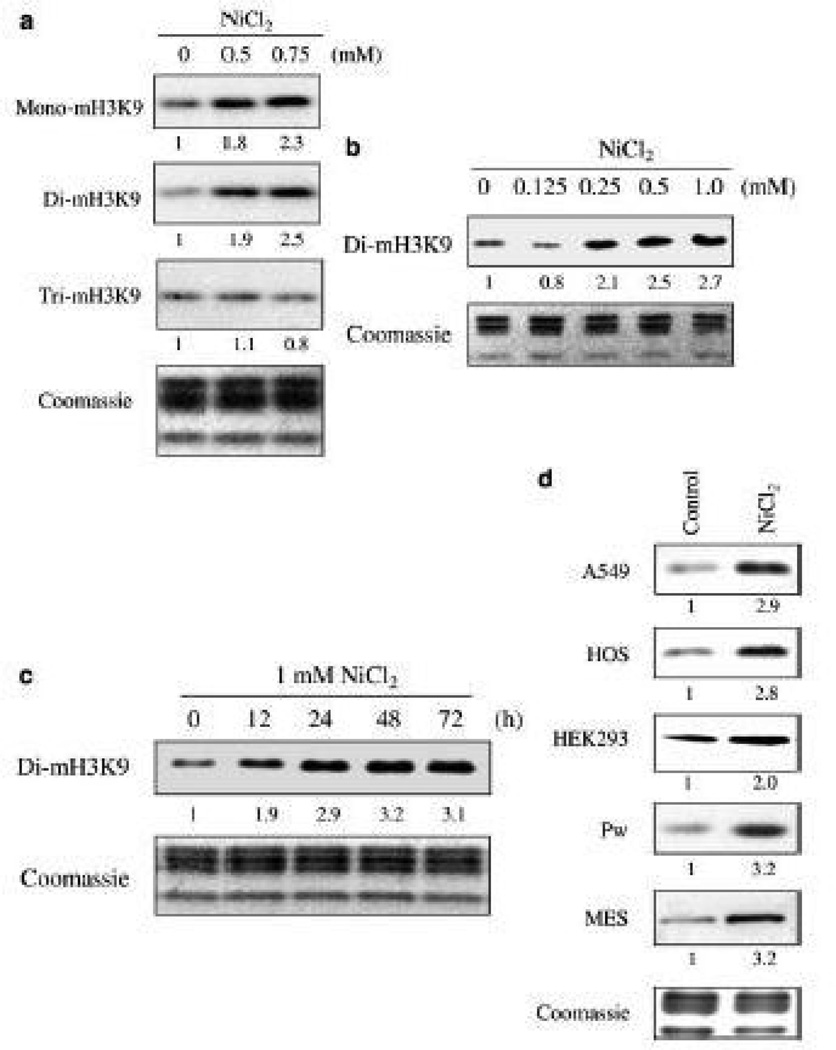
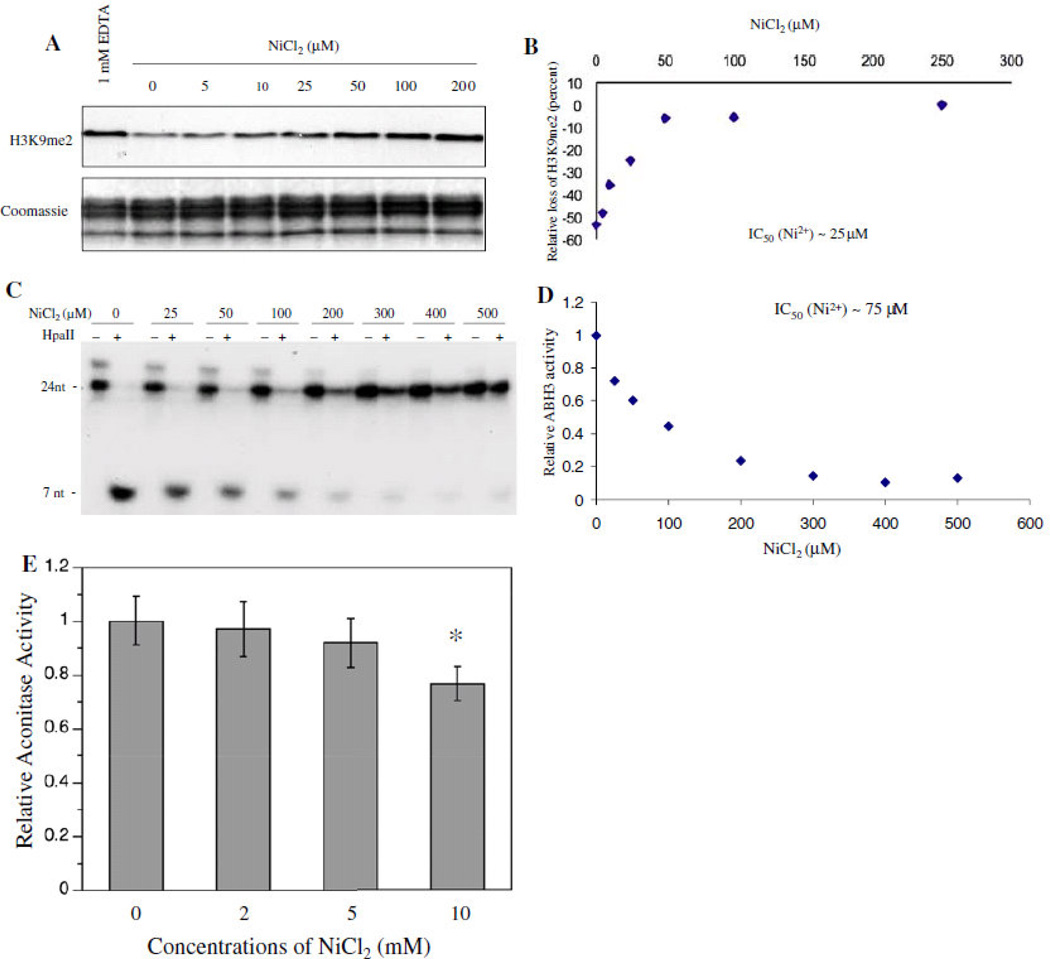
Our studies on epigenetic effects of nickel led to the discovery of a new class of enzymes that belong to the same dioxygenase family as HIF-prolyl hydroxylases, H3K9 demethylases 29. Nickel was able to selectively inhibit this family of enzymes, because the Fe that it replaces is bound to two histidine and a carboxylic acid facial triad at the active site of this enzyme 25, 27.Figure 4 shows that acute Ni ion exposure resulted in an increase in global H3K9me1 and H3K9me2, both of which are critical marks for DNA methylation and long-term gene silencing, in several different cell lines 29, 30. Given these marks’ role in DNA methylation and gene silencing, we also examined the affinity of nickel ions for other deoxygenases, such as JHDM2A/JMJD1A and ABH2 and ABH3, human homologues of bacterial DNA repair enzymes Alkb (ABH1-9). We found that both JHDM2A/JMJD1A and ABH2/3 activities are highly sensitive to nickel inhibition 31, 32. Once bound, the nickel ions replaced the ferrous iron in the catalytic center of these enzymes and inhibited their activity in intact cells (Figure 5) 33.
Figure 4.
The changes of global histone H3K9 methylation following nickel ion exposure. (a) A549 cells were exposed to 0.5 mM or 0.75 mM NiCl2 for 24 h. (b) A dose-dependent increase of global H3K9 dimethylation by nickel ions. A549 cells were exposed to various concentrations of NiCl2 for 24 h. (c) A time course study on global H3K9 dimethylation following nickel ion exposure. A549 cells were exposed to 1 mM NiCl2 for selected time intervals as indicated. (d) Exposure to 1 mM NiCl2 for 24 h increased H3K9 dimethylation in different cell types. HOS, human osteosarcoma; MES, murine embryonic stem. Histones were extracted and separated in a 15% SDS-polyacrylamide gel and immunoblotted with various antibodies as indicated. Loading of the histones in all gels was assessed using Coomassie blue staining. From “Ions Increase Histone H3 Lysine 9 Dimethylation and Induce Transgene Silencing” vol. 26(10) by Haobin Chen et al. Copyright 2006 by Molecular and Cellular Biology. Reproduced with permission of Molecular and Cellular Biology via Copyright Clearance Center
Figure 5.
A kinetic study on Ni inhibition of JHDM2A and ABH3 demethylase activity. (a) Purified Flag- JHDM2A was assayed for its demethylase activity in the presence of different concentrations of Ni(II) ions as indicated. The assay with addition of EDTA, a chelator of divalent metals, was performed in parallel as a negative control. (a) Data quantification of (a). (c) Purified 69His-tagged ABH3 was assayed for its demethylase activity in the presence of different concentrations of Ni(II) ions as indicated. (d) Data quantification of (c). (e) Purified aconitase, an Fe–S cluster-containing enzyme, was incubated with different concentrations of Ni(II) ions for 4 h. After incubation, the aconitase activity was measured immediately as previously described. Aconitase activity is presented as that relative to levels in the control samples. Each bar represents the mean (±SD) from three samples per treatment. *Statistically significant change (P<0.05) compared to control samples. From “Iron- and 2-oxoglutarate-dependent Dioxygenases:an emerging group of molecular targets for nickel toxicity and carcinogenicity” vol. 22 by Chen H. and Costa M. Copyright 2009 by Biometals. Reproduced with permission of Biometals via Copyright Clearance Center
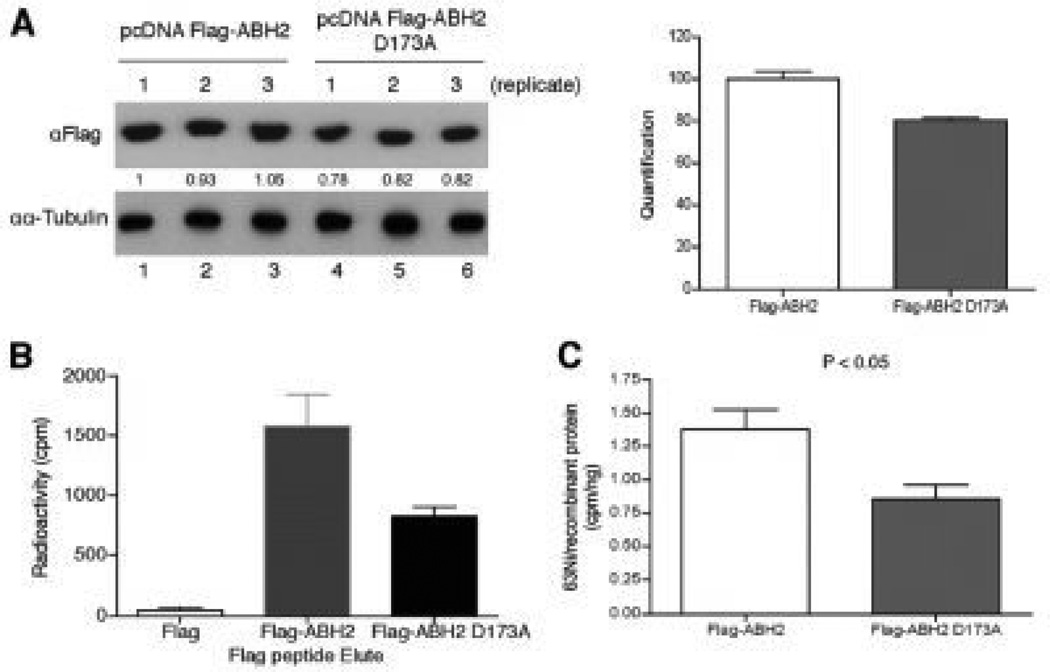
Using purified JHDM2A and ABH3 recombinant proteins we were able to show that in the presence of iron, nickel was able to inhibit the demethylase activities of both JHDM2A and 69His-ABH3 in a dose-dependent manner. On the other hand, the purified aconitase, a Krebs cycle enzyme that binds iron in a form of iron-sulfur cluster, remained uninhibited by up to 5 mM nickel choride (NiCl2)31 (Figure 6). We also showed that Ni preferentially binds to ABH2 with a dissociationconstant of 1.7 µM, compared to 4.5 µM for Fe. Furthermore, x-ray absorption spectroscopy studies have shown that Ni binds to the dioxygenases at the same sites as iron, but unlike iron, nickel assumes a 6 coordinate geometry preventing oxygen binding, while iron binds with a 5 coordinate geometry allowed oxygen to bind 34. The consequences of nickel’s ability to interact with the active sites of these enzymes and inhibit their activity can, as in the case of JHMD2A/ JMJD1A, result in the accumulation of the silencing mark H3K9me2 in promoters of genes and loss of their transcription 29, 32.
Figure 6.
Nickel ions bind to the iron-binding site ofABH2in cells. (a) measurement of FLAG-ABH2 and ABH2(D173A) expression levels in the nickel-treated 293T cells. 293T cells were transiently transfected with FLAG-ABH2 and FLAG-ABH2(D173A) expression vectors and then treated with 1mMNiCl2 that contained 0.22 mCi of 63NiCl2. Expression of FLAG-ABH2 or ABH2(D173A) in cell lysates was measured by Western blot using anti-FLAG antibody. The intensity of bands was quantified using ImageJ software and marked below the graph. The quantification results were graphed on the right. (b), cell lysates collected in (a) were subject to IP with anti-FLAG resin. The FLAG-tagged recombinant proteins were eluted with FLAG peptide, and their associated radioactivity was measured. (c) 63Ni-specific radioactivity associated with FLAG-ABH2 or ABH2(D173A) was calculated. The experiment was conducted in triplicate, and values are means_S.D. for triplicates. The difference in 63Ni-specific radioactivity between FLAG-ABH2 and FLAG-ABH2(D173A) samples is statistically significant because a two-tailed Student t test analysis gives a p value of 0.044. From “Nickel Ions Inhibit Histone Demethylase JMJD1A and DNA Repair Enzyme ABH2 by Replacing the Ferrous Iron in the Catalytic Centers” vol. 285(10) by Chen et al. Published, JBC Papers in Press 2009.
Having seen the effects that nickel can exert on the global post-translational histone modifications, as well as, on the enzymes that modulate them, we wanted to investigate its effects on a gene specific level. Therefore, we performed Chip-on-chip experiments on nickel chloride (NiCl2) treated human bronchial epithelial cells and found that both H3K9me2, a mark of transcriptional repression, and H3K4me3, a mark of transcriptional activation, were increased in the promoter regions of several genes (Table 1). The increase of H3K9me2 in gene promoters of IL12B, C10orf86, CD3EAP, and CCT7, was associated with a decrease in gene expression of these genes. CD3EAP and IL12B, are of particular interest, since CD3EAP is a DNA-dependent RNA polymerase that catalyzes the transcription of DNA into RNA, and IL12B is a cytokine involved in the immune response. An increase of H3K4me3 in the promoters of genes such as NDRG1, CA9, STC2 and EGLN3, however, was associated with increased expression. Interestingly, both NRDG1 and CA9 are hypoxia inducible genes that have been implicated in nickel carcinogenesis, tumor-associated cell migration and invasion, tumor microenvironment and survival, and whose increased expression can be found in variety of cancers 35–39. Moreover, our most recent work on genome-wide mapping of H3K4me3 by chromatin immunoprecipitation and direct genome sequencing (ChIP-seq) and transcriptome genome-wide mapping of RNA transcripts by massive parallel sequencing of cDNA (RNA-seq) in nickel chloride (NiCl2) treated A549 cells, has also identified CA9 and NDRG1 as two of highest nickel-induced genes that had an increased level of H3K4me3 in both the promoters and coding regions 37.
Table 1.
| (a) Correlation of increases of H3K9me2 in gene promoter and Gene expression | ||||||
|---|---|---|---|---|---|---|
| Strand | Annotation | Symbol | Chromosome | Start | End | Expression |
| − | IL12B | NM_002187 | 5 | 158694368 | 158694540 | ↓8.4 |
| − | C10orf86 | NM_01761 | 10 | 123730785 | 123730944 | ↓3.3 |
| + | CD3EAP | NM_01209 | 5 | 96056126 | 96056327 | ↓3.5 |
| + | CCT7 | NM_01590 | 7 | 100312376 | 100312519 | ↓3.00 |
| (b) Correlation of increases of H3K4me3 in gene promoter and Gene expression | ||||||
|---|---|---|---|---|---|---|
| strand | annotation | Symbol | Chromosome | Start | End | Expression |
| + | NDRG1 | NM_006096 | 8 | 134376200 | 134377500 | ↑21.2 |
| + | CA9 | NM_001216 | 9 | 35663914 | 35671151 | ↑6.5 |
| + | STC2 | NM_003714 | 5 | 172685450 | 172685600 | ↑3.5 |
| + | EGLN3 | NM_022073 | 14 | 33463173 | 33490036 | ↑3.3 |
In addition to inducing a myriad of epigenetic effects in vitro, nickel can also altered epigenetic modifications and subsequent effects on gene transcription in a human population. In a study of 120 males subjects with occupation and environmental exposures to nickel, we found that urinary nickel was elevated in subjects with occupational exposure (p = 0.0006). We also found that the global levels of H3K4me3 were elevated (0.25%±0.11%, 0.15%±0.04%, p=0.0004) and H3K9me2 were decreased (0.11%±0.05%, 0.15%±0.04%, p=0.003) in Ni-exposed subjects. H3K4me3 was positively (r=0.4, p=0.0008) and H3K9ac was negatively (r=0.1, p=0.01) associated with urinary nickel. Furthermore, our results indicated that temporal variability within individuals was relatively small, compared to variability between subjects, suggesting that global H3K4me3, H3K9ac, and H3K9me2 histone modifications are relatively stable over time in human peripheral blood mononuclear cells (PBMC)s from both nickel-exposed and referent subjects40.
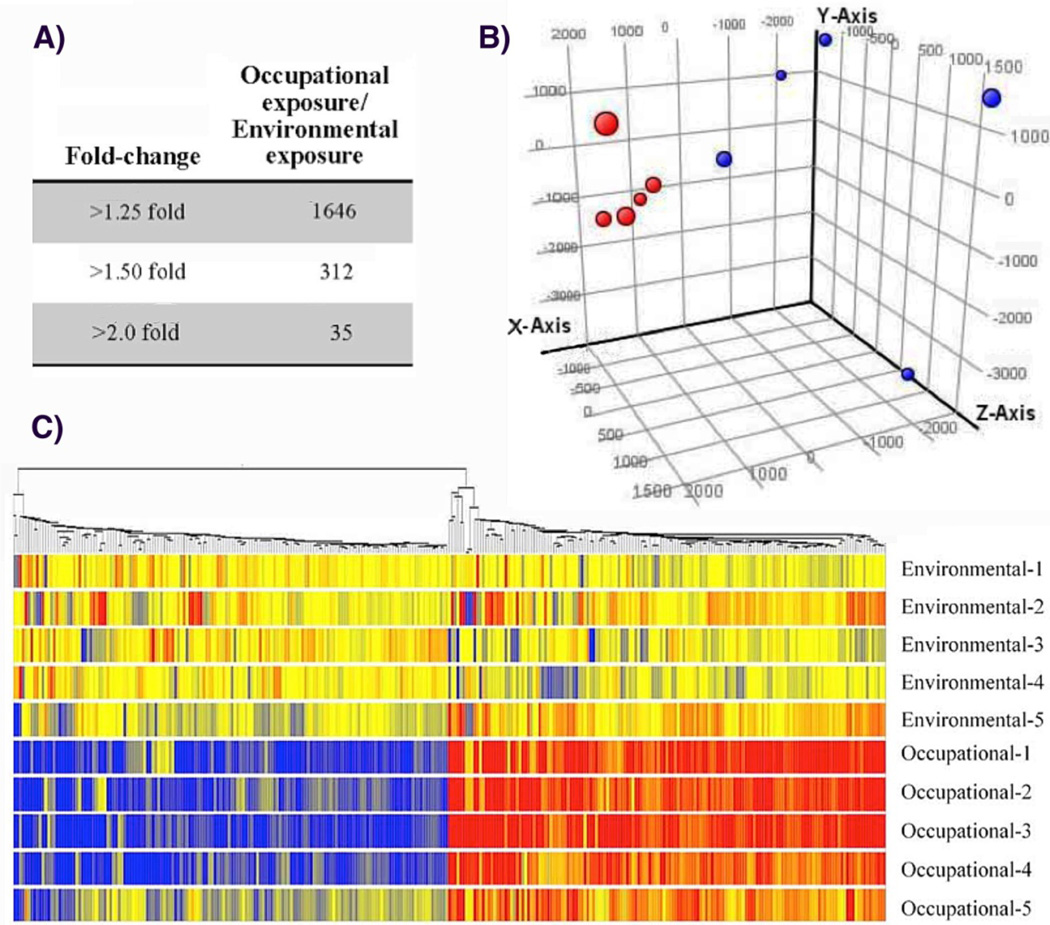
Having found statistically significant differences in nickel associated post-translational histone modifications in environmentally and occupationally exposed subjects, we also wanted to in investigate their gene expression profiles. Therefore, we selected a total of 10 subjects (5 high / 5 low) based on urinary nickel, with no significant differences in age, self-reported, data on smoking habits, or urinary cotinine. We analyzed the gene expression profiles of PBMCs from both exposure groups using Affymetrix Human Gene 1.0 ST Array containing 28,869 well-annotated genes. As shown in Figure 7A, a total of 1646 genes in PBMCs from subjects with occupational exposure displayed a greater than 1.25-fold difference in all subjects when compared with the expression in PBMCs of subjects with environmental exposure. The numbers of gene entities decreased to 312 and 35 when the cut-off threshold was increased to 1.50 and 2.0-fold difference, respectively. The gene expression profiles of subjects with occupational exposure to nickel showed a clear separation from subjects with environmental exposure, the gene expression profiles in subjects with occupational exposure clustered more closely with other subjects with occupational exposure to nickel than subjects with environmental exposure. This separation was observed with Principal Component Analysis (PCA) of the microarray data (Figure 7B, red vs. blue). A similar separation was observed with hierarchical clustering analysis of genes changed more than 1.5-fold in all subjects with occupational exposure, in which samples were sorted based on the similarity of gene expression (Figure 7C). After elimination of the probe sets that represented unannotated genes, a total of 31 genes were changed more than 2- fold in all subjects with occupational exposure compared with subjects with environmental exposure, including 16 down-regulated and 15 up-regulated genes. The function of the majority of the nickel-induced and repressed genes by in vivo exposure to nickel is related to cytokine-cytokine interaction and chemokine signaling (CCL20, CCR2, CX3CR1, IL1A, IL6, IL1RN, TNFSF10, IL8RB, and IL8RA).
Figure 7.
Gene expression profiles of PBMCs of subjects with occupational exposure compared to subjects with environmental exposure to nickel. (a) The number of entities with more than 1.25-, 1.5-, and 2-fold change in expression level with pairwise comparison. (b) Principal component analysis revealed distinct separation between subjects with environmental exposure to low levels of nickel (red circle0 and subjects with occupational exposure to high levels of nickel (blue circle). (c) Hierarchical cluster analysis of genes with more than 1.5-fold changes expression in PBMCs of all 5 subjects occupationally exposed to nickel compared to subjects with environmental exposure. The color bar related color code to the expression value determined after quantile normalization and baseline transformation to the median levels of all samples.
Chromium
Chromium (Cr) is a well-established carcinogen that is a contaminant at half of the EPA Superfund sites in the United States41. Hexavalent chromium compounds are well established human respiratory carcinogens and have also been shown to elevate other types of human cancers 42–44. Hexavalent chromium, Cr(VI), is one of the few carcinogenic metals that can actually react with DNA, form adducts, and induce mutations45, 46 47, 48. It is an oxyanion that resembles a sulphate and phosphate and is actively transported into all cells of the body by the oxyanion transporters44, 49. However, the hexavalent chromium inside the cell is rapidly reduced by ascorbic acid and other intracellular reductants to trivalent Cr (CrIII), which reacts with proteins and DNA to produce toxicity and carcinogenicity 44.
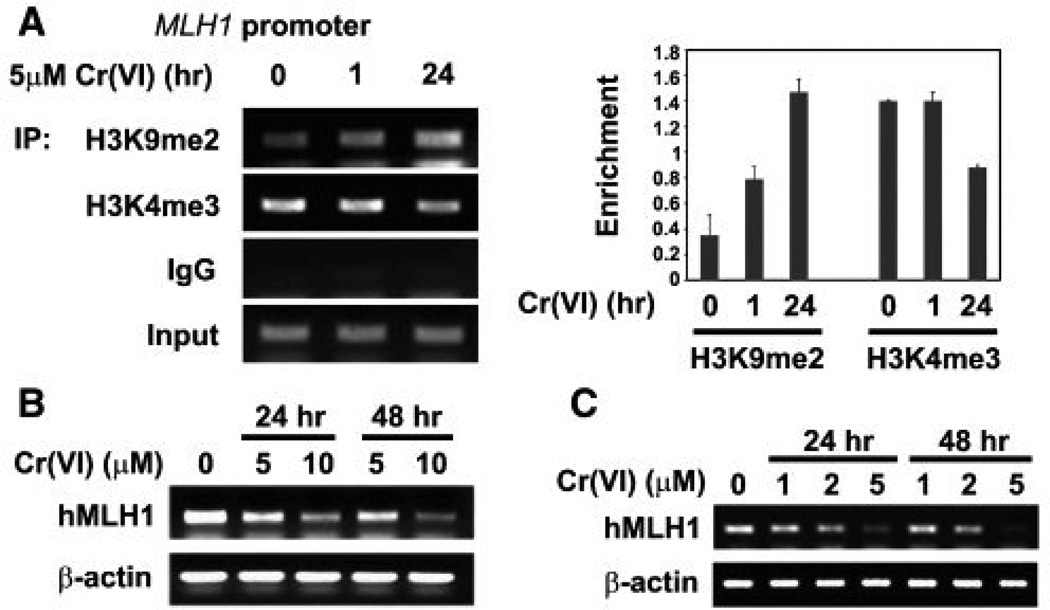
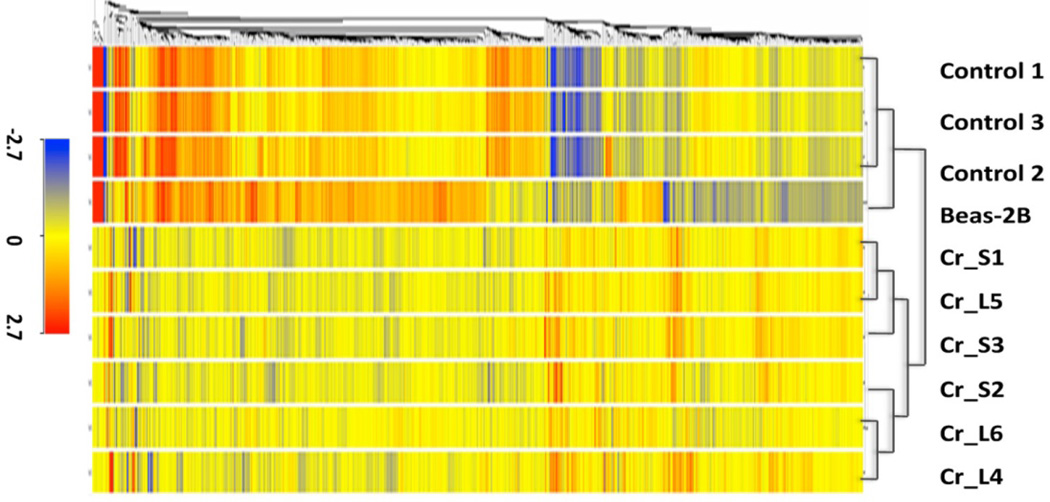
Like nickel, chromium also appears to have epigenetic effects. When we exposed human lung cells to hexavalent chromium, we observed an increase in the global levels of H3K9me2, H3K9me3 and H3K4me3 50, 51. Hexavalent chromium exposure can also decrease gene expression via post-translational histone modifications. Specifically, we found that hexavalent chromium exposure decreases the expression of mismatch repair gene, MLH1, by increasing the levels of the silencing mark H3K9me2, while decreasing the activating mark H3K4me3 in the promoter of MLH1 in A549 cells and BEAS-2B cells (Figure 8) 51. Furthermore, we examined the gene expression patters of chromium-transformed normal human bronchial epithelial BEAS-2B cells and identified 409 differentially expressed in hexavalent chromium transformed cells compared to control cells. As can be seen in Figure 9, the gene expression profiles of six hexavalent chromium transformed cell lines were remarkably similar to each other yet markedly differed from that of either control clones from untreated BEAS-2B cells that spontaneously grew in soft agar or normal BEAS-2B cells. Genes that were either up-regulated or down-regulated in all hexavalent chromium transformed cells were either associated with cell-to-cell junction or interaction between cells and their extracellular matrices, respectively. Expression of genes involved in cell proliferation and apoptosis were also changed 52.
Figure 8.
Cr(VI) exposure increased G9a protein levels. A549 cells were treated with 5 or 10 µM of Cr(VI) for 24 hours. Total protein lysates were extracted and analyzed with antibody against G9a. Antibody against tubulin was used to assess the loading of proteins. (b) A549 cells were exposed to 5 or 10 µM Cr(VI) for 24 hours. The global levels of H3K9me2 were measured using specific antibodies. Coomassie blue staining was used to assess the equal loading of the histones. (C and D) Chromate exposure increased G9a mRNA levels after 24 hr exposure. A549 cells were treated with 5 or 10 µM Cr(VI) for 1 (d) or 24 (c) hours. G9a mRNA levels were analyzed by Northern blotting. The ethidium bromide staining of 28S and 18S RNA was performed to assess the loading of RNA samples. The relative intensity of the bands was measured and plotted as the mean ratio of G9a mRNA to 18S RNA ± SE (error bars). * P < 0.05, ** P < 0.01, *** P < 0.001. From “Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium” vol. 237(3) by Sun et al. Copyright 2009 Toxicology and Applied Pharmacology. Reproduced with permission of Toxicology and Applied Pharmacology via Copyright Clearance Center.
Figure 9.
Gene expression profiles of control, Cr(VI) transformed and parental BEAS-2B cells. Hierarchical cluster analysis of genes with more than 2-fold changed expression in one out of three groups (control, Cr_small, Cr_large) compared to parental BEAS-2B cells. The color bar related color code to the expression value determined after quantile normalization and baseline transformation to the median levels of all samples. From “Comparison of gene expression profiles in chromate transformed BEAS-2B cells” vol. 6(3) by Sun et al. Published in PloS One in 2011.
Arsenic
Arsenic and arsenic compounds have been classified as group 1 human carcinogens by IARC7. Given its natural abundance and industrial use, arsenic’s environmental impact is felt by nearly 150 million people in at least 70 countries 53. Arsenic exposure is associated with the development of human skin, lung bladder, liver, and prostate cancers, as well as some non-carcinogenic health outcomes, including cardiovascular disease, neurologic deficits, neuro-developmental deficits in childhood, and hypertension 54–59.
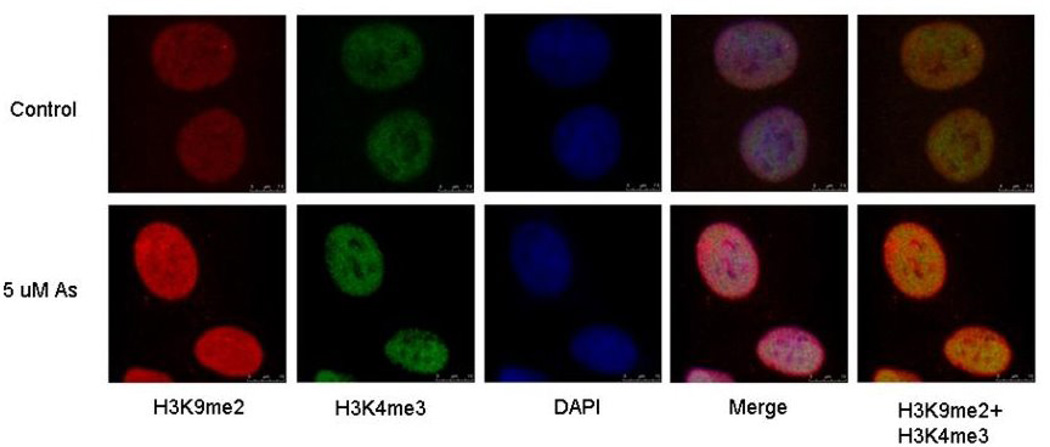
Although not as extensive as our work on nickel and chromium, our arsenic work also demonstrates that arsenic is capable of inducing epigenetic effects (Figure 10). When we exposed A549 cells to arsenite we observed a loss of H3K27me3 and a global increase of H3K9me2 and H3K4me3. The observed increase in H3K9me2 was mediated by an increase in G9a, a histone methyltransferases responsible for specifically methylating H3K9 60. Later, we also showed that 1 µM sodium arsenite treatment produced a significant increase in H3K4me3 after 24-hour or 7 day exposure in A549 cells, which was maintained a week after the removal of arsenite, suggesting that this epigenetic effect was inherited through cell division 61. Moreover, our most recent work has demonstrated that chronic exposure to inorganic arsenic via drinking water can induce statistically significant alterations in global post translational histone modifications in a human population. Specifically, our results showed that chronic As exposure in adults from Araihazar, Bangladesh, increases H3K9me2 and H3K27me3 (females only), which are marks of transcriptional repression, and a decrease in H3K9 acetylation and H3K4me3 (males only), marks usually associated with relaxed chromatin structure and active chromatin, in PBMC. The combination of these observed post-translational histone modifications, maybe associated with global transcriptional repression and may be used as biomarker of arsenic exposure (Chervona et al. AACR Annual Meeting, 2011, Abstract 4221).
Figure 10.
Distinct localization of H3K9 di-methylation (H3K9me2) and H3K4 tri-methylation (H3K4me3) in arsenite exposed cells. A549 cells were exposed 5 µM arsenite for 24 h. After exposure, cells were co-stained with di-methylated H3K9 (red) and tri-methylated H3K4 (green) antibodies. The nucleus was counterstained with DAPI (blue). Merge images show a merge of the red, green and blue staining. The pictures were taken using a confocal microscope. From “Effects of nickel, chromate, and arsenite on histone 3 lysine methylation” vol. 236(1) by Zhou X. et al. Copyright 2009 Toxicology and Applied Pharmacology. Reproduced with permission of Toxicology and Applied Pharmacology via Copyright Clearance Center.
Results summary
Nickel ions inhibit the dioxygenase histone demethylases leading to increased levels of global H3K4me3 and H3K9me2, which increases or decreases the expression of specific genes, respectively. Nickel ions also bind and displace iron in the histidine-histidine carboxylic acid facial triad at the active site of dioxygenases, such as ABH2. Mapped genomic positions of H3K4 tri and H3K9 di methylation changes induced by nickel using Chip-on-chip and Chip-Seq technologies correlate with gene expression changes. Nickel exposure in vivo is able to induce or repress a number of genes related to cytokine-cytokine interaction and chemokine signaling. Furthermore, gene expression changes are very metal specific in normal human bronchial epithelial cells transformed by nickel and chromate. Nickel, chromium and arsenic are able to induce changes in the global levels of post-translational histone modifications in vitro and in vivo. Histone tail modifications in peripheral blood monocytes can serve as potential biomarkers for human exposure to nickel and arsenic.
Conclusion
Our work on carcinogenic metals, such as, nickel, chromium and arsenic, has demonstrated that all three are capable of perturbing effects on the epigenome and draws attention to the possibility that these carcinogenic metals can exert their effect by manipulating the state of chromatin and gene expression. The ramifications of metal exposure on post-translational histone modifications, as well as, on the enzymes that modulate them are pretty remarkable. Members of the iron- and 2-oxoglutarate-dependent dioxygenase family of enzymes, in particular, have emerged as credible molecular targets for metal toxicity and carcinogenicity. Because these enzymes are involved in many different biological processes in cells, their perturbation or inhibition by carcinogenic metals, such as nickel, chromium and arsenic, could have an extensive and multifaceted impact on cells. Future research should aim to further elucidate the mechanism by which carcinogenic metals inhibits these enzymes as well as to understand the biological consequences of their inhibition.
References
- 1.Nawrot TS, Staessen JA, Roels HA, Munters E, Cuypers A, Richart T, Ruttens A, Smeets K, Clijsters H, Vangronsveld J. Biometals. 2010;23:769–782. doi: 10.1007/s10534-010-9343-z. [DOI] [PubMed] [Google Scholar]
- 2.Smith AH, Hopenhayn-Rich C, Bates MN, Goeden HM, Hertz-Picciotto I, Duggan HM, Wood R, Kosnett MJ, Smith MT. Environ Health Perspect. 1992;97:259–267. doi: 10.1289/ehp.9297259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AH, Steinmaus CM. Annu Rev Public Health. 2009;30:107–122. doi: 10.1146/annurev.publhealth.031308.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doll R, Morgan LG, Speizer FE. Br J Cancer. 1970;24:623–632. doi: 10.1038/bjc.1970.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibb HJ, Lees PS, Pinsky PF, Rooney BC. Am J Ind Med. 2000;38:115–126. doi: 10.1002/1097-0274(200008)38:2<115::aid-ajim1>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.IARC. Monograph on the Evaluation of Carcinogenic Risks to Humans. Chromium, Nickel and Welding. Lyon, France: World Health Organization; 1990. [PMC free article] [PubMed] [Google Scholar]
- 7.IARC. Monographs on the Evaluation of Carcinogenic Risk to Humans. Some Drinking-water Disinfectants and Contaminants, including Arsenic. Lyon, France: World Health Organization; 2004. [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YW, Klein CB, Kargacin B, Salnikow K, Kitahara J, Dowjat K, Zhitkovich A, Christie NT, Costa M. Mol Cell Biol. 1995;15:2547–2557. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossman T. Mutat Res. 2003;533:37–65. doi: 10.1016/j.mrfmmm.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Kargacin B, Klein CB, Costa M. Mutat Res. 1993;300:63–72. doi: 10.1016/0165-1218(93)90141-y. [DOI] [PubMed] [Google Scholar]
- 11.Berger SL. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, Kallin EM, Zhang Y. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 14.Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ. J Inorg Biochem. 2006;100:644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Norseth T. Handbook on the Toxicology of Metals. Elsevier/North-Holland Biomedical Press; Amsterdam: 1979. [Google Scholar]
- 16.Kasprzak KS, Sunderman FW, Salnikow K. Mutat Res. 2003;533:67–97. doi: 10.1016/j.mrfmmm.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Basketter DA, Angelini G, Ingber A, Kern PS, Menné T. Contact Dermatitis. 2003;49:1–7. doi: 10.1111/j.0105-1873.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 18.Sunderman FW, Hopfer SM, Plowman MC, Knight JA. Res Commun Chem Pathol Pharmacol. 1990;70:103–113. [PubMed] [Google Scholar]
- 19.Costa M, Mollenhauer HH. Carcinogenic activity of particulate nickel compounds is proportional to their cellular uptake. Science. 1980;209(4455):515–517. doi: 10.1126/science.7394519. [DOI] [PubMed] [Google Scholar]
- 20.Costa M, Abbracchio MP, Simmons-Hansen J. Toxicol Appl Pharmacol. 1981;60:313–323. doi: 10.1016/0041-008x(91)90234-6. [DOI] [PubMed] [Google Scholar]
- 21.Abbracchio MP, Simmons-Hansen J, Costa M. J Toxicol Environ Health. 1982;9:663–676. doi: 10.1080/15287398209530194. [DOI] [PubMed] [Google Scholar]
- 22.Abbracchio MP, Heck JD, Costa M. Carcinogenesis. 1982;3:175–180. doi: 10.1093/carcin/3.2.175. [DOI] [PubMed] [Google Scholar]
- 23.Evans RM, Davies PJ, Costa M. Cancer Res. 1982;42:2729–2735. [PubMed] [Google Scholar]
- 24.Costa M, Heck JD. Adv Inorg Biochem. 1984;6:285–309. [PubMed] [Google Scholar]
- 25.Davidson T, Chen H, Garrick MD, D'Angelo G, Costa M. Mol Cell Biochem. 2005;279:157–162. doi: 10.1007/s11010-005-8288-y. [DOI] [PubMed] [Google Scholar]
- 26.Ke Q, Davidson T, Kluz T, Oller A, Costa M. Toxicol Appl Pharmacol. 2007;219:18–23. doi: 10.1016/j.taap.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 27.Davidson TL, Chen H, Di Toro DM, D'Angelo G, Costa M. Mol Carcinog. 2006;45:479–489. doi: 10.1002/mc.20176. [DOI] [PubMed] [Google Scholar]
- 28.Ke Q, Costa M. Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Ke Q, Kluz T, Yan Y, Costa M. Mol Cell Biol. 2006;26:3728–3737. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson JP, Johnson L, Jasencakova Z, Zhang X, PerezBurgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, Jacobsen SE. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Costa M. Biometals. 2009;22:191–196. doi: 10.1007/s10534-008-9190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H, Kluz T, Zhang R, Costa M. Carcinogenesis. 2010;31:2136–2144. doi: 10.1093/carcin/bgq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Giri NC, Zhang R, Yamane K, Zhang Y, Maroney M, Costa M. J Biol Chem. 2010;285:7374–7383. doi: 10.1074/jbc.M109.058503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giri NC, Sun H, Chen H, Costa M, Maroney MJ. Biochemistry. 2011;50:5067–5076. doi: 10.1021/bi101668x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choschzick M, Oosterwijk E, Müller V, Woelber L, Simon R, Moch H, Tennstedt P. Virchows Arch. 2011;459:193–200. doi: 10.1007/s00428-011-1105-y. [DOI] [PubMed] [Google Scholar]
- 36.Shin HJ, Rho SB, Jung DC, Han IO, Oh ES, Kim JY. J Cell Sci. 2011;124:1077–1087. doi: 10.1242/jcs.072207. [DOI] [PubMed] [Google Scholar]
- 37.Tchou-Wong KM, Kiok K, Tang Z, Kluz T, Arita A, Smith PR, Brown S, Costa M. PLoS One. 2011;6:e17728. doi: 10.1371/journal.pone.0017728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellen TP, Ke Q, Zhang P, Costa M. Carcinogenesis. 2008;29:2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- 39.Genega EM, Ghebremichael M, Najarian R, Fu Y, Wang Y, Argani P, Grisanzio C, Signoretti S. Am J Clin Pathol. 2010;134:873–879. doi: 10.1309/AJCPPPR57HNJMSLZ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arita A, Niu J, Qu Q, Zhao N, Ruan Y, Nádas A, Chervona Y, Wu F, Sun H, Hayes RB, Costa M. Environ Health Perspect. 2011 doi: 10.1289/ehp.1104140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aldrich MV, Gardea-Torresdey JL, Peralta-Videa JR, Parsons JG. Environ Sci Technol. 2003;37:1859–1864. doi: 10.1021/es0208916. [DOI] [PubMed] [Google Scholar]
- 42.Birk T, Mundt KA, Dell LD, Luippold RS, Miksche L, Steinmann-Steiner-Haldenstaett W, Mundt DJ. J Occup Environ Med. 2006;48:426–433. doi: 10.1097/01.jom.0000194159.88688.f8. [DOI] [PubMed] [Google Scholar]
- 43.Park RM, Bena JF, Stayner LT, Smith RJ, Gibb HJ, Lees PS. Risk Anal. 2004;24:1099–1108. doi: 10.1111/j.0272-4332.2004.00512.x. [DOI] [PubMed] [Google Scholar]
- 44.Costa M, Klein CB. Crit Rev Toxicol. 2006;36:155–163. doi: 10.1080/10408440500534032. [DOI] [PubMed] [Google Scholar]
- 45.Venier P, Montaldi A, Majone F, Bianchi V, Levis AG. Carcinogenesis. 1982;3:1331–1338. doi: 10.1093/carcin/3.11.1331. [DOI] [PubMed] [Google Scholar]
- 46.Kortenkamp A, O'Brien P, Beyersmann D. Carcinogenesis. 1991;12:1143–1144. doi: 10.1093/carcin/12.6.1143. [DOI] [PubMed] [Google Scholar]
- 47.Zhitkovich A, Voitkun V, Costa M. Biochemistry. 1996;35:7275–7282. doi: 10.1021/bi960147w. [DOI] [PubMed] [Google Scholar]
- 48.Zhitkovich A, Voitkun V, Costa M. Carcinogenesis. 1995;16:907–913. doi: 10.1093/carcin/16.4.907. [DOI] [PubMed] [Google Scholar]
- 49.Costa M. Crit Rev Toxicol. 1997;27:431–442. doi: 10.3109/10408449709078442. [DOI] [PubMed] [Google Scholar]
- 50.Zhou X, Li Q, Arita A, Sun H, Costa M. Toxicol Appl Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun H, Zhou X, Chen H, Li Q, Costa M. Toxicol Appl Pharmacol. 2009;237:258–266. doi: 10.1016/j.taap.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun H, Clancy HA, Kluz T, Zavadil J, Costa M. PLoS One. 2011;6:e17982. doi: 10.1371/journal.pone.0017982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ravenscroft P, Brammer H, Richards K. Arsenic pollution: a global synthesis. Wiley-Blackwell, U.K.: 2009. [Google Scholar]
- 54.Some Drinking-water Disinfectants and Contaminants, including Arsenic. vol. 84. Lyon, France: 2004. Editon edn. [PMC free article] [PubMed] [Google Scholar]
- 55.Schuhmacher-Wolz U, Dieter H, Klein D, Schneider K. Crit Rev Toxicol. 2009;39:271–298. doi: 10.1080/10408440802291505. [DOI] [PubMed] [Google Scholar]
- 56.Chiu H, Ho S, Wang L, Wu T, Yang C. J Toxicol Environ Health A. 2004;67:1491–1500. doi: 10.1080/15287390490486806. [DOI] [PubMed] [Google Scholar]
- 57.Benbrahim-Tallaa L, Waalkes M. Environ Health Perspect. 2008;116:158–164. doi: 10.1289/ehp.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rahman M, Ng J, Naidu R. Environ Geochem Health. 2009;31(Suppl 1):189–200. doi: 10.1007/s10653-008-9235-0. [DOI] [PubMed] [Google Scholar]
- 59.IARC. Some drinking water disinfectants and contaminants including arsenic. vol. 84. World Health Organization; Lyon, France: 2004. Editon edn. [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou X, Sun H, Ellen T, Chen H, Costa M. Carcinogenesis. 2008;29:1831–1836. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou X, Li Q, Arita A, Sun H, Costa M. Toxicol Appl Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]