Abstract
Malignant migrating partial seizures of infancy (MMPSI) is a rare epileptic encephalopathy of infancy that combines pharmacoresistant seizures with developmental delay1. We performed exome sequencing in 3 probands with MMPSI and identified de novo gain-of-function mutations in the C-terminal domain of the KCNT1 potassium channel. We sequenced KCNT1 in 9 additional patients with MMPSI and identified mutations in 4 of them, in total identifying mutations in 6 out of 12 unrelated patients. Functional studies showed that the mutations led to constitutive activation of the channel, mimicking the effects of phosphorylation of the C-terminal domain by protein kinase C. In addition to regulating ion flux, KCNT1 has a non conducting function as its C terminus interacts with cytoplasmic proteins involved in developmental signaling pathways. These results provide a target for future diagnostic approaches and research in this devastating condition.
First described in 1995, Malignant migrating partial seizures of infancy (MMPSI) is characterized by polymorphous focal seizures and arrest of psychomotor development in the first 6 months of life1. Seizures are pharmacoresistant and ictal EEG discharges arise randomly from various areas of both hemispheres and “migrate” from one brain region to another, conferring to this syndrome its main feature and denomination. Brain magnetic resonance imaging (MRI) is generally normal at the onset of the disease. To date, approximately 80 MMPSI patients have been reported with males and females being equally affected1–17.
MMPSI belongs to the group of early-onset epileptic encephalopathies (EOEE) defined as severe age-related disorders where cognitive, sensory, and motor impairment is caused by recurrent clinical seizures or prominent interictal epileptiform discharges18. There is a growing evidence of genetic etiology in EOEE with a prevalence ranging from 10% for STXBP1 gene mutations in Ohtahara syndrome to 70% for the SCN1A gene in Dravet syndrome19. The screening for voltage-gated ion channel genes already reported in epilepsies (KCNQ2, KCNQ3, SCN1A, SCN2A and CLCN2) failed to detect mutations in MMPSI2. SCN1A mutations were later reported in 2 patients15,16, however screening for this gene was negative in 13 MMPSI patients15.
We collected DNA samples from 12 individuals fulfilling the criteria for MMPSI (supplementary Table 1). Ictal EEG showed “migrating” seizures (Supplementary Fig. 1) and brain MRI showed delayed myelination with an extremely thin corpus callosum (Supplementary Fig. 2). Clinical and neuroradiological data are detailed in supplementary Table 1. Molecular screening for SCN1A was negative in all cases.
To identify the disease-causing gene, we performed exome sequencing in 3/12 probands and their unaffected parents. Six to eight Gb of sequence were produced for each sample. The mean exome coverage was 63 –fold, with 82% of target sequence covered at least 15 times. Considering that all reported cases are sporadic, we searched for de novo dominant mutations and focused primarily on nonsynonymous (NS) variants, splice acceptor and donor site mutations (SS) and coding insertions/deletions (indels). We regarded variants as previously unidentified if they were absent from control populations and from in-house exome data and all publicly available data sets, including those of dbSNP135, the 1000 Genomes Project and the NHLBI Exome Sequencing Project. Three to four de novo variants fulfilling these criteria were identified per proband (supplementary Table 2). A single gene, KCNT1 (MIM 608167, NM_020822.2), was affected by distinct heterozygous missense variants in two unrelated probands (c.1283G>A p.Arg428Gln and c.2800G>A p.Ala934Thr). Both mutations were confirmed by Sanger sequencing and none was present in the unaffected parents, showing de novo occurrence (Supplementary Fig. 3a). Both occurred on a highly conserved aminoacid residue (Supplementary Fig. 3b) and were absent in 200 healthy controls. We sequenced the KCNT1 coding regions in our 9 additional patients and their parents. We identified the same de novo p.Arg428Gln missense mutation in two further patients and two additional distinct de novo mutations in the KCNT1 gene (c.1421G>A p.Arg474His and c.2280C>G p.Ile760Met) (Table 1) (Supplementary Table 2).
Table 1.
KCNT1 mutations identified in patients with MMPSI
| Patient | sexe | Ethnic origin |
Genomic position |
cDNA position |
Protein change |
Polyphen-2 prediction |
SIFT prediction |
|---|---|---|---|---|---|---|---|
| 1 | M | European (France) | 138671275 | c.2800G>A | p.Ala934Thr | Possibly damaging | Deleterious |
| 2 | M | European (France) | 138657552 | c.1283G>A | p.Arg428Gln | Probably damaging | Deleterious |
| 3 | M | European (France) | 138657552 | c.1283G>A | p.Arg428Gln | Probably damaging | Deleterious |
| 4 | M | European (France) | 138657552 | c.1283G>A | p.Arg428Gln | Probably damaging | Deleterious |
| 5 | M | European (France) | 138660694 | c.1421G>A | p.Arg474His | Probably damaging | Deleterious |
| 6 | F | European (Ukraine) | 138667192 | c.2280C>G | p.Ile760Met | Probably damaging | Deleterious |
KCNT1 (also known as SLACK, SLO2.2 or KCa4.1) encodes a sodium-activated potassium (KNa) channel20. KCNT1 is widely expressed in the nervous system. Its activity contributes to the slow hyperpolarization that follows repetitive firing. KCNT1 regulates the rate of bursting and enhances the accuracy with which action potentials lock to incoming stimuli21,22. KNa channels also play a role in protecting cells from injury under ischemic conditions23. The KCNT1 protein represents the largest known potassium channel subunit. The C-terminal cytoplasmic domain interacts with a protein network including FMRP (Fragile-X Mental Retardation Protein) that is a potent stimulator of KCNT1 channel activity24. The two aminoacid residues mutated in MMPSI lie within this functionally important cytoplasmic C-terminal domain (Fig. 1a). A second gene, KCNT2 (known as SLICK, SLO2.1 or KCa4.2, MIM 610044), also encodes a KNa channel. Moreover, KCNT1 and KCNT2 coassemble to form heteromeric channels that differ from the homomers in their unitary conductance, kinetic behavior, subcellular localization and response to activation by protein Kinase C25. We thus hypothesized that KCNT2 mutations may account for some of the KCNT1-negative MMPSI cases. Direct sequencing of coding exons of KCNT2 failed to reveal pathogenic variants. Although murine Kcnt1 channels are known to be expressed in neurons of the adult central nervous system21, immunostaining experiments have shown that Kcnt1 is also abundantly expressed in embryonic hippocampal and cortical murine neurons, suggesting their contribution to early excitability (Supplementary Fig. 4).
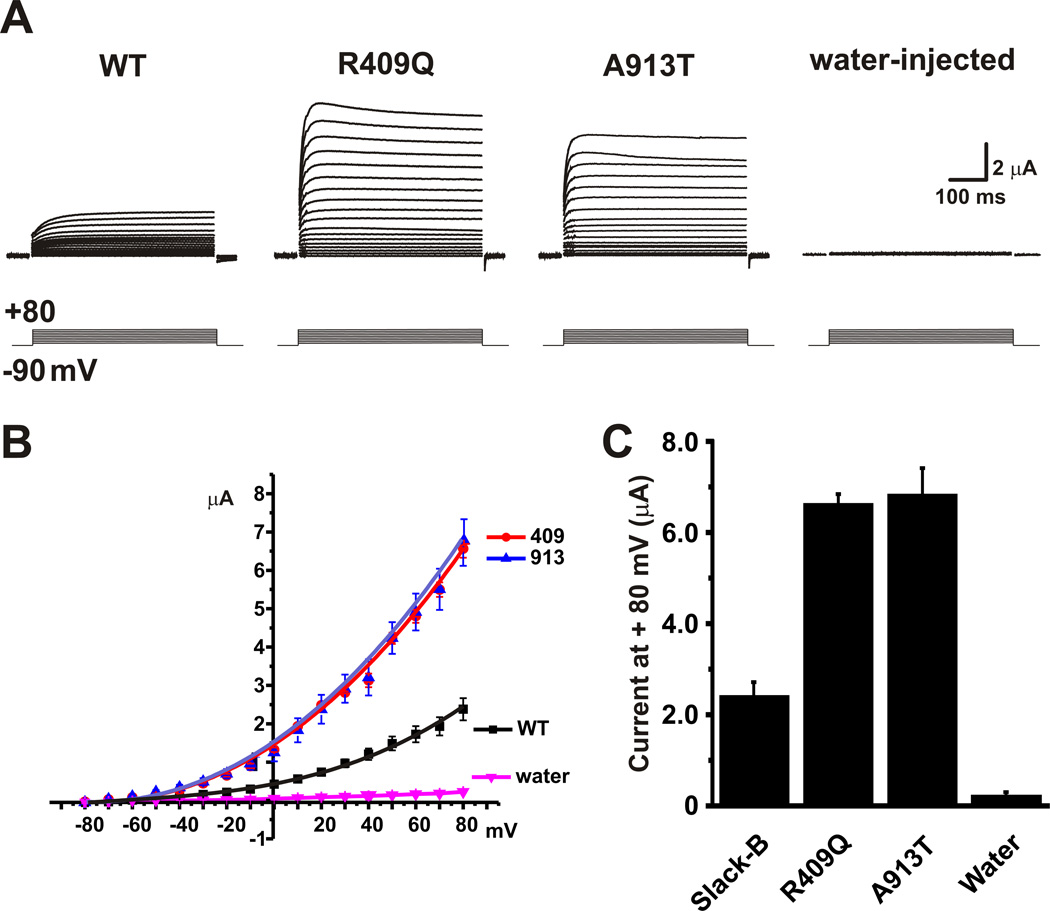
Figure 1. MMPSI mutations increase amplitude of Kcnt1 currents.
Equal amounts (20 ng) of cRNA for wild-type rKcnt1, Arg409Gln or Ala913Thr mutant constructs, or water alone were injected into Xenopus oocytes and two-electrode whole cell voltage clamp were performed 5 days post-injection. (a) Representative families of whole-oocyte currents evoked by stepping from −80 mV to +80 mV in 10 mV increments in oocytes expressing wild-type (WT) rKcnt1 or mutant channels and control oocytes. (b) Mean current-voltage relationship ± SEM for oocytes expressing wild-type rKcnt1 or mutant channels, and control oocytes (N= 5,5,5,7 respectively). Currents were measured at the end of the test pulse. (c) Relative currents (± SEM) at +80 mV for the four groups of oocytes in B. Both MMPSI mutations had significantly increased currents compared to wild-type Kcnt1 or water-injected oocytes as analyzed by ANOVA p<0.05 followed by Tukey’s test at p< 0.001. The two MMPSI mutants were not significantly different from each other.
The transmembrane and C-terminal regions of human and rat Kcnt1 proteins are 92.0% identical. In order to evaluate the impact of the human p.Arg428Gln and p.Ala934Thr mutations, we injected the wild type and mutant rat Kcnt1 constructs (p.Arg409Gln and p.Ala913Thr) in Xenopus oocytes. Both mutations gave rise to Kcnt1 currents that resembled those of wild-type Kcnt1 channels in voltage-dependence and kinetic behavior (Fig. 1.a,b). However, the amplitude of currents produced by the two mutants was 2–3 fold greater than that of wild-type Kcnt1 currents (Fig. 1c, n = 5, p<0.001).
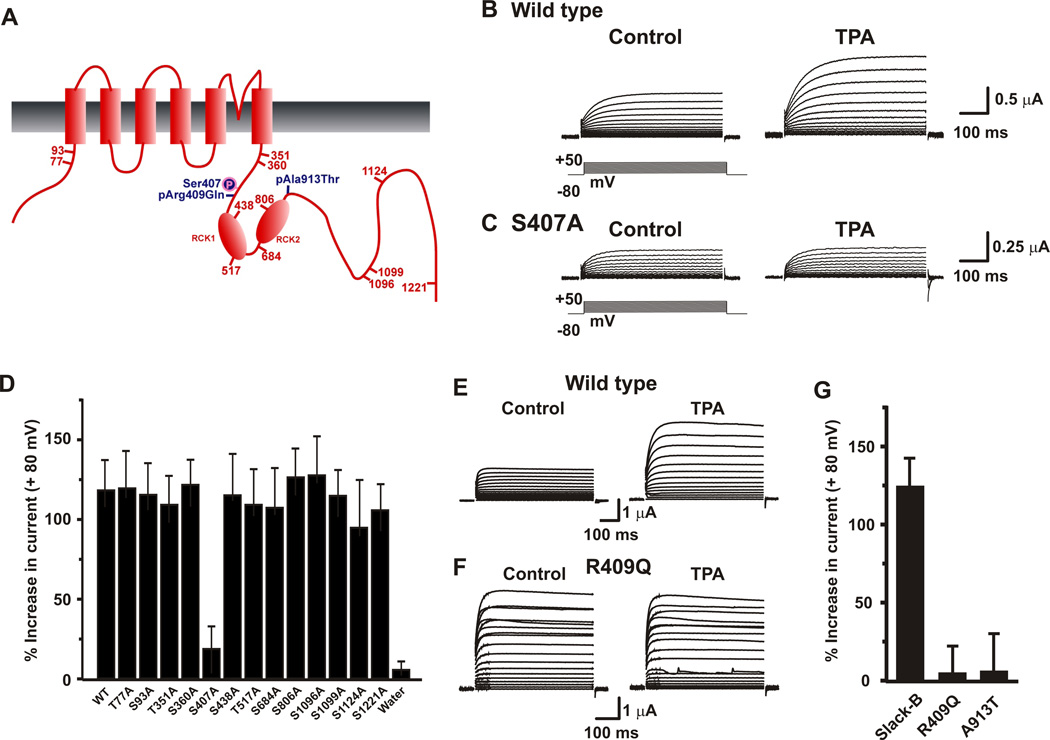
The amplitude of Kcnt1 currents is regulated by protein kinase C (PKC), which, when activated, produces a ~2–3 fold increase in current amplitude, similar to that induced by the p.Arg409Gln and p.Ala913Thr mutations26. The p.Arg409Gln mutation alters one of the 13 predicted PKC phosphorylation sites on the extended C-terminus of Kcnt1 (Fig. 2 a)27. To determine whether any of these consensus sites regulates Kcnt1 current amplitude, we generated mutants in which the serines or threonines were replaced by an alanine at each individual site. Each of the mutant channels was then expressed in Xenopus oocytes and tested for its electrophysiological response to the PKC activator, 12-O-tetradecanoyl-phorbol-13 acetate (TPA, 100 nM). All 13 mutants produced normal Kcnt1-like currents. By contrast, all but one (Ser407) responded to TPA with an increased current comparable to that of wild-type channels (Fig. 2b, 2d). Indeed, mutation of the Ser407 residue, which is adjacent to the Arg409 mutated in MMPSI, completely abolished TPA response (Fig. 2c,d). The vicinity of rat Ser407 and Arg409 residues raised the possibility that the p.Arg409Gln mutation may enhance current amplitude by locking the channel into a state similar to that produced by PKC activation. If this were the case, activation of PKC in mutant channels would not be expected to produce further current increase. Consistent with this hypothesis, we found that currents of both p.Arg409Gln and p.Ala913Thr channels were not significantly increased in response to TPA (Fig. 2e,f,g). These findings demonstrate that both mutant channels are constitutively activated, mimicking and occluding the effects of PKC activation.
Figure 2. MMPSI mutations mimic and occlude the effects of phosphorylation of rKcnt1 at Ser407.
(a) Schematic diagram of rKcnt1-B, including the relative locations of 13 consensus PKC phosphorylation sites and the p.Arg409Gln and p.Ala913Thr mutations. Representative families of whole-oocyte currents evoked by stepping from −80 mV to +50 mV in 10 mV increments in oocytes expressing wild-type rKcnt1 (b) or S407A mutant channels (c) before and after application of the PKC activator TPA (100 nM). (d) Relative increases in currents at +80 mV for wild-type rKcnt1 or Serine to Alanine mutations of the 13 consensus PKC phosphorylation sites mutants, measured 20 minutes after application of TPA (N=3–5 for all mutants except for wild-type and Ser407Ala for which N= 8 each). Representative families of whole-oocyte currents evoked by stepping from −80 mV to +80 mV in 10 mV increments in oocytes expressing wild-type rKcnt1 (e) or Arg409Gln mutant channels (f) before and after application of TPA (100 nM). (g) Relative increases in currents (± SEM) at +80 mV for wild-type rKcnt1 the Arg409Gln and Ala913Thr mutations, 20 minutes after application of TPA (100 nM) (N=8 for wild-type and N=4 for mutants).
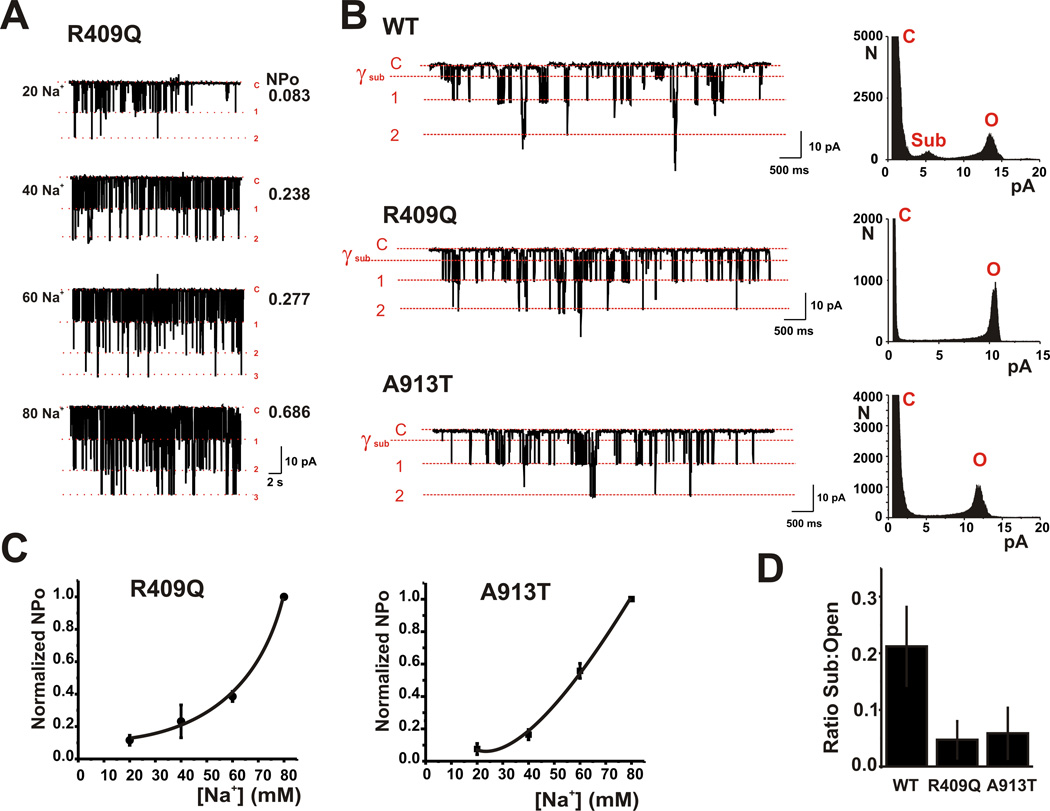
To determine whether the p.Arg409Gln and p.Ala913Thr Kcnt1 mutations produced any other changes in the biophysical behavior of the channels, we carried out single channel recordings in excised inside-out patches. The unitary conductance of both mutant channels, determined from the size of single channel openings at different potentials, was ~140 pS in symmetrical 140 mM [K+], and was no different from that of the wild type channels27. Moreover the opening probability of the mutant channels was increased by elevations of Na+ at the cytoplasmic face of the patches (Fig. 3a,3c) and their dependence on Na+ was indistinguishable from that previously reported for wild type rat Kcnt1 channels21,28. A previous study has demonstrated that C-terminal activation of Kcnt1 is associated with decreased openings to a state with a lower conductance than that of the fully open channel (subconductance state). Quantification of the openings of the p.Arg409Gln and p.Ala913Thr channels revealed that both had reduced openings to subconductance states compared to wild-type channels (Fig. 3b,3d), consistent with the increased macroscopic currents observed with these mutations.
Figure 3. rKcnt1 mutations do not alter Na+ sensitivity but suppress channel subconductance states.
(a) Inside-out recordings of an excised membrane patch containing several R409Q mutant channels with various concentrations of Na+ (20, 40, 60 or 80 mM) at the cytoplasmic face of the channels. The closed state (C) and openings to the level of one or more channels are indicated in red. Values of NPo (Number of channels × Open Probability) are shown at right. Patch was held at −80 mV. (b) Traces at left show excised inside-out single channel recordings from patches containing (WT) rKcnt1 and the Arg409Gln and Ala913Thr mutant channels and held at −80 mV with 20 mM Na+. The closed state and openings to one or more fully open channel levels are shown in red. Also shown is the level of the most prominent subconductance state (γsub). All corresponding point histograms at right show the distribution of openings from the closed state (C) to the first open state (O) or to the subconductance state (Sub). (c) Mean Na+ Concentration-response relationships for R409Q and A914T mutant channels. NPO values are normalized to the NPO obtained at 80 mM Na+. (n = 4 for each mutant, data are shown + SEM). (d) Group data for the mean proportion of time that (WT) rKcnt1 and the Arg409Gln and Ala913Thr mutant channels spent in the subconductance state over the fully open state (n = 12 for all conditions, data are shown + SEM).
Our data show that KCNT1 is a major disease gene for MMPSI and demonstrate that the pathophysiological mechanism underlying this disease is a constitutive hyperactivation of KNa channels. This is the first epilepsy gene identified by exome sequencing in sporadic patients. This strategy will likely emerge as a powerful approach for unraveling rare epilepsies where familial recurrence is sparse or nonexistent, provided preliminary phenotyping enables one to identify homogeneous cohorts.
Our findings further confirm the genetic etiology of MMPSI and ascribe MMPSI to the large family of channelopathies. A few patients with MMPSI were reported with SCN1A mutations. SCN1A is the major gene in Dravet syndrome, another severe childhood encephalopathy phenotypically distinct from MMPSI and characterized by seizure-onset before one year of age with high fever sensitivity, frequent status epilepticus, hemiclonic seizures and later development of various seizure types and mental delay29. MMPSI has been included therefore in the SCN1A phenotype spectrum15. However, no further SCN1A mutations or deletions were found in MMPSI patients15,16 or in this current series. While previous studies have clearly shown that mutations in potassium channel subunits can underlie epilepsies, the consequences of the KCNT1 mutations described here appear substantially more severe than those reported for other potassium channels. Voltage-gated potassium channels KCNQ2, KCNQ3 have been reported in 20–30% of benign neonatal familial seizures, a syndrome characterized by seizures-onset before 2 months with cessation before one year and a good cognitive outcome30. Yet, this correlation between voltage-gated K channels and benign epilepsies has been recently questioned by the report of a series of EOEE patients with KCNQ2 mutations31. However, these patients do not present the clinical and EEG characteristics of MMPSI. In addition, KCNT1 belongs to a KNa channel family distinct from the voltage-gated potassium channels in which mutations have been previously reported in epilepsies. KCNMA1, a distant ortholog of KCNT1 with only 7% homology32, has been shown to account for generalized adult onset epilepsy and dyskinesia in one family. Interestingly, the KCNMA1 mutation also led to a gain-of-function with an increase in channel opening probability33.
The difference in clinical outcome between KCNT1 and the other potassium channels mutations may not simply reflect the effects of the mutations on neuronal excitability. Recent studies have established that, in addition to regulating ion flux, a number of channels have “non-conducting” functions that regulate biochemical functions independently of ion flux34–36. This is likely to be the case for the KCNT1 channel, which in its C-terminus domain interacts with the mRNA-binding protein FMRP, and may therefore participate in transduction pathways that link neuronal activity to the stimulation of protein synthesis. Thus it is likely that KCNT1 mutations reported here may alter the conformation of the C-terminus region of the protein, impairing not only the gating of the channel but also its ability to interact with developmental proteins such as FMRP and other cytoplasmic signaling molecules. Psychomotor outcome in EOEE is usually severe and the role of epilepsy in this poor outcome remains open to question. Along these lines, the expanding phenotypic spectrum of mutations in two genes (the homeobox transcription factor ARX and the STXBP1 gene) from severe early onset epilepsy with mental delay to isolated cognitive delay without epilepsy, support the view of a proper developmental role for these genes37,38. Similarly, in Tuberous Sclerosis Complex – a specific type of infantile seizure disorder with developmental delay and autism spectrum disorders - cognitive prognosis is worsened by infantile spasms. However, patients with no or rapidly resolving infantile spasms are also predisposed to cognitive delay and autism spectrum disorders39.
Our data identify KCNT1 as a major disease gene in MMPSI. They also suggests that defects in KCNT1 may alter developmental signaling pathways coupled to the C-terminus of this channel linking dysfunction of firing, thus epilepsy, to impaired function of proteins causing arrest of psychomotor development.
ONLINE METHODS
Patients
We selected patients presenting with clinical and EEG features of MMPSI and followed in the last 10 years at our Institution. Twelve patients, 5 females and 7 males fulfilled the criteria of MMPSI1,2 (Supplementary Table 1). All were born full-term and none had a history of fetal distress. One patient was born to first-cousin parents of Moroccan origin. The others were born from non consanguineous healthy parents of European origin. The average age at seizure onset was 2 months (range: 2 hours – 7 months). All patients presented polymorphous and focal motor seizures at onset with an autonomic components including cyanosis or bradycardia in 4. Seizures were refractory to various antiepileptic drugs used mostly in combination (Supplementary Table 1). EEG disclosed multifocal paroxysmal abnormalities affecting alternatively both hemispheres and video-EEG recordings showed in all patients the specific migrating feature.
All patients underwent blood and cerebrospinal fluid (CSF) diagnostic workup including extensive studies for inborn errors of metabolism included blood and CSF lactic acid, serum ammonia and lactate, plasma aminoacid and urine organic acid chromatographies, and southern blot analysis for congenital disorder of glycosylation. Four patients had a negative study of respiratory chain enzymes on muscle biopsy in 6 and on liver biopsy in 3. Brain MRI was normal at onset and showed later a severe delay in myelinization in 7 patients with an extremely thin corpus callosom. No calcification was reported on brain CT. All children developed severe neurological impairment with severe hypotonia and microcephaly ranging from −2.5 to −5 DS. They showed profound developmental delay. All children display social impairment, they acquired eye contact but no other communication skills (Supplementary Table 1). Molecular screening excluded SCN1A mutations, deletion or duplication at this locus.
Informed consent was obtained in accordance with the ethical standards of the Institutional Review Board (IRB) on Human Experimentation.
Exome sequencing
We collected blood samples from affected individuals and performed massively parallel sequencing. DNA (3 ug) was extracted from leukocyte cells from the cases and was sheared with a Covaris S2 Ultrasonicator. An adaptor-ligated library was prepared with the Paired-End Sample Prep kit V1 (Illumina). Exome capture was performed with the SureSelect Human All Exon kit (Agilent)40,41. Paired-end sequencing was carried out on an Illumina HiSeq 2000 that generated 100-bp reads. For sequence alignment, variant calling and annotation, the sequences were aligned to the human genome reference sequence (hg19 build) using the Burrows-Wheeler Aligner (BWA)42. Downstream processing was carried out with the Genome analysis toolkit (GATK)43, SAMtools44 and Picard [http://picard.sourceforge.net]. Variant calls were made with a GATK Unified Genotyper. All calls with a read coverage ≤2× and a Phred-scaled SNP quality of ≤20 were removed from consideration. All variants were annotated using an annotation software system that was developed in-house.
Because MMPSI is likely to be genetically heterogeneous, and therefore not all affected individuals will carry mutations in the same gene, we looked for candidate genes shared among subsets of affected individuals. Specifically, we searched for subsets of 2/3 exomes having ≥ 1 novel variant in the same gene.
Mutation detection
We designed a series of 29 intronic primers to amplify the 31 coding exons of KCNT1. We purified the amplicons and sequenced them using the fluorescent dideoxy-terminator method on an automatic sequencer (ABI 3100; Applied Biosystems).
Site-directed mutagenesis
Site-directed mutagenesis of the Kcnt1 construct was performed using the QuikChange kit (Stratagene, La Jolla, CA, USA) and the primers described in the Supplementary Table 3.
Mutations and construct fidelity were confirmed by DNA sequencing.
Electrophysiological characterization in X. laevis oocytes
cRNA was created from rat Kcnt1 wild-type and mutant channel cDNA in the pOX expression vector with a mMessage mMachine T3 kit (Ambion) and aliquoted in sterile water. Voltage-clamp recordings were carried out as described previously 45 using Xenopus laevis oocytes that were defolliculated by collagenase treatment and injected with 100 nl of sterile water containing 20 ng cRNA encoding wild type Kcnt1, Kcnt1 mutants or water alone. Oocytes were incubated at 18° C and recordings were carried out 4 – 5 days post-injection. Whole-oocyte currents were measured by a two-electrode voltage clamp amplifier (Warner Instruments Inc.). Electrodes were filled with 3 M KCl and had resistance 0.1–1.0 MΩ. Data were sampled at 1 kHz and filtered at 0.25 kHz. Standard bath solution was MND-96 containing (mM): 88 NaCl, 1 KCl, 2 MgCl2, 1.8 CaCl2, 5 glucose, 5 HEPES, 5 sodium pyruvate, and 50 µg/µLGentamycin (Gibco), pH 7.4. For measurement of channel activation, oocytes were depolarized by 400 ms pulses from a holding potential of −90 mV to test voltages between –80 mV and +80 mV in 10 mV increments every 5 s. Phorbol 12-myristate 13-acetate (TPA) was purchased from Sigma Aldrich (St. Louis, MO). To evaluate the effects of TPA, cells were clamped for 20 minutes, and every 10 minutes the response of the cells to 400 ms depolarizing pulses from a holding potential of −90 mV to test pulses of between −80 and +80 mV in 10 mV increments was recorded. In order to exclude any cells with leak current, only cells in which the current at +80 mV remained unchanged during this time were included in analysis. TPA was then bath applied to the cell, and the protocol repeated after 20 minutes.
For patch-clamp recording, injected Xenopus oocytes were manually devitellinized in a hypertonic solution containing the following (in mM): 220 Na aspartate, 10 KCl, 2 MgCl2, 10 HEPES, and then incubated in the MND-96 solution for recovery. Excised inside-out patch recordings were performed using a symmetrical 140 [K+] solutions containing the following (in mM): 100 K gluconate, 40 KCl, 20 NaCl, 1 MgCl2, 1 CaCl2, 5 EGTA, and 10 HEPES pH 7.6. Single-channel currents in Xenopus oocytes were recorded using an Axopatch 1D amplifier (Molecular Devices). Currents were filtered at 1 kHz and data were acquired at 10 kHz. Data recording and analysis were performed using pClamp (Molecular Devices) and Origin 8.0 (MicroSoft). To determine Na+ concentration-response relationships, patches were perfused with solutions containing concentrations of Na+ from 20 to 80 mM. [K+] was kept constant at 140 mM, and [Cl−] was kept constant at 40 mM. NPO values were calculated using single channel search in clampfit. NPO values were then normalized to the NPO obtained at 80 mM Na+.
Immunostaining of embryonic neuronal cultures
Rodents were handled in accordance with protocols approved by the Yale University institutional animal care committee. Primary hippocampal cultures were prepared from embryonic day 17.5 rat brains as described previously 46 and grown in Neurobasal medium supplemented with B27 (Invitrogen). Neurons were plated on coverslips coated with poly-D-lysine (30 µg/ml) and laminin (2 µg/ml) at a density of 50,000 cells per well. After 3 days in vitro, cells were washed twice with 1x PBS with 1% BSA, fixed in 4% paraformaldehyde in phosphate buffer, pH 7.4, for 20 min, and then blocked with blocking solution for 1 h at room temperature or at 4°C overnight. After adding the primary antibodies against Kcnt1 (chicken anti-Kcnt1 (800 ng/ml), cultures were agitated either for 1 h at room temperature or at 4°C overnight. Cultures were then washed three times 10 min each, and fluorescent labeled secondary antibodies were added for 30 min (Alexa Fluor donkey anti chicken 488 at 1:400). Cover glasses were washed first with 1x PBS with 1% BSA three times 10 min each, and then mounted on glass slides with anti-fade 2.5% PVA-DABCO solution. Images were taken immediately with a Zeiss laser scanning microscope (LSM 510 META, Germany). DAB staining was performed on hippocampal cultures to confirm the staining patterns. No staining was observed in control experiments, when primary antibodies were omitted and donkey anti-chicken Alexa Fluor 488 was used. Nuclei were stained with propidium iodide.
Mouse cortical cell cultures were prepared from mice at embryonic day 14.5 as described previously 46,47. Dissociated cortical cells were plated on 24-well plates coated with poly-D-lysine and laminin in Eagles’ minimal essential medium supplemented with 5% heat-inactivated horse serum, 5% fetal bovine serum, 2 mM glutamine, and glucose (total of 21 mM). Cultures were maintained in a humidified 5% CO2 incubator at 37°C for 2 days. Staining was carried out as for the rat hippocampal cultures, except that Kcnt1 was labeled using a donkey anti-chicken Cy3 secondary antibody (Red) and nuclei were stained with Topro-3 (Blue).
Reagents for immunocytochemistry was as follows: chicken anti-rKcnt1 antibody (previously described from Aves labs); donkey anti-chicken Alexa Fluor (Invitrogen); donkey anti-chicken Cy3 (Jackson Immuno Research); Propidium Iodide and Topro-3 (Invitrogen).
Supplementary Material
ACKOWLEDGEMENTS
We are grateful to the patients and their family for their participation in the study. L. Colleaux’s team was supported in part by the Centre National de la Recherche Scientifique and the French National Research Agency (N° ANR-08-MNP-010).Work by L. K. Kaczmarek’s team is supported by NIH grants HD067517, DC01919, NS073943 and a grant from the FRAXA foundation. The Laboratory of Human Genetics of Infectious Diseases is supported in part by grants from the St. Giles Foundation, the Rockefeller University Center for Clinical and Translational Science grant number 5UL1RR024143, and the Rockefeller University.
Footnotes
URLs:
Integrative Genomics Viewer (IGV) browser for data visualization, http://www.broadinstitute.org/igv; Variant Effect Predictor, http://www.ensembl.org/tools.html; Exome Variant Server, NHLBI Exome Sequencing Project (ESP), http://evs.gs.washington.edu/EVS/; Online Mendelian Inheritance in Man (OMIM), http://www.omim.org
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
AUTHOR CONTRIBUTIONS
R.N. designed the study. G.B. and L.C. designed and performed the genetics experiments and wrote sections related to sequence analysis. J.L.C. and A.A. performed the exome study and wrote the section related to exome sequencing. P.N. developed the web interface allowing the exome data analysis. GB and L.C. analysed the exome data. M.L. contributed to genetic experiments. L.K.K. supervised electrophysiological experiments. M.R.F. and L.K. designed the electrophysiology experiments, coordinated the recordings analysis and wrote sections related to electrophysiology. M.R.F. performed macroscopic current electrophysiology recordings and J.K. carried out the single channel recordings. J.K., M.B. and H.C. participated in the design and analysis of the electrophysiological data. V.R.G. performed and wrote the section related to immunohistochemistry. R.N., O.D., I.D., A.D., A.K. and R.C. recruited and evaluated the study subjects. N.B. performed and analysed the brain imaging. O.D. and A.M. participated in revising the manuscript. R.N. and L.C. supervised G.B., wrote and revised the manuscript.
References
- 1.Coppola G, Plouin P, Chiron C, Robain O, Dulac O. Migrating partial seizures in infancy: a malignant disorder with developmental arrest. Epilepsia. 1995;36:1017–24. doi: 10.1111/j.1528-1157.1995.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 2.Coppola G, et al. Mutational scanning of potassium, sodium and chloride ion channels in malignant migrating partial seizures in infancy. Brain Dev. 2006;28:76–9. doi: 10.1016/j.braindev.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Gerard F, Kaminska A, Plouin P, Echenne B, Dulac O. Focal seizures versus focal epilepsy in infancy: a challenging distinction. Epileptic Disorders. 1999;1:135–139. [PubMed] [Google Scholar]
- 4.Okuda K, et al. Successful control with bromide of two patients with malignant migrating partial seizures in infancy. Brain Dev. 2000;22:56–9. doi: 10.1016/s0387-7604(99)00108-4. [DOI] [PubMed] [Google Scholar]
- 5.Wilmshurst JM, Appleton DB, Grattan-Smith PJ. Migrating partial seizures in infancy: two new cases. J Child Neurol. 2000;15:717–22. doi: 10.1177/088307380001501102. [DOI] [PubMed] [Google Scholar]
- 6.Veneselli E, Perrone MV, Di Rocco M, Gaggero R, Biancheri R. Malignant migrating partial seizures in infancy. Epilepsy Res. 2001;46:27–32. doi: 10.1016/s0920-1211(01)00197-8. [DOI] [PubMed] [Google Scholar]
- 7.Gross-Tsur V, Ben-Zeev B, Shalev RS. Malignant migrating partial seizures in infancy. Pediatr Neurol. 2004;31:287–90. doi: 10.1016/j.pediatrneurol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Marsh E, Melamed SE, Barron T, Clancy RR. Migrating partial seizures in infancy: expanding the phenotype of a rare seizure syndrome. Epilepsia. 2005;46:568–72. doi: 10.1111/j.0013-9580.2005.34104.x. [DOI] [PubMed] [Google Scholar]
- 9.Hmaimess G, Kadhim H, Nassogne MC, Bonnier C, van Rijckevorsel K. Levetiracetam in a neonate with malignant migrating partial seizures. Pediatr Neurol. 2006;34:55–9. doi: 10.1016/j.pediatrneurol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Zamponi N, Rychlicki F, Corpaci L, Cesaroni E, Trignani R. Vagus nerve stimulation (VNS) is effective in treating catastrophic 1 epilepsy in very young children. Neurosurg Rev. 2008;31:291–7. doi: 10.1007/s10143-008-0134-8. [DOI] [PubMed] [Google Scholar]
- 11.Caraballo RH, et al. Migrating focal seizures in infancy: analysis of the electroclinical patterns in 17 patients. J Child Neurol. 2008;23:497–506. doi: 10.1177/0883073807309771. [DOI] [PubMed] [Google Scholar]
- 12.Lee EH, Yum MS, Jeong MH, Lee KY, Ko TS. A case of malignant migrating partial seizures in infancy as a continuum of infantile epileptic encephalopathy. Brain Dev. 2011 doi: 10.1016/j.braindev.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Gilhuis HJ, Schieving J, Zwarts MJ. Malignant migrating partial seizures in a 4- month-old boy. Epileptic Disord. 2011;13:185–7. doi: 10.1684/epd.2011.0424. [DOI] [PubMed] [Google Scholar]
- 14.Djuric M, Kravljanac R, Kovacevic G, Martic J. The efficacy of bromides, stiripentol and levetiracetam in two patients with malignant migrating partial seizures in infancy. Epileptic Disord. 2011;13:22–6. doi: 10.1684/epd.2011.0402. [DOI] [PubMed] [Google Scholar]
- 15.Carranza Rojo D, et al. De novo SCN1A mutations in migrating partial seizures of infancy. Neurology. 2011;77:380–3. doi: 10.1212/WNL.0b013e318227046d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freilich ER, et al. Novel SCN1A mutation in a proband with malignant migrating partial seizures of infancy. Arch Neurol. 2011;68:665–71. doi: 10.1001/archneurol.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vendrame M, et al. Treatment of malignant migrating partial epilepsy of infancy with rufinamide: report of five cases. Epileptic Disord. 2011;13:18–21. doi: 10.1684/epd.2011.0406. [DOI] [PubMed] [Google Scholar]
- 18.Nabbout R, Dulac O. Epileptic syndromes in infancy and childhood. Curr Opin Neurol. 2008;21:161–6. doi: 10.1097/WCO.0b013e3282f7007e. [DOI] [PubMed] [Google Scholar]
- 19.Poduri A, Lowenstein D. Epilepsy genetics--past, present, and future. Curr Opin Genet Dev. 2011;21:325–32. doi: 10.1016/j.gde.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan A, et al. The sodium-activated potassium channel is encoded by a member of the Slo gene family. Neuron. 2003;37:765–73. doi: 10.1016/s0896-6273(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharjee A, Kaczmarek LK. For K+ channels, Na+ is the new Ca2+ Trends Neurosci. 2005;28:422–8. doi: 10.1016/j.tins.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Brown MR, et al. Amino-termini isoforms of the Slack K+ channel, regulated by alternative promoters, differentially modulate rhythmic firing and adaptation. J Physiol. 2008;586:5161–79. doi: 10.1113/jphysiol.2008.160861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruffin VA, et al. The sodium-activated potassium channel Slack is modulated by hypercapnia and acidosis. Neuroscience. 2008;151:410–8. doi: 10.1016/j.neuroscience.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown MR, et al. Fragile X mental retardation protein controls gating of the sodiumactivated potassium channel Slack. Nat Neurosci. 2010;13:819–21. doi: 10.1038/nn.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. The N-terminal domain of Slack determines the formation and trafficking of Slick/Slack heteromeric sodium-activated potassium channels. J Neurosci. 2009;29:5654–65. doi: 10.1523/JNEUROSCI.5978-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Santi CM, et al. Opposite regulation of Slick and Slack K+ channels by neuromodulators. J Neurosci. 2006;26:5059–68. doi: 10.1523/JNEUROSCI.3372-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joiner WJ, et al. Formation of intermediate-conductance calcium-activated potassium channels by interaction of Slack and Slo subunits. Nat Neurosci. 1998;1:462–9. doi: 10.1038/2176. [DOI] [PubMed] [Google Scholar]
- 28.Yang YY, et al. [Effects of tetramethylpyrazine on large-conductance Ca(2+)- activated potassium channels in porcine coronary artery smooth muscle cells.] Sheng Li Xue Bao. 2006;58:83–9. [PubMed] [Google Scholar]
- 29.Nabbout R, et al. Spectrum of SCN1A mutations in severe myoclonic epilepsy of infancy. Neurology. 2003;60:1961–7. doi: 10.1212/01.wnl.0000069463.41870.2f. [DOI] [PubMed] [Google Scholar]
- 30.Steinlein OK, Conrad C, Weidner B. Benign familial neonatal convulsions: always benign? Epilepsy Res. 2007;73:245–9. doi: 10.1016/j.eplepsyres.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 31.Weckhuysen S, et al. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann Neurol. 2012;71:15–25. doi: 10.1002/ana.22644. [DOI] [PubMed] [Google Scholar]
- 32.Wei AD, et al. International Union of Pharmacology. LII. Nomenclature and molecular relationships of calcium-activated potassium channels. Pharmacol Rev. 2005;57:463–72. doi: 10.1124/pr.57.4.9. [DOI] [PubMed] [Google Scholar]
- 33.Du W, et al. Calcium-sensitive potassium channelopathy in human epilepsy and paroxysmal movement disorder. Nat Genet. 2005;37:733–8. doi: 10.1038/ng1585. [DOI] [PubMed] [Google Scholar]
- 34.Kaczmarek LK. Non-conducting functions of voltage-gated ion channels. Nat Rev Neurosci. 2006;7:761–71. doi: 10.1038/nrn1988. [DOI] [PubMed] [Google Scholar]
- 35.Fleming MR, Kaczmarek LK. Use of optical biosensors to detect modulation of Slack potassium channels by G protein-coupled receptors. J Recept Signal Transduct Res. 2009;29:173–81. doi: 10.1080/10799890903056883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2012;44:471. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamdan FF, et al. Intellectual disability without epilepsy associated with STXBP1 disruption. Eur J Hum Genet. 2011;19:607–9. doi: 10.1038/ejhg.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friocourt G, Parnavelas JG. Mutations in ARX Result in Several Defects Involving GABAergic Neurons. Front Cell Neurosci. 2010;4:4. doi: 10.3389/fncel.2010.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolton PF, Park RJ, Higgins JN, Griffiths PD, Pickles A. Neuro-epileptic determinants of autism spectrum disorders in tuberous sclerosis complex. Brain. 2002;125:1247–55. doi: 10.1093/brain/awf124. [DOI] [PubMed] [Google Scholar]
- 40.Byun M, et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J Exp Med. 2010;207:2307–12. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolze A, et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am J Hum Genet. 2010;87:873–81. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen W, Han Y, Chen Y, Astumian D. Electric field-induced functional reductions in the K+ channels mainly resulted from supramembrane potentialmediated electroconformational changes. Biophysical Journal. 1998;75:196–206. doi: 10.1016/S0006-3495(98)77506-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized Survival of Hippocampal-Neurons in B27-Supplemented Neurobasal(Tm), a New Serum-Free Medium Combination. Journal of Neuroscience Research. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 47.Lobner D. Saturation of neuroprotective effects of adenosine in cortical culture. Neuroreport. 2002;13:2075–2078. doi: 10.1097/00001756-200211150-00017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.