SUMMARY
Atria and ventricles exhibit distinct molecular profiles that produce structural and functional differences between the two cardiac compartments. However, factors that determine these differences remain largely undefined. Cardiomyocyte-specific COUP-TFII ablation produces ventricularized atria that exhibit ventricle-like action potentials, increased cardiomyocyte size, and development of extensive T-tubules. Changes in atrial characteristics are accompanied by alterations of 2584 genes, in which 81% of them were differentially expressed between atria and ventricles, suggesting that a major function of myocardial COUP-TFII is to determine the atrial identity. Chromatin immunoprecipitation assays using E13.5 atria identified classic atrial-ventricular identity genes Tbx5, Hey2, Irx4, MLC2v, MLC2a and MLC1a, among many other cardiac genes, as potential COUP-TFII direct targets. Collectively, our results reveal that COUP-TFII confers the atrial identity through direct binding and modulating expression of a broad spectrum of genes that have an impact on atrial development and function.
INTRODUCTION
Atria and ventricles display distinct gene expression profiles that consist of thousands of differentially expressed genes (Barth et al., 2005; McGrath and de Bold, 2009; Tabibiazar et al., 2003). The distinct profiles not only reflect the unique chamber identities but also serve as the basis to create profound differences in structural properties, electric activation, excitation-contraction coupling and neurohormonal functions between the two cardiac compartments (de Bold et al., 1981; Ng et al., 2010). As a result, the ventricle consists of more mitochondria and an extensive transverse tubule (T-tubule) system to provide energy and calcium flux for force generation while the atria have more Golgi complexes and endoplasmic reticulum for their role as source and target of neurohormonal signaling (Barth et al., 2005; McGrath and de Bold, 2009; Tabibiazar et al., 2003).
Atrial and ventricular cardiomyocytes develop their unique identities at an early embryonic stage, partly through regulation of the expression of distinct genes in each chamber by transcription factors such as Tbx5, Hey2 and Irx4 (Bao et al., 1999; Bruneau et al., 2001a; Bruneau et al., 2001b; Evans et al., 2010; Koibuchi and Chin, 2007; Xin et al., 2007). Tbx5 promotes the expression of both atrial natriuretic factor (Nppa) and connexin 40 (Gja5) that are preferentially expressed in the atria (Bruneau et al., 2001b; Mori et al., 2006). In contrast, Irx4 promotes expression of ventricular genes Hand1 and Hand2 and suppresses the atrial reporter SmyHC3-HAP (Bao et al., 1999; Bruneau et al., 2001a). Whilst Irx4 is important for the regulation of ventricular genes, it is not sufficient to confer ventricular identity (Bruneau et al., 2001a). The ventricular Hey2 is known to maintain ventricular identity by suppressing the atrial genes Myl4 (MLC1a), Myl7 (MLC2a), Sacrolipin (Sln), Gja5 and Nppa, but loss of Hey2 does not alter the expression of ventricular genes (Koibuchi and Chin, 2007; Xin et al., 2007). Collectively, these findings reveal a transcription network that controls a set of marker genes for atrial/ventricular identity. However, the aforementioned insufficiency of Tbx5, Hey2 and Irx4 in directing major atrial/ventricular gene expression also suggests that additional major regulator(s), yet to be identified, are responsible for controlling a large number of atrial and ventricular genes (Bruneau et al., 2001a; Bruneau et al., 2001b; Xin et al., 2007) to set up atrial/ventricular identity.
Chicken Ovalbumin Upstream Promoter Transcription Factor II (COUP-TFII) is an orphan nuclear receptor that belongs to the steroid receptor superfamily (Tsai and Tsai, 1997). The highly conserved COUP-TFII controls cell proliferation, differentiation and fate determination by modulating transcriptional activities of a large number of target genes (Lin et al., 2010; Pipaon et al., 1999; Qin et al., 2010a; Qin et al., 2010b; Tang et al., 2012; Tang et al., 2010; Xie et al., 2011; Yu et al.). In the cardiovascular system, COUP-TFII is expressed in the venous/lymphatic endothelium, vascular smooth muscle cells, the endocardium, and the epicardium (Lin et al., 2010; Lin et al., 2011; Pereira et al., 1999; You et al., 2005). Most interestingly, we found that COUP-TFII is expressed in atrial cardiomyocytes, but not in the ventricular myocardium (Lin et al., 2012). Further, germ-line deletion of COUP-TFII results in defective vascular remodeling and heart development (Pereira et al., 1999), and conditional ablation of COUP-TFII in endothelial cells determined that COUP-TFII confers vein identity by suppressing Notch signaling in the venous endothelium (Chen et al., 2012; You et al., 2005). The specific COUP-TFII expression pattern in the atrial myocardium prompted us to investigate the role of COUP-TFII in the specification of atrial identity.
Based on our understanding of COUP-TFII’s role in cell fate determination and its atrial specific expression pattern, we hypothesized that COUP-TFII determines atrial identity through regulation of genes necessary for conferring atrial/ventricular characteristics. In the present study, by in vivo manipulation of COUP-TFII levels in the developing cardiomyocytes, we show that COUP-TFII is necessary and sufficient to confer atrial identity. Our data further shows that COUP-TFII regulates the expression of a wide spectrum and a large number of differentially expressed genes between atria and ventricles, including known transcription regulators Tbx5, Hey2 and Irx4. Moreover, chromatin immunoprecipitation assays and subsequent sequencing analysis reveal that many COUP-TFII potential target genes are associated with cardiac development and physiology. Taken together, the present work demonstrates that COUP-TFII is a major regulator to determine the atrial identity during cardiac development.
RESULTS
Ablation of COUP-TFII in the myocardium ventricularizes the atrium
To specifically dissect the role of COUP-TFII in the developing myocardium, the COUP-TFII flox allele was crossed with Myh6-cre mice to generate myocardial-specific COUP-TFII deficient (CKO) mice. Diminished COUP-TFII protein levels were first observed in the myocardium of free wall regions in E9.5 CKO atria (Figures S1A–G) while its expression in the endocardial cells remained the same (Figures S1D–G). Importantly, COUP-TFII protein levels in the Isl1-demarcated zone of progenitors and the adjacent newly differentiated α smooth muscle actin (αSMA) positive cardiomyocytes remained comparable between control and CKO mice (Figures S1H–S). By E10.5, COUP-TFII is absent in most CKO myocytes (Figures S1T–Y). Deletion of the COUP-TFII gene was further confirmed by expression of recombinant β-galactosidase (Figures S1C and S1Z) that is activated only upon successful recombination (Takamoto et al., 2005). These findings indicate that COUP-TFII was efficiently deleted in cardiomyocytes of CKO mice.
The resulting adult CKO atria showed a gain of expression of the ventricular marker gene Myl2 (MLC2v) in both sides of the atria (Figures 1A, 1B, 1D, 1E, S1AB and S1AC), but a loss of expression of the atrial markers MLC2a and Gja5 (Figures 1C, 1F, S1AA, S1AC and S1AD). In addition, the size of the CKO atria is significantly larger than the atria of control mice (Figures 1A and 1D). A similar phenotype was also observed in the SM22-Cre mediated myocardial COUP-TFII deletion mutant (CKOSM22-cre) mice that exhibited gain of MLC2v, loss of MLC2a, and an increase in atrial size (Figures 1G–J). The fact that ventricularized atria are observed in two independent models strongly suggests an indispensable role of COUP-TFII in determining the atrial identity.
Figure 1. Morphology of the ventricularized atria.
(A–J) Hearts of 2 months old adult mice staining for MLC2a and MLC2v in denoted genotypes. Hematoxylin was utilized for counterstaining. Scale bars indicate 500 µm. (K–L) Bar graphs showing mean length (K) and width (L) of isolated cardiomyocytes. (M–O) Confocal fluorescent images of representative di-8-ANEPPS-stained myocytes with genotypes denoted at side. (P) Index of the spatial integrity of T-Tubules (TT-power). Each genotype comprised of 3 animals at 2 months old. Error bars denote standard error of mean. ***, p<0.001. ra, right atria; rv, right ventricles; la, left atria; lv, left ventricles. See also Figure S1.
To examine the morphological and structural alterations in the COUP-TFII deficient cardiomyocytes, we isolated individual cardiomyocytes from adult atria and ventricles. We found that atrial CKOA myocytes were significantly longer than myocytes from control atria (CTRLA), reaching the length of ventricular (CTRLV) myocytes of wild type control mice (Figures 1K and S1AE-AH). Also, the width of the mutant atrial myocytes was thicker than the width of control cells (Figures 1L and S1AE-AH).
In small mammals, a distinct feature of adult ventricular myocytes is the T-tubule system that is an extension of narrow and inward projection of sacrolemma, which is not commonly observed in the atrium (Orchard and Brette, 2008; Smyrnias et al., 2010). In contrast to the absence of fully developed T-tubules in CTRLA myocytes (Figure 1M), CKOA myocytes displayed organized T-tubules that crossed and extended deep into the cells (Figure 1N), similar to CTRLV myocytes (Figure 1O). Quantitative analysis of spatial integrity of T-tubules, represented by the TT power index, further confirmed the increase of T-tubule development in CKOA myocytes (Figure 1P). These morphological assessments together indicate that myocardial deletion of COUP-TFII allows the atrial myocytes to acquire ventricular cellular morphology.
Alteration in the molecular profile of COUP-TFII depleted atria
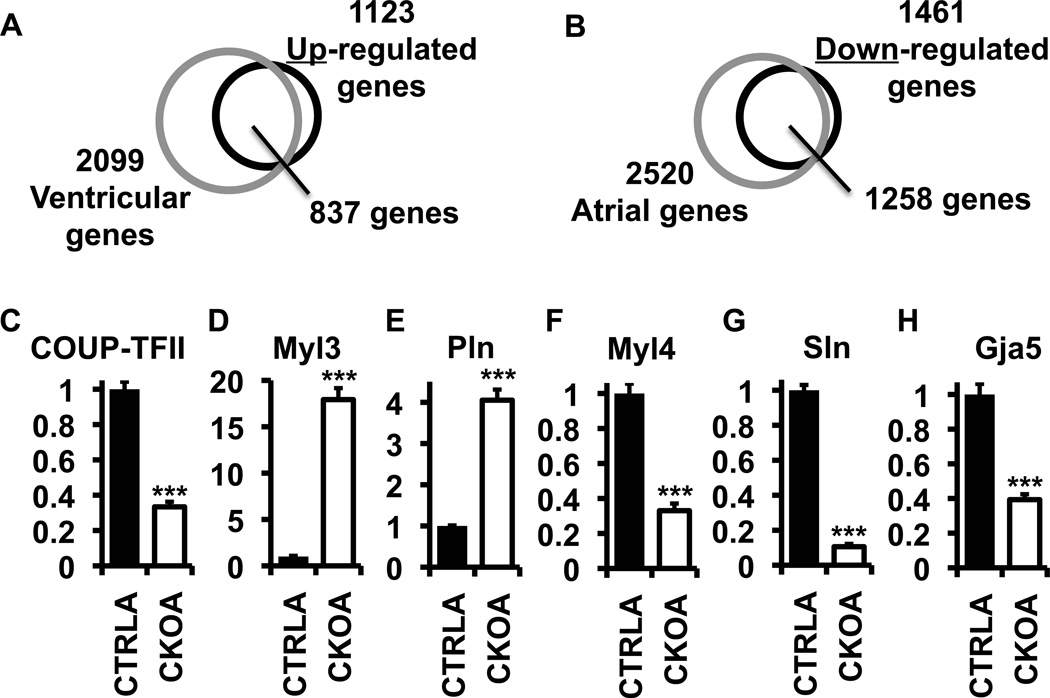
To gain insight into the molecular changes displayed by the ventricularized CKO atria, we performed gene expression profiling on the CKOA, CTRLA and CTRLV tissues using microarray analysis (Figure 2A, 2B and Table S1). Expression profiling between CTRLA and CTRLV identified 2099 and 2520 genes preferentially expressed in ventricles (ventricular genes) and atria (atrial genes), respectively (Grey circles in figures 2A and 2B), while COUP-TFII ablation resulted in 1123 up- and 1461 down-regulated genes in CKOA in comparison to the CTRLA (Black circles in figures 2A and 2B). Among the COUP-TFII regulated genes, 75% (837 out of 1123) of up-regulated genes belonged to ventricular genes and 86% (1258 out of 1461) of down-regulated genes were atrial genes. Together, COUP-TFII controlled 45.4% (2095 out of 4619) of genes that were differentially expressed between the atrial and ventricles. Transcript levels measured by qPCR further confirm the results of microarray analysis, in which myocardial ablation of COUP-TFII caused increased expression of ventricular marker genes Myl3 (MLC1v) and Phospholamban (Pln) (Figures 2D and 2E), and decreased expression of the atrial marker genes MLC1a, Sln and Gja5 (Figures 2F–H) (Babu et al., 2007; Xin et al., 2007). These changes in gene expression indicate that the main function of COUP-TFII in the myocardium is to promote atrial and to suppress ventricular gene expression.
Figure 2. Molecular profile of ventricularized atria in CKO hearts.
(A) Venn diagram to compare the genes up-regulated in CKO atria (black circle) and ventricular genes (grey circle). (B) Venn diagram to compare the genes down-regulated in CKO atria (black circle) and atrial genes (grey circle). (C–H) Relative mRNA levels of marker genes in adult atria (n=4 for each genotype). Error bars denote standard error of mean. ***, p<0.001. See also Tables S1, S2 and S3.
Functional classification of the atrial/ventricular genes revealed that COUP-TFII controls a wide-range of molecular functions (Table S2). Further analysis of these genes in the category of cellular components indicated that genes associated with mitochondria and the endoplasmic reticulum/Golgi apparatus were overrepresented in both up- or down-regulated genes, respectively (Table S3). In the category of biological processes, terms associated with energy production are overrepresented in the group of up-regulated genes while many down-regulated genes are associated with protein/vesicle transportation. The altered genes in CKOA correspond to the unique neurohormonal character of atria and efficient energy production apparatus of the ventricles (Barth et al., 2005; McGrath and de Bold, 2009; Tabibiazar et al., 2003). Together, our results demonstrate that the main function of COUP-TFII in myocardium is to confer atrial identity by promoting atrial and suppressing ventricular gene expression in a broad range of gene targets.
COUP-TFII deficient atrial myocytes exhibit ventricular electrical activities
Our gene expression analyses identified a ventricularized expression pattern of many ion channel genes in CKOA myocytes (Table S4). This altered molecular pattern and the development of organized T-tubules together suggest that COUP-TFII deficient atrial myocytes may also exhibit ventricularized electrical activities. To investigate this possibility, we perform patch clamp analysis to measure action potentials on isolated adult cardiomyocytes.
In control mice, the atrial action potential had a triangle shape (Figure 3A), while the ventricular action potential comprised a more pronounced plateau phase with a longer action potential duration at 90% recovery of depolarization (APD90) (Figures 3C, 3D and S2A). Strikingly, the action potential of CKOA myocytes exhibited a pronounced plateau phase and had comparable APD90 to the CTRLV myocytes (Figures 3B, 3D and S2A). This altered pattern of action potential indicates that CKOA myocytes gained functional electrical characteristics of ventricular myocytes.
Figure 3. Electric properties of adult CKO myocytes and hearts.
(A–C) Representative diagrams of action potential recording from isolated CTRLA (A), CKOA (B) and CTRLV (C) myocytes. (D) Mean action potential durations of isolated cardiomyocytes at APD90. (E&F) Representative surface and intra-atrial ECG during programmed induction of AF in CTRL (E) and CKO (F) mice. N (numbers of cells) =15 for CTRLA, 15 for CKOA and 7 for CTRLV within each bar from 3 animals per group. Error bars denote standard error of mean. ***, p<0.001; mV, millivolt; ms, millisecond. See also Figure S2 and Tables S4 and S5.
To examine whether the ventricularized atria still possess atrial electrical characteristics, adult hearts were subject to a stress test using rapid pacing to induce atrial fibrillation (AF) (Li and Wehrens, 2010). Whereas 6 out of 11 control mice developed at least 1 episode of AF (Figures 3E and Figure S2B), none of the 7 CKO mice developed AF (Figures 3F and Figure S2B). We also measured the proportion of attempts that resulted in successful induction of AF and termed it as the AF score. Among the control mice, 4 mice (36%) had an AF score of 33%, one mouse (9%) had an AF score of 67%, and one had an AF score of 100%. In contrast, all the CKO mice had an AF score of 0% (Figure S2C). These results indicate that whereas mice with normal atria were susceptible to AF induction, the ventricularized CKO atrial myocardium lost the AF susceptibility as part of atrial electrical characteristics and prevented induction of AF following programmed electrical stimulation.
To exclude the possibility that a change of AF susceptibility is secondary to ventricular dysfunction, we performed 2D echocardiography on both CKO and control mice to examine ventricular morphology and pump functions. We found that the ventricle size and mass, as reflected by left ventricle internal diameter in diastole (LVID;d) and left ventricular posterior wall thickness in diastole (LVPWD;d), respectively, were both similar between the groups (Table S5). The left ventricle pump function measured as fractional shortening and ejection fraction was also similar between both the groups (Table S5). We thus concluded that the altered electrical properties in CKO mice were primarily due to the effect of loss of COUP-TFII in atrial myocardium.
COUP-TFII determines the atrial identity during embryonic heart development
The COUP-TFII dependent identity switch as revealed by positive immunostaining of MLC2v in atrial cardiomyocytes can be detected as early as E10.5 (compare Figure 4C with 4D) and continues to be observed at later stages of embryonic development (Figures 4E–H). The temporal increase in mRNA expression of MLC2v in the mutant atrium as compared to the control atrium from E9.5 to adult is further confirmed by qRTPCR analysis (Figure 4I). While an increase in the expression of ventricular markers, MLC1v and Pln, and a reduction in the expression of atrial markers, Sln, Kcnj3 and Gja5, could be easily seen in the E14.5 CKO atria (Figures S3A–E), the CKO atria have yet to achieve a complete ventricular identity change at E14.5, as evidenced by the facts that the mutant atria still display the mosaic MLC2v staining pattern (Figure 4H) and the expression of MLC2v mRNA has yet to reach the maximal levels as seen in the adult (Figure 4I).
Figure 4. Temporal analysis of identity switch.
(A–H) Heart regions of CKO and control mice stained for MLC2v with age denoted on the side. (I) qRTPCR for MLC2v mRNA levels in atria from E9.5 to 2 months old adult mice. Fold changes of CKO are normalized to the corresponding control at the same age. Asterisks denote significant differences between genotypes of the same age. (J–M) Heart regions E18.5 of CKOMyh6-MCM and corresponding control mice stained for MLC2v. E18.5TamE12.5 and E18.5TamE15.5, embryos received tamoxifen at E12.5 and E15.5, respectively. Inserts in (K&M), double staining of MLC2v (red) and β-gal (green) with DAPI for nuclei staining on an adjacent section of (K&M), respectively. Hematoxylin was utilized for counterstaining in all sections except inserts in (K& M). Error bars denote standard error of mean. ***, p<0.001. ra, right atria; rv, right ventricles; la, left atria; lv, left ventricles. See also Figure S3.
To determine whether COUP-TFII is required for identity switch at later stages of development, we used an inducible-Cre (Myh6-MerCreMer) system to ablate COUP-TFII (CKOMyh6-MCM) in cardiomyocytes at later stages. Tamoxifen was administered to embryos at E12.5 (E18.5TamE12.5) or E15.5 (E18.5TamE15.5) to induce COUP-TFII ablation and cardiac sections from embryos were then used to examine the expression of ventricular marker MLC2v at E18.5 (Figures 4J–M). It is clear that ablation of COUP-TFII at E12.5 results in ectopic expression of MLC2v in the mutant atria (Figure 4K). Using β-gal staining to mark COUP-TFII-ablated cells in atria, we also found that β-gal positive cells (green in Figure 4K insert) often co-express with the ventricle marker MLC2v (red in Figure 4K insert) in embryos that received tamoxifen at E12.5 (Figure 4K). The above results indicate that atrial myocytes can switch identity when COUP-TFII is deleted at E12.5. In contrast, both immunopositive MLC2v myocytes and co-expression of MLC2v (red in figure 4M insert) and β-gal (green in figure 4M insert) mycocytes are no longer seen from atrial cardiomyocytes when COUP-TFII was ablated at E15.5, indicating that the plasticity of switching identity is lost when COUP-TFII is ablated at later embryonic stages (Figure 4M). In addition, no apparent alteration of atrial size was observed in mice with COUP-TFII deletion after E15.5 (data not shown), suggesting that the atrial enlargement phenotype is only observed in COUP-TFII deletion prior to E15.5. Collectively, these results suggest that COUP-TFII is required for the specification of atrial cardiomyocyte identity at early stages of development, but not for maintenance of the identity when atrial fate is already determined.
COUP-TFII directly regulates expression of genes for atrial/ventricular identities
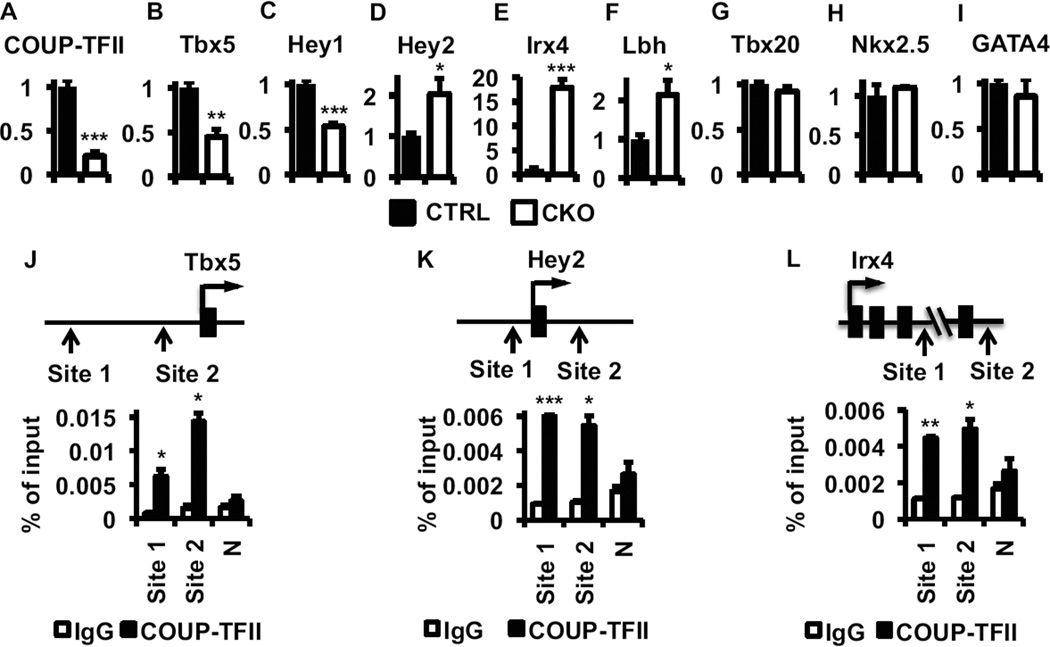
Cardiac morphogenesis requires coordination of multiple transcription factors to control differentially expressed genes in each individual compartment (Evans et al., 2010; Small and Krieg, 2004). We next examined whether and to what extend the expression of cardiac transcription factors were affected in E14.5 CKO atria. Expression levels of Tbx5 and Hey1, two transcription factors preferentially expressed in the atrial compartment (Bruneau et al., 2001b; Leimeister et al., 1999; Nakagawa et al., 1999), decreased subsequent to COUP-TFII deletion (Figures 5A–C). In contrast, the expression of Hey2, Irx4, and Lbh transcription factors that are restrictively expressed in ventricles (Briegel and Joyner, 2001; Bruneau et al., 2000; Koibuchi and Chin, 2007; Nakagawa et al., 1999), significantly increased in CKO atria (Figures 5D–F), while the expression of general cardiac transcription factors Tbx20, Nkx2.5 and Gata4 remained unchanged (Figures 5G–I) (Kraus et al., 2001; Nemer and Nemer, 2002; Tanaka et al., 1999).
Figure 5. Regulation of atrial/ventricular identity transcription factors by COUP-TFII.
(A–I) Relative mRNA levels of denoted genes from pooled E14.5 CTRL (solid bar) and CKO (open bar) atria measured by qPCR. Each group comprised two independent pools of atrial samples. Levels were shown as relative folds over control. (J–L) ChIP-PCR assays on pooled E13.5 atria using anti-COUP-TFII antibody (solid bar) or IgG (open bar). Diagrams on the top indicated COUP-TFII binding sites. Bar graphs show enrichment of DNA fragments pulled down by antibodies. N indicates a region in the Hbb locus without COUP-TFII or Sp1 binding sites served as negative control. Error bars denote standard error of mean. *, p<0.05; **, p<0.01; ***, p<0.001. See also Figure S4.
Reduction of Tbx5 mRNA in E14.5 COUP-TFII mutant atria suggests that COUP-TFII promotes Tbx5 expression. Others and we have shown previously (Chen et al., 2012; Kim et al., 2009; Pipaon et al., 1999; Qin et al., 2010a; Rohr et al., 1997; Tang et al., 2012; Tang et al., 2010; Yu et al., 2012) that COUP-TFII positively regulates its target gene expression through tethering to the Sp1 factor at Sp1-binding sites. To identify Sp1 binding sites in the Tbx5 locus, we used ECR browser/multiTF software at the NCBI’s dcode.org and found 3 potential Sp1 sites in the evolutionarily conserved regions (Figure S4A). ChIP-qPCR assays found binding of COUP-TFII at sites 1 and 2 (Figure 5J) while no enhanced COUP-TFII recruitment was found in site 3 (data not shown). To test the functionality of identified Sp1 binding sites, a 2-kb Tbx5 upstream sequence that harbors the −6.3 kb Sp1 site (Site 1) was cloned and placed in front of a SV40 promoter driven-luciferase reporter (Figure S4B). Results of luciferase assays, performed in the HL-1 atrial cardiomyocytes, showed that this sequence enhanced luciferase reporter activities and the enhancement was greatly reduced upon deletion of the Sp1 site (Figure S4B). Taken together, the results indicate that the region containing the Sp1 binding site can mediate COUP-TFII dependent Tbx5 expression.
qPCR analysis also suggests that COUP-TFII suppresses Hey2 and Irx4 expression (Figures 5D & E). We showed previously that COUP-TFII represses its target gene expression through direct binding to the COUP-TFII response elements, a direct repeat (DR) of AGGTCA sequences with different spacing between the half sites (Tsai and Tsai, 1997). Based on previously identified COUP-TFII binding motifs (Chen et al., 2012; Qin et al., 2010b; Tang et al., 2010; Tsai and Tsai, 1997; Xie et al., 2011), we deduced a consensus sequence and use it to identify candidate COUP-TFII binding sites in the evolutionarily conserved regions listed in the phastConsElements30way table of the USCS genome browser. We found 10 regions that harbor potential COUP-TFII binding motifs with variable homology to the consensus sequences in the Hey2 genomic locus (marked blue regions in Figure S4C). ChIP-qPCR assays found COUP-TFII binding only at sites 1 and 2 (Figures 5K and S4C), while no enhanced COUP-TFII recruitment was found in other regions with less homology to the consensus sequences (other Blue regions in Figure S4C). This finding confirms our previous observation that COUP-TFII suppresses Hey2 expression in HUVEC cells (Chen et al., 2012), supporting the notion that COUP-TFII utilizes a similar mechanism to regulate Hey2 expression in cardiomyocytes. Using the same approach, we found that COUP-TFII is recruited to sites 1 and 2 among the 5 conserved regions identified in the Irx4 genomic locus (Figures 5L and S4D). Taken together, our results suggest that COUP-TFII represses Hey2 and Irx4 gene expression in cardiomyocytes via direct binding to COUP-TFII response elements at the Hey2 and Irx4 genomic loci.
To globally identify potential COUP-TFII direct targets, we immuno-precipitated E13.5 atrial chromatin and the subsequent deep sequencing indentified 2863 COUP-TFII binding sites. 5982 genes were found to have COUP-TFII binding sites within 50-kb of their gene margins Functional annotation analysis on those genes revealed enrichments for cardiac morphogenesis, heart physiology, and cardiomyopathy (Figure 6A and Table S6), suggesting that COUP-TFII directly regulates expression of numerous genes involved in heart development and function. Notably, COUP-TFII binds to genomic loci of classic atrial-ventricular identity genes MLC2v, MLC2a, MLC1a and Irx4 (Bruneau et al., 2000; Xin et al., 2007) and modulates their expression (Figures 6B–D & 5E), supporting that COUP-TFII directly regulates these atrial/ventricular identity genes at the time when COUP-TFII confers the atrial identity. In addition to genes important for atrial specification, COUP-TFII also binds to and controls the expression of many genes that participate in various aspects of cardiac function, such as Kcne1, a voltage gated potassium channel, Id2, a cardiac transcription factor, and Fgf1, a growth factor (Figures 6E–G). Therefore, our results show that COUP-TFII confers the atrial identity through direct binding to promoter/enhancer regions to modulate the expression of a broad spectrum of genes that play key roles in atrial development and function.
Figure 6. Identification of global COUP-TFII binding targets in atria.
(A) Enriched functional annotation terms by DAVID Bioinformatics Sources from ChIP-seq identified genes that contain COUP-TFII binding sites in E13.5 atria. (B–G) Expression levels of denoted genes in E14.5 CKO atria compared with control. (H) The ChIP-seq result of COUP-TFII binding in the MLC2v (Myl2) locus. COUP-TFII binding motifs are shown in capital letters in boxes. (I) ChIP-qPCR results on pooled E13.5 atria using anti-COUP-TFII antibody (solid bar) or IgG (open bar). Bar graphs show enrichment of DNA fragments pulled down by antibodies. N indicates a region in the Hbb locus without COUP-TFII binding sites served as negative control. Error bars denote standard error of mean. *, p<0.05; **, p<0.01; ***, p<0.001. See also Figure S5 and Table S6.
To test the functionality of the identified COUP-TFII binding sites, we took advantage of a defined MLC2v enhancer, the mm77 regulatory sequence located upstream of the MLC2v gene that was previously validated in vivo for recapitulating the endogenous MLC2v expression pattern (Blow et al., 2010). ChIP-seq identified one binding site that has a DR2 COUP-TFII binding motif within its regulatory sequence (Site 1 in figure 6H), which we confirmed by ChIP-qPCR (Site 1 in Figure 6I). In addition to this site, ChIP-qPCR also identified a COUP-TFII-binding region that contained a DR0 motif sequence (Site 2 in Figures 6H and 6I). This second COUP-TFII binding site was not detected by ChIP-seq analysis, which may be due to the different sensitivities of the detection methods we used. Indeed, ChIP-qPCR analysis showed that the recruitment of COUP-TFII to both sites is significantly biased for the site identified by ChIP-seq (Figure 6I). Luciferase reporter assays in the HL-1 atrial cardiomyocytes revealed that either one site alone is sufficient to suppress reporter activity (Figure S5A), indicating that COUP-TFII represses MLC2v gene expression through binding to multiple chromatin sites. These results confirm our previous findings that COUP-TFII can repress target gene expression through direct binding to DR COUP-TF response elements to negatively regulate transcription (Chen et al., 2012; Qin et al., 2010b; Tang et al., 2010; Tsai and Tsai, 1997; Xie et al., 2011).
Atrialization of ventricular cardiomyocytes by ectopic COUP-TFII expression
Loss-of-function studies indicated that COUP-TFII is essential to determine atrial cardiomyocyte identity. To confirm that COUP-TFII is sufficient to confer atrial identity, we ectopically expressed COUP-TFII in ventricular myocardium by using a SM22-cre mediated COUP-TFII overexpression (OE) system (Qin et al., 2013; Wu et al., 2010). The mosaicism produced by the transient SM22-cre expression in myocardium provided an advantage in comparing COUP-TFII expressing and non-expressing cells side by side. At E17.5, a subpopulation of OE ventricular cardiomyocytes expressed atrial marker MLC2a instead of the ventricular marker MLC2v (Figure 7A–D). Double immunostaining showed that MLC2a-positive myocytes were only co-localized with COUP-TFII (Figure 7E), while other MLC2v expressing myocytes did not show COUP-TFII expression (Figure 7F). Moreover, compared with the control, COUP-TFII overexpressing ventricles exhibited increased levels of Tbx5 for atrial gene expression and decreased Hey2 and Irx4 levels that promote ventricular gene expression (Figures 7G–I). Collectively, our results demonstrate that COUP-TFII is sufficient to confer the atrial identity in a cell autonomous manner.
Figure 7. Atrialized ventricular myocytes by COUP-TFII overexpression.
Adjacent coronal sections of E17.5 control (A & B) and COUP-TFII overexpression (C & D) hearts stained for MLC2a (A & C) and MLC2v (B & D). (E & F) High power view of adjacent section corresponding to areas being bracketed in (C and D) that are double-stained for COUP-TFII (green) with MLC2a (E) or MLC2v (F) in red. Nuclei were stained in blue by DAPI. (G–I) Relative mRNA levels of Tbx5 (G), Hey2 (H) and Irx4 (I) from E17.5 CTRL (black bar) and COUP-TFII overexpression (OE, grey bar) ventricles measured by qRTPCR. Each groups consisted of 3 individual ventricles. Levels are shown as relative folds over control. Error bars denote standard error of mean. **, p<0.01; ***, p<0.001.
Increased proliferation in COUP-TFII deficient cardiomyocytes
The increase in CKO atrial chamber size is first apparent at E14.5 (Figures S3F–G). To determine whether cardiomyocyte proliferation contributes to the atrial enlargement, we examined mitogenic activities by counting phosphohistone H3 positive, Nkx2.5 positive cardiomyocytes at E12.5 atria. Mutant atria contained a significantly higher number of phosphorylated histone H3, Nkx2.5-positive cells (Figure S3H), suggesting that an increase in myocyte proliferation contributes to the increase of atrial chamber size. Interestingly, mutant atrial cardiomyocytes exhibit a slightly but significantly smaller estimated cell size compared with the control (Figure S3J) at both E14.5 and E16.5. Our data suggests that increased myocyte proliferation serves as the primary cause of enlarged atrial size during embryonic development while the increase of cell size occurs after E16.5.
DISCUSSION
Previous works by others have shown that the atrial and ventricular myocytes are originated from different sets of cardiac progenitor cells pre-allocated at specific anatomical positions. Yet, it is unclear at which stage the atrial/ventricular identities are determined. Using two independent myocardial-restrictive Cre lines, the Myh6-cre and SM22-cre, to abate COUP-TFII, we show that the atrial/ventricular identities can be switched in differentiating cardiomyocytes. The temporal study using Myh6-MerCreMer mice as the cre driver further reveals that the plasticity of identity switch remains subsequent to initial cardiomyocyte differentiation but lost at later embryonic stages. These results suggest that the differentiating cardiomyocytes have the plasticity of changing identity. Our results also demonstrate that ablation of COUP-TFII in cardiomyocytes is sufficient to produce ventricularized atria in which myocytes increase in size, develop organized t-tubules, acquire ventricular electrical properties, and exhibit a molecular profile resembling the ventricular myocytes. These structural, electrical and molecular alterations strongly suggest that the mutant atrial myocyte switched its identity from atria to ventricles.
Based on our results, COUP-TFII could be considered as a key component within the regulatory network that determines atrial-ventricular identity. In this network, Tbx5 is known to be essential for atrial development and promotes expression of several atrial genes (Bruneau et al., 2001b). However, its ventricular expression and its propensity in promoting ventricular gene expression and morphogenesis reduce its merit as a specific determinant of atrial identity (Bruneau et al., 2001b; Mori et al., 2006). In contrast, myocardial COUP-TFII expression is strictly restricted to the atria, rendering it capable of promoting atrial genes while suppressing ventricular gene expression. As expected, myocardial ablation of COUP-TFII only affects atrial specification without obvious impacts on ventricular morphology and function, indicating that COUP-TFII is a critical factor in conferring atrial identity. On the other hand, specification of ventricles requires promoting ventricular and suppressing atrial genes. Ventricular Hey2 has been shown to suppress several atrial genes including MLC1a, MLC2a, Sln, Gja5 and Nppa, but has no effect on the expression of ventricular genes, such as MLC1v, MLC2v and Pln (Koibuchi and Chin, 2007; Xin et al., 2007). These findings suggest that Hey2 is required but not sufficient to confer ventricular identity. Irx4 also promotes ventricular Hand1 and Hand2 and suppresses the atrial reporter SmyHC3-HAP expression (Bao et al., 1999; Bruneau et al., 2001a), yet it does not regulate many atrial/ventricular maker genes such as MLC1a, MLC2a, MLC1v and MLC2v (Bruneau et al., 2001a). Interestingly, Irx4 ablation does not affect ventricular formation, indicating that it is not essential for atrial/ventricular specification during embryonic development (Bruneau et al., 2001a). In contrast, COUP-TFII is both sufficient and necessary in conferring atrial while suppressing ventricular characteristics. Thus, COUP-TFII is the only factor that is currently known to be essential and sufficient in determining atrial/ventricular identity.
Based on our findings, COUP-TFII confers atrial identity through direct regulation of a broad spectrum of genes that play roles in atrial development and function. In addition to the known transcription regulators Tbx5, Hey2 and Irx4, silencing Id2 expression may also be a way for COUP-TFII to confer the atrial identity. In the heart, a group of specialized cardiomyocytes that are functionally distinct from the atrial and ventricular myocytes form the cardiac conduction system (Chien et al., 2008). It has been shown that Id2 is prominently expressed in the ventricular conduction system, including the atrioventricular bundle and the bundle branches, and is required for the development of the ventricular conduction system (Moskowitz et al., 2007). However, Id2 is largely absent in atrial myocardium (Jongbloed et al., 2011; Moskowitz et al., 2007). Our ChIP-seq and qRTPCR results suggest a direct repression role of COUP-TFII on the expression of Id2 (Figure 6F). Thus, it is possible that COUP-TFII directly suppresses Id2 expression to prevent abnormal development of ventricular conduction systems in the atria, which would be another means to establish atrial identity. Further studies are needed to test this hypothesis.
Results from our ChIP-seq assay also implicate that COUP-TFII may regulate cell differentiation through the HDAC-MEF2 pathway. COUP-TFII binding sites are found in genomic loci of Hdac9, Mef2d and Camk2d. The MEF2 transcription factors have been shown to be critical for regulation of differentiation in the heart (Potthoff and Olson, 2007). In addition, emerging evidences indicate the capacity of Hdac9, Mef2d and Camk2d in controlling cell fate decision (Della Gaspera et al., 2012; Sagasti et al., 2001; Tao et al., 2007). These results implicate potential functional interactions between COUP-TFII, Mef2d, Hdac9 and Camk2d during heart development. Furthermore, we also found COUP-TFII binding sites in genomic loci of many genes in the calcium signaling pathway (Figure S5B), an important pathway that transduces signals into the HDAC-MEF2 pathway to regulate heart development (Potthoff and Olson, 2007). Whether the calcium signaling, HDAC-MEF2 and COUP-TFII act jointly to specify atrial fate decision awaits future experimentation.
Our results indicate that COUP-TFII may directly modulate expression of many genes that encode immediate physiological regulators for the atrial function (Table S6). The importance of direct regulation of expression of physiological regulators is exemplified by the regulation of the MLC2v gene by COUP-TFII. MLC2v is not only a classic ventricular identity gene, but also encodes an essential component in muscle fibers. Simple ectopic expression of the MLC2v gene in atria can reduce protein levels of MLC2a, the atrial-specific form of myosin light chain, with subsequent changes in mechanical properties of the transgenic atrial myocytes to a level similar of ventricular myocytes (Buck et al., 1999; Pawloski-Dahm et al., 1998). Therefore, silencing the MLC2v gene in atrial myocytes is critical to maintain the correct atrial mechanical properties, and we show that COUP-TFII directly suppresses transcriptional activities of the MLC2v gene to ensure the establishment of the atrial identity (Figures 6B). This further demonstrates that COUP-TFII confers the atrial identity via direct control of genes that have a critical impact on the atrial function.
Among the 837 COUP-TFII regulated ventricular genes that are over-expressed in mutant atria, 286 genes (34%) contain COUP-TFII binding sites in the promoter or enhancer of the gene loci. On the other hand, among the 1258 COUP-TFII regulated atrial genes that exhibit under-expression in mutant atria, 389 genes (31%) harbor COUP-TFII binding sites in their gene loci. Together, these 675 genes constitute 32% of the COUP-TFII regulated atrial/ventricular genes. Our finding suggests that COUP-TFII may control the atrial/ventricular gene expression pattern through direct and indirect actions of COUP-TFII.
EXPERIMENTAL PROCEDURES
Mice
Mice carrying the COUP-TFII null, COUP-TFII flox and CAG-S-COUP-TFII allele were described previously (Pereira et al., 1999; Qin et al., 2013; Takamoto et al., 2005). Myh6-cre (Agah et al., 1997), SM22-cre (Boucher et al., 2003) and Myh6-MerCreMer mice (Sohal et al., 2001) were acquired from the Jackson Laboratory (stock# 011038, 004746 and 005650, respectively). All animal experiments adhered to guidelines of the Institutional Animal Care and Use Committee of the Baylor College of Medicine and conducted within the scope of approved animal protocols.
Expression array analysis
Cardiac tissues were collected from 2-month old animals. The control atria group consists of 3 pools of right atria, 6 atria per pool from COUP-TFIIflox/flox mice while the mutant atria group is made of 3 individual right atria from Myh6-cre; COUP-TFIIflox/flox mice. The control ventricles were collected from 3 individual whole ventricles of COUP-TFIIflox/flox mice. Affymetrix mouse genome 430 2.0 chips were used in this study. Raw data will be submitted to the NCBI GEO.
Electrocardiogram and echocardiogram
Transthoracic echocardiography, electrocardiogram and programmed electrical stimulation on mice were performed as described previously (Li and Wehrens, 2010; Respress and Wehrens, 2010; van Oort et al., 2010). Detailed information is included in the supplementary experimental procedures.
Ventricular and atrial cell action potential recordings
Current clamp recordings were performed using a conventional whole-cell mode patch clamp as described previously (Hamill et al., 1981). Detailed information can be found in the supplemental experimental procedures. APD were determined at different repolarization levels, with full repolarization defined as 100%.
T-tubule imaging and analysis
T-tubules of cardiomyocytes were visualized by di-8-aminoaphthylethenylpyridinium (di-8-ANEPPS) staining. More information is included in the supplementary experimental procedures.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation analysis (ChIP) on fresh cardiac tissues is performed according to a previously described method (Lee et al., 2006). Chromatin/DNA complexes were collected from E13.5 atria of 50 mouse embryos. The mouse monoclonal anti-COUP-TFII (R&D Systems PP-H7147-00) and normal mouse IgG (Millipore 12–371) were used for the ChIP assay. DNA released from the precipitation was subject to qPCR analysis for quantification of presence of specific loci. Primer sequences can be found in the supplemental experimental procedures.
ChIP-Seq
A pool of atrial tissues from 57 E13.5 C57BL/6J mouse embryos snap-frozen immediately after harvest was sent to Active Motif (Calsbad, CA) for ChIP, library preparation, sequencing and initial data analysis. The Active Motif Rabbit anti-NR2F2 polyclonal antibody (61214) was used for ChIP-Seq. Peak calling was done with MACS 1.4.2 with a cutoff p value set at 10−5 and produces a FDR of 1.78%. Gene calling was based on the presence of COUP-TFII binding sites within 50-kb of the gene margin. The raw datasets will be deposited to NCBI GEO.
Supplementary Material
Highlights.
Differentiating cardiomyocytes have the plasticity of changing identity.
COUP-TFII is essential for conferring the identity of atrial cardiomyocytes.
COUP-TFII promotes atrial genes while suppresses ventricular genes expression.
COUP-TFII controls a broad range of functions in cardiomyocytes.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. A.J. Marian (University of Texas Health Science Center at Houston) for providing the Myh6-cre mice before the line was available from the Jackson Laboratory. We also thank W. Qian and X.F. Tong for excellent technical support and J.R. Hebert for editing. This work is supported by grants from National Institute of Health HL76448 (SYT), DK45641 (MJT), DK62434, DK59820 (SYT and MJT) and DRC Center P30 DK079638 for core lab services.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu GJ, Bhupathy P, Carnes CA, Billman GE, Periasamy M. Differential expression of sarcolipin protein during muscle development and cardiac pathophysiology. J Mol Cell Cardiol. 2007;43:215–222. doi: 10.1016/j.yjmcc.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Pfeufer A, Uberfuhr P, Dugas M, Steinbeck G, Nabauer M. Functional profiling of human atrial and ventricular gene expression. Pflugers Arch. 2005;450:201–208. doi: 10.1007/s00424-005-1404-8. [DOI] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J. LRP: role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–332. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- Briegel KJ, Joyner AL. Identification and characterization of Lbh, a novel conserved nuclear protein expressed during early limb and heart development. Dev Biol. 2001;233:291–304. doi: 10.1006/dbio.2001.0225. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D, Maguire CT, Berul CI, Kass DA, Kuroski-de Bold ML, de Bold AJ, Conner DA, Rosenthal N, Cepko CL, Seidman CE, Seidman JG. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol. 2001a;21:1730–1736. doi: 10.1128/MCB.21.5.1730-1736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL, Seidman JG, Seidman CE. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev Biol. 2000;217:266–277. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, Seidman JG. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001b;106:709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Buck SH, Konyn PJ, Palermo J, Robbins J, Moss RL. Altered kinetics of contraction of mouse atrial myocytes expressing ventricular myosin regulatory light chain. Am J Physiol. 1999;276:H1167–H1171. doi: 10.1152/ajpheart.1999.276.4.H1167. [DOI] [PubMed] [Google Scholar]
- Chen X, Qin J, Cheng CM, Tsai MJ, Tsai SY. COUP-TFII Is a Major Regulator of Cell Cycle and Notch Signaling Pathways. Mol Endocrinol. 2012;26:1268–1277. doi: 10.1210/me.2011-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science. 2008;322:1494–1497. doi: 10.1126/science.1163267. [DOI] [PubMed] [Google Scholar]
- de Bold AJ, Borenstein HB, Veress AT, Sonnenberg H. A rapid and potent natriuretic response to intravenous injection of atrial myocardial extract in rats. Life Sci. 1981;28:89–94. doi: 10.1016/0024-3205(81)90370-2. [DOI] [PubMed] [Google Scholar]
- Della Gaspera B, Armand AS, Lecolle S, Charbonnier F, Chanoine C. Mef2d Acts Upstream of Muscle Identity Genes and Couples Lateral Myogenesis to Dermomyotome Formation in Xenopus laevis. PLoS One. 2012;7:e52359. doi: 10.1371/journal.pone.0052359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]
- Jongbloed MR, Vicente-Steijn R, Douglas YL, Wisse LJ, Mori K, Yokota Y, Bartelings MM, Schalij MJ, Mahtab EA, Poelmann RE, Gittenberger-De Groot AC. Expression of Id2 in the second heart field and cardiac defects in Id2 knock-out mice. Dev Dyn. 2011;240:2561–2577. doi: 10.1002/dvdy.22762. [DOI] [PubMed] [Google Scholar]
- Kim BJ, Takamoto N, Yan J, Tsai SY, Tsai MJ. Chicken Ovalbumin Upstream Promoter-Transcription Factor II (COUP-TFII) regulates growth and patterning of the postnatal mouse cerebellum. Dev Biol. 2009;326:378–391. doi: 10.1016/j.ydbio.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koibuchi N, Chin MT. CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circ Res. 2007;100:850–855. doi: 10.1161/01.RES.0000261693.13269.bf. [DOI] [PubMed] [Google Scholar]
- Kraus F, Haenig B, Kispert A. Cloning and expression analysis of the mouse T-box gene tbx20. Mech Dev. 2001;100:87–91. doi: 10.1016/s0925-4773(00)00499-8. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimeister C, Externbrink A, Klamt B, Gessler M. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- Li N, Wehrens XH. Programmed electrical stimulation in mice. J Vis Exp. 2010 doi: 10.3791/1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Chen X, Qin J, Hong YK, Tsai MJ, Tsai SY. Direct transcriptional regulation of neuropilin-2 by COUP-TFII modulates multiple steps in murine lymphatic vessel development. J Clin Invest. 2010;120:1694–1707. doi: 10.1172/JCI40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Qin J, Tang K, Tsai SY, Tsai MJ. Coup d’Etat: an orphan takes control. Endocr Rev. 2011;32:404–421. doi: 10.1210/er.2010-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, You LR, Yu CT, Hsu W, H Tsai MJ, Tsai SY. Endocardial Cushion Morphogenesis and Coronary Vessel Development Require Chicken Ovalbumin Upstream Promoter-Transcription Factor II. Arteriosclerosis, thrombosis, and vascular biology. 2012:000. doi: 10.1161/ATVBAHA.112.300255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath MF, de Bold AJ. Transcriptional analysis of the mammalian heart with special reference to its endocrine function. BMC Genomics. 2009;10:254. doi: 10.1186/1471-2164-10-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, Henkelman RM, Bruneau BG. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol. 2006;297:566–586. doi: 10.1016/j.ydbio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, Seidman JG, Seidman CE. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129:1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- Nemer G, Nemer M. Cooperative interaction between GATA5 and NF-ATc regulates endothelial-endocardial differentiation of cardiogenic cells. Development. 2002;129:4045–4055. doi: 10.1242/dev.129.17.4045. [DOI] [PubMed] [Google Scholar]
- Ng SY, Wong CK, Tsang SY. Differential gene expressions in atrial and ventricular myocytes: insights into the road of applying embryonic stem cell-derived cardiomyocytes for future therapies. Am J Physiol Cell Physiol. 2010;299:C1234–1249. doi: 10.1152/ajpcell.00402.2009. [DOI] [PubMed] [Google Scholar]
- Orchard C, Brette F. t-Tubules and sarcoplasmic reticulum function in cardiac ventricular myocytes. Cardiovasc Res. 2008;77:237–244. doi: 10.1093/cvr/cvm002. [DOI] [PubMed] [Google Scholar]
- Pawloski-Dahm CM, Song G, Kirkpatrick DL, Palermo J, Gulick J, Dorn GW, Robbins J, 2nd, Walsh RA. Effects of total replacement of atrial myosin light chain-2 with the ventricular isoform in atrial myocytes of transgenic mice. Circulation. 1998;97:1508–1513. doi: 10.1161/01.cir.97.15.1508. [DOI] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev. 1999;13:1037–1049. doi: 10.1101/gad.13.8.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipaon C, Tsai SY, Tsai MJ. COUP-TF upregulates NGFI-A gene expression through an Sp1 binding site. Mol Cell Biol. 1999;19:2734–2745. doi: 10.1128/mcb.19.4.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Qin J, Chen X, Xie X, Tsai MJ, Tsai SY. COUP-TFII regulates tumor growth and metastasis by modulating tumor angiogenesis. Proc Natl Acad Sci U S A. 2010a;107:3687–3692. doi: 10.1073/pnas.0914619107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Chen X, Yu-Lee LY, Tsai MJ, Tsai SY. Nuclear receptor COUP-TFII controls pancreatic islet tumor angiogenesis by regulating vascular endothelial growth factor/vascular endothelial growth factor receptor-2 signaling. Cancer Res. 2010b;70:8812–8821. doi: 10.1158/0008-5472.CAN-10-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wu SP, Creighton CJ, Dai F, Xie X, Cheng CM, Frolov A, Ayala G, Lin X, Feng XH, Ittmann MM, Tsai SJ, Tsai MJ, Tsai SY. COUP-TFII inhibits TGF-beta-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Respress JL, Wehrens XH. Transthoracic echocardiography in mice. J Vis Exp. 2010 doi: 10.3791/1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr O, Aunis D, Schaeffer E. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J Biol Chem. 1997;272:31149–31155. doi: 10.1074/jbc.272.49.31149. [DOI] [PubMed] [Google Scholar]
- Sagasti A, Hisamoto N, Hyodo J, Tanaka-Hino M, Matsumoto K, Bargmann CI. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell. 2001;105:221–232. doi: 10.1016/s0092-8674(01)00313-0. [DOI] [PubMed] [Google Scholar]
- Small EM, Krieg PA. Molecular regulation of cardiac chamber-specific gene expression. Trends Cardiovasc Med. 2004;14:13–18. doi: 10.1016/j.tcm.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Smyrnias I, Mair W, Harzheim D, Walker SA, Roderick HL, Bootman MD. Comparison of the T-tubule system in adult rat ventricular and atrial myocytes, and its role in excitation-contraction coupling and inotropic stimulation. Cell Calcium. 2010;47:210–223. doi: 10.1016/j.ceca.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- Tabibiazar R, Wagner RA, Liao A, Quertermous T. Transcriptional profiling of the heart reveals chamber-specific gene expression patterns. Circ Res. 2003;93:1193–1201. doi: 10.1161/01.RES.0000103171.42654.DD. [DOI] [PubMed] [Google Scholar]
- Takamoto N, You LR, Moses K, Chiang C, Zimmer WE, Schwartz RJ, DeMayo FJ, Tsai MJ, Tsai SY. COUP-TFII is essential for radial and anteroposterior patterning of the stomach. Development. 2005;132:2179–2189. doi: 10.1242/dev.01808. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Wechsler SB, Lee IW, Yamasaki N, Lawitts JA, Izumo S. Complex modular cis-acting elements regulate expression of the cardiac specifying homeobox gene Csx/Nkx2.5. Development. 1999;126:1439–1450. doi: 10.1242/dev.126.7.1439. [DOI] [PubMed] [Google Scholar]
- Tang K, Rubenstein JLR, Tsai SY, Tsai M-J. COUP-TFII controls amygdala patterning by regulating neuropilin expression. Development. 2012;139:1630–1639. doi: 10.1242/dev.075564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Xie X, Park JI, Jamrich M, Tsai S, Tsai MJ. COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development. 2010;137:725–734. doi: 10.1242/dev.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, Wells AD, Hancock WW. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- Tsai SY, Tsai MJ. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- van Oort RJ, Respress JL, Li N, Reynolds C, De Almeida AC, Skapura DG, De Windt LJ, Wehrens XH. Accelerated development of pressure overload-induced cardiac hypertrophy and dysfunction in an RyR2-R176Q knockin mouse model. Hypertension. 2010;55:932–938. doi: 10.1161/HYPERTENSIONAHA.109.146449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SP, Lee DK, Demayo FJ, Tsai SY, Tsai MJ. Generation of ES cells for conditional expression of nuclear receptors and coregulators in vivo. Mol Endocrinol. 2010;24:1297–1304. doi: 10.1210/me.2010-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Qin J, Lin SH, Tsai SY, Tsai MJ. Nuclear receptor chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) modulates mesenchymal cell commitment and differentiation. Proc Natl Acad Sci U S A. 2011;108:14843–14848. doi: 10.1073/pnas.1110236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Small EM, van Rooij E, Qi X, Richardson JA, Srivastava D, Nakagawa O, Olson EN. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci U S A. 2007;104:7975–7980. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Yu CT, Tang K, Suh JM, Jiang R, Tsai SY, Tsai MJ. COUP-TFII is essential for metanephric mesenchyme formation and kidney precursor cell survival. Development. 2012;139:2330–2339. doi: 10.1242/dev.076299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.