Abstract
RNA viruses of filamentous fungi fall into two broad categories, those that contain double-stranded RNA (dsRNA) genomes in rigid particles and those that are more closely related to positive-sense, single-stranded RNA viruses with dsRNA replicative intermediates found within lipid vesicles. Effective infectivity systems have been described for the latter, using RNA transcripts, but not for the former. We report the characterization of a reovirus from Cryphonectria parasitica, the filamentous fungus that causes chestnut blight disease. The virus substantially reduces the virulence of the fungus and results in dramatically altered colony morphology, as well as changes in other associated fungal traits, relative to the virus-free isogenic strain. Virus particles from infected mycelium contained 11 segments of dsRNA and showed characteristics typical of the family Reoviridae. Sequences of the largest three segments revealed that the virus is closely related to the Coltivirus genus of animal pathogens, which includes the human pathogen Colorado tick fever virus. The introduction of purified virus particles into protoplasts from virus-free isolates of the fungus resulted in a newly infected mycelium with the same morphology and virus composition as the original virus-infected isolate. This represents the completion of Koch's postulates for a true dsRNA virus from a filamentous fungus and the description of a definitive fungal member of the family Reoviridae.
Reoviruses constitute one of the largest virus families in terms of numbers of genera and breadth of host range, which includes plants, arthropods, fish, and mammals (21; see reference 25 for review). They have been of considerable importance as pathogens and, particularly, as tools for virologists and molecular biologists. Much of our knowledge of eukaryotic mRNA synthesis and capping mechanisms was developed from reovirus models (see reference 12 for review). In the plant kingdom, the first demonstration of transmission of a plant virus by arthropods was done with Rice dwarf virus, an important plant reovirus (10). Their visually arresting architecture and complex particle construction make reoviruses popular subjects for structural studies (31).
Among fungal viruses, double-stranded RNA (dsRNA) genomes are the rule rather than the exception (21). Fungal viruses classified as dsRNA viruses fall within two broad categories. The “true” dsRNA viruses have rigid particles containing genomic dsRNA that is not polyadenylated on either strand. The particle itself is generally known to be critical for viral transcription and replication processes but is usually not infectious (see reference 41 for review). Other viruses that are classified as dsRNA viruses show more affinity with positive-sense RNA viruses and are not associated with particles that have a rigid capsid structure. Members of the family Hypoviridae, which falls into this second category, have been shown to infect protoplasts as RNA transcripts synthesized in vitro (4). This characteristic has not been demonstrated for particle-associated dsRNA viruses. Fungal colonies resulting from transfection with viral RNA are identical to their naturally infected counterparts in terms of morphology and genomic dsRNA composition (4). This latter system is relevant to the present study because viruses of the family Hypoviridae also cause hypovirulence of the chestnut blight fungus, Cryphonectria parasitica, and they constitute the most thoroughly studied hypovirulence-associated viruses (26).
The lack of infectivity systems for fungal viruses has greatly hampered progress in the study of true dsRNA viruses, such as the Totiviridae family of viruses, which are particularly well studied in yeast (41), and the Partitiviridae family, whose members are ubiquitous among filamentous fungi (13). In some cases, infectivity with purified particle preparations has been reported, but only rarely and with considerable difficulty (7, 37). dsRNA viruses of plants also are not easily infectious as whole-particle preparations (14).
Reoviruses represent the prototypical particle-associated dsRNA viruses, and viruses from two filamentous fungi have been suggested to be reovirus-like. In the early 1990s, two viruses from Cryphonectria parasitica were found to have 11 dsRNA segments and properties similar to those of reoviruses (8, 9). Though no sequence data were reported, it was hypothesized that these might be true reoviruses (8, 9). Recently, sequences of dsRNA segments isolated directly from the distantly related ascomycete Rosellinia necatrix were found to have homology to two members of the Coltivirus genus of the family Reoviridae (28), and reovirus-like particles were recently identified in association with that virus (39).
In this paper, we describe the purification and partial characterization of one of the viruses associated with C. parasitica. Sequence analysis of the three largest dsRNA segments of the virus indicates that it is closely related to members of the Coltivirus genus of mammalian pathogens. The introduction of purified virus particles into fungal protoplasts and subsequent regeneration of colonies resulted in stable infection of a fungal strain that had not previously harbored the virus. This virus is therefore a true reovirus and is unusual among fungal viruses in that it is infectious as particles.
MATERIALS AND METHODS
Fungal isolate maintenance and properties of isolates.
Virus-containing isolate 9B21 (8) was originally obtained from William MacDonald (West Virginia University, Morgantown). It was maintained by subculturing or as a frozen dried mycelium stock as described previously (16). Virus-free single conidial isolates were obtained by germination of a dilution of conidia on potato dextrose agar (PDA). Several single conidial isolates with morphological characteristics typical of wild-type C. parasitica were subcultured, and the absence of dsRNA was confirmed. Isolate 9B21ss1, chosen as a representative, was confirmed to be isogenic by hyphal anastomosis with strain 9B21, with the attendant acquisition of virus and virus-associated characteristics following anastomosis.
Growth, sporulation, pigment accumulation, and virulence of C. parasitica isolates were measured as described previously (15, 30). Fungal colonies were cultured on PDA under benchtop conditions (22 to 25°C) for 7 days (colony morphology evaluation) or 30 days (measurement of sporulation levels). Apples inoculated with freshly grown mycelia were also incubated on the laboratory benchtop for 7 days for virulence assays. Laccase activity was determined by use of modified Bavendamm's malt-tannic acid medium and quantified spectrophotometrically as described previously (36).
dsRNA extraction and purification from mycelium.
dsRNA was extracted from the fungal mycelium essentially as described previously (15), using CF11 cellulose and the centrifugation method of Morris and Dodds (23). Residual DNA and single-stranded RNA were removed by treatment with DNase I and S1 nuclease and then were purified with RNaid (Qbiogene). dsRNA segments were purified with RNaid after separation of the total dsRNA through agarose gels and excision of the individual segments with a razor.
cDNA library preparation.
Four cDNA libraries representing groups of dsRNA segments were synthesized. Approximately 5 μg of dsRNA was purified as described above, except that groups of segments were used to initiate cDNA synthesis. In four separate reaction sets, randomly primed cDNA was synthesized from dimethyl sulfoxide-denatured RNA (2) by use of a Time Saver kit from Amersham. After second-strand DNA synthesis, DNA was purified by use of GeneClean (Qbiogene), and a 3′-terminal dA residue was added by use of Taq polymerase (Sigma-Aldrich), as described by Zhang and Rowhani (43). The modified cDNA was ligated to the vector pGEM-T Easy (Promega) and introduced into DH5α cells (Promega). White ampicillin-resistant colonies were identified on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) plates and screened for insert size.
5′ RACE.
5′ rapid amplification of cDNA ends (RACE) reactions were performed essentially as described previously (30), with the following exception: after specifically primed first-strand cDNA synthesis, a portion of each reaction set was subjected to the addition of a tail for 2 min at 37°C with either dA or dT residues. The dA- or dT-tailed products were purified by use of GeneClean. The untailed, dA-tailed, and dT-tailed cDNAs were then subjected to the addition of a tail for 10 min with dC residues and to PCR using the 5′ abridged anchor primer 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′ and an appropriate nested primer specific to that segment terminus. Products were purified by use of GeneClean and cloned as described above.
Alternatively, 5′ RACE clones were made by the oligo-capping method (19). Ligation of the oligoribonucleotide GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA, cDNA synthesis, and PCR were carried out by use of the Ambion First Choice RLM-RACE kit as described by the manufacturer.
Oligonucleotides.
The oligonucleotides used in this study were synthesized by Sawady Technology, Inc. (Tokyo, Japan) or the Rutgers DNA Synthesis facility. All sequences not specifically given here are available upon request.
Sequencing, sequence analysis, and phylogenetic analysis.
Sequencing reactions were performed by use of a Big Dye kit from Applied Biosystems (ABI) with DNA prepared in Qiagen spin columns. Most sequencing was performed on an ABI model 2100 sequencer; some reactions were run on an ABI model 377 sequencer. Sequence analysis was performed with the DNAstar suite of programs (Lasergene). Database searches were performed with the BLAST suite of programs from the National Center for Biotechnology Information. Sequence alignments were performed with ClustalX (38). Bootstrapped neighbor-joining trees were drawn with the ClustalX tree-drawing program.
Virus particle extraction and purification.
Virus particles were extracted by a modification of the method described by Enebak et al. (9). Fungal mycelia grown in four to eight flasks, each containing 500 ml of potato dextrose broth, were dried to dampness, ground to a fine powder in liquid nitrogen, and then homogenized in a blender in 4 volumes of 0.1 M NaPO4, pH 7.0, and 1 volume of CCl4 or Freon. Extracts were clarified by low-speed centrifugation (2,200 × g for 20 min) in 50-ml screw-cap tubes in a swinging bucket rotor. The clear aqueous phase was centrifuged for 3 h at 82,700 × g (rav) in a Beckman SW28 swinging bucket rotor. Each high-speed pellet fraction was resuspended in 200 μl of 0.1 M NaPO4, spun for 2 min at 9,000 × g in a microcentrifuge, and loaded directly onto linear-log or 20 to 50% sucrose gradients poured in 5-ml tubes for use in a Beckman SW50.1 rotor. Gradients were centrifuged for 25 min at 120,000 × g (rav), and fractions were collected. Fractions were subjected to additional high-speed centrifugation at 190,000 × g (rav) for 2 h in an SW50.1 rotor. Fraction pellets were resuspended in 50 to 100 μl of 0.1 M NaPO4.
Electron microscopy.
Purified virus particles from sucrose density gradient fractions were negatively stained with 2% uranyl acetate, examined in a Hitachi model H-7100 transmission electron microscope, and photographed.
Virus transfections of fungal protoplasts and regeneration of infected colonies.
Preparation of fungal protoplasts and introduction of purified virus into the protoplast preparations by use of polyethylene glycol were performed according to the methods described by Churchill et al. for basic fungal transformation (6). Plating of transfected protoplasts, regeneration, and subculturing of infected colonies on PDA were performed essentially as described by Chen et al. (4).
Nucleotide sequence accession numbers.
The sequences of the C. parasitica 9B21 virus have been deposited under accession numbers AY277888 (segment 1), AY277889 (segment 2), and AY277890 (segment 3).
RESULTS
Biological properties of C. parasitica strain 9B21.
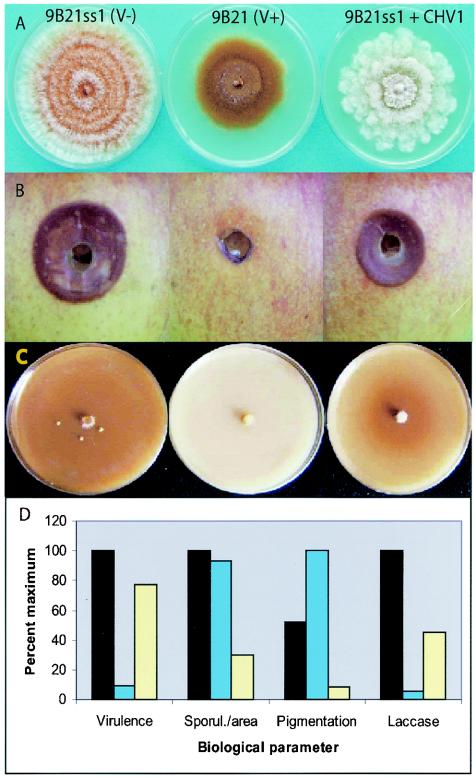
The colony morphology of isolate 9B21 was distinctive and unlike other virus-infected C. parasitica isolates described. Colonies grown on PDA were deep orange in color and had little aerial mycelium compared to their isogenic virus-free counterparts (Fig. 1). In contrast, infection of the same strain with the well-studied hypovirus CHV1-EP713 resulted in a white phenotype and fluffy aerial mycelium (Fig. 1A, right panel) (also see reference 26). When inoculated into apple fruits, a rapid measure of C. parasitica virulence (11), the lesions resulting from 9B21 were very small, indicating that its virulence was very low (Fig. 1B). The virulence of 9B21 was comparable to that of the least virulent C. parasitica isolates examined in our laboratory to date (Fig. 1D) (B. I. Hillman, unpublished results). Sporulation was not substantially reduced for 9B21 compared to that for its virus-free counterpart, and orange pigmentation was higher in the virus-infected isolate (Fig. 1D). This is in stark contrast to CHV1-EP713-infected isolates, which are white and sporulate poorly (Fig. 1A and D) (also see reference 15). Production of extracellular laccase, which is significantly down-regulated by infection with CHV1-EP713 (34), was almost completely eliminated by infection with the 9B21 virus (Fig. 1C and D).
FIG. 1.
Effects of the 9B21 reovirus on morphology and selected biological traits of C. parasitica. Comparisons in this figure are to the virus-free (V−) single conidial isolate derived from 9B21 (9B21ss1) and the same virus-free isolate transfected with RNA of CHV1-EP713, as described by Chen et al. (4). (A) Colony morphology of cultures grown on the laboratory benchtop for 7 days on PDA. (B) Lesions on apple fruits 7 days after inoculation. (C) Colony morphology of cultures grown in darkness on Bavendamm's malt-tannic acid medium. (D) Quantified results of virulence, sporulation, pigmentation, and laccase assays of the three isolates (black, 9B21SS1; blue, 9B21; yellow, 9B21SS1+CHV1), performed as described in the text.
Reovirus-like particles associated with strain 9B21.
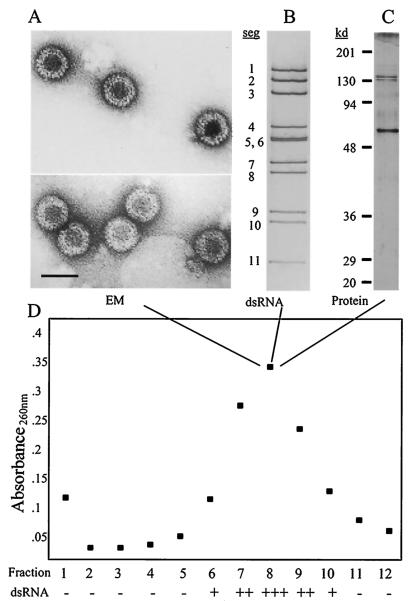
A fraction that sedimented 2/3 of the way down a linear-log sucrose gradient (Fig. 2D) contained particles of approximately 80 nm in diameter that were morphologically typical of reoviruses (Fig. 2A). Similar results were obtained whether CCl4 or Freon was used as the organic solvent. The yield of particles was very low: 3 to 6 μg of purified virus was obtained from each purification reaction, representing 2 liters of fungal culture. RNA extracted from this particle-containing fraction contained the same complement of 11 dsRNA segments that had been identified in the total dsRNA preparation from infected fungal tissue (Fig. 2B). Two of the dsRNA segments, 5 and 6, comigrated in most gels and appeared as a single, more intense band; segments 7 and 8 comigrated in some gels as well.
FIG. 2.
Properties of particles purified by sucrose gradient centrifugation. (A) Electron micrographs of uranyl acetate-stained particles. Bar = 100 nm. (B) Silver-stained 11% polyacrylamide gel with dsRNA. (C) Silver-stained 10% polyacrylamide gel with proteins from the peak fraction, fraction 8, of a sucrose gradient (D) used to purify virus isolated from C. parasitica strain 9B21. Prior to examination, all fractions were concentrated by ultracentrifugation. Absorbance values (at 260 nm) and relative amounts of dsRNA from the individual fractions are shown.
Examination of virion proteins from the peak fraction revealed three clear and several lighter bands on silver-stained polyacrylamide gels (Fig. 2C). The three more intense bands, with migration positions of approximately 65, 130, and 140 kDa, were clearly visible on gels stained with Coomassie brilliant blue; the lighter bands were faintly visible. The sizes and pattern of these proteins are generally consistent with those of reoviruses (21). Taken together, these data indicate that the virus in strain 9B21 is reovirus-like in its morphology and general physicochemical properties.
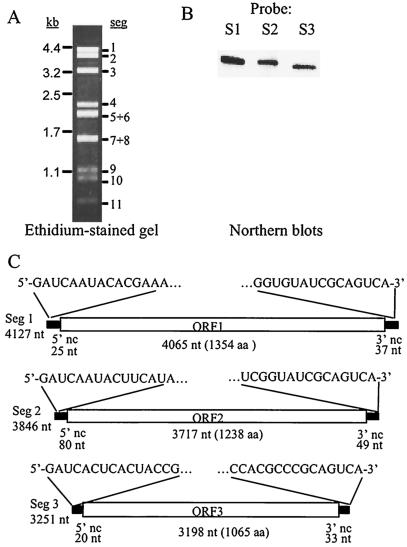
Nucleotide sequence analysis of segments 1 to 3.
We reasoned that the possible relationship of the 9B21 virus to other reoviruses would most likely be revealed in the largest genome segments, which would be predicted to encode conserved RNA polymerase and modifying functions (25). From cDNA libraries representing the largest dsRNAs, segments 1 to 3 were sequenced and their termini were determined from 5′ RACE clones. Northern blots were used to confirm that clones represented those specific segments (Fig. 3A and B). Maps of these segments, including the 5′- and 3′-terminal residues, are provided in Fig. 3. As is the case with other reoviruses, the terminal sequences of the 9B21 dsRNA segments were conserved among the segments examined. The 5′ pentamer 5′GAUCA and 3′ octamer CGCAGUCA3′ were conserved among the three sequenced segments of 9B21. The 3′ termini were similar to those described for the R. necatrix W370 virus (UGCAGAC3′) (28) and for coltiviruses (A/UUGYAGUG/C3′) (3), while the 5′ termini were more similar to those of the genus Oryzavirus (5′GAU) (21). Base pairing of sequences near the 5′ and 3′ termini was inferred from folding of the termini of the coding strand of each genome segment with the program mFold (20, 44), but the extreme terminal residues were not predicted to base pair into a classic panhandle structure as first described by Xu et al. (42) (not shown).
FIG. 3.
Structure of genomic segments 1 to 3 of the 9B21 reovirus. (A) Ethidium bromide-stained 1% agarose gel with dsRNA purified from C. parasitica strain 9B21. (B) Northern blots of individual lanes of a 1% agarose gel containing 9B21 dsRNA, blotted and probed with digoxigenin-labeled PCR products representing segment 1 (nucleotides [nt] 2256 to 2988), segment 2 (nt 261 to 920), and segment 3 (nt 1728 to 2523). (C) Overall organization based on sequences from randomly primed clones made from gel-separated genome segments and 5′ RACE clones made for each terminus, as described in the text. The terminal 15 residues of the coding strand of each segment are shown.
Analysis of the open reading frames predicted from the sequences of segments 1 to 3 revealed that they were homologous to the corresponding segments of several other reoviruses. The relationship between 9B21 and other reoviruses was particularly evident for segments 1 and 2. The deduced amino acid sequence of segment 1, VP1, contained the GDD motif that is highly conserved among viral RNA-dependent RNA polymerases (positions 760 to 762) and aligned robustly with other reovirus VP1 sequences; thus, it very likely encodes the viral polymerase (data not shown; see below).
Alignments of the VP2 deduced amino acid sequences of the 9B21 virus (1,238 residues), Colorado tick fever virus (CTFV) (1,209 residues), and Eyach virus (EYAV) (1,242 residues) were strong and continuous from amino acid F146 of 9B21, Y174 of CTFV, and Y182 of EYAV until a few residues from the deduced C termini (data not shown). Thus, VP2 of 9B21 is homologous to VP2 of these coltiviruses. The putative methyltransferase domain identified near the N termini of CTFV and EYAV VP2 (3) was not included in the strongly aligned region, but a homologue of the downstream region recently identified by Wei et al. at residues L510 to R539 of the R. necatrix W370 virus VP2 as a possible methyl transferase domain (39) was present in the 9B21 amino acid sequence at positions L519 to Q548. The RGD motif identified in VP1 and VP2 of CTFV and EYAV, speculated to be involved in cell binding (3), was not present in either segment of 9B21. This may reflect the lack of a requirement of cell binding in a fungal virus, which likely moves with the growing hyphal tip.
Only the VP3 sequences of the two coltiviruses were identified in BLASTX searches with the 9B21 segment 3 sequence. The levels of similarity were low, with 34% identity and 50% similarity over a 50-amino-acid stretch, from Y206 of 9B21, Y251 of CTFV, and Y251 of EYAV. Little is known about this genome segment of coltiviruses; it is thought to be involved in RNA replication (3).
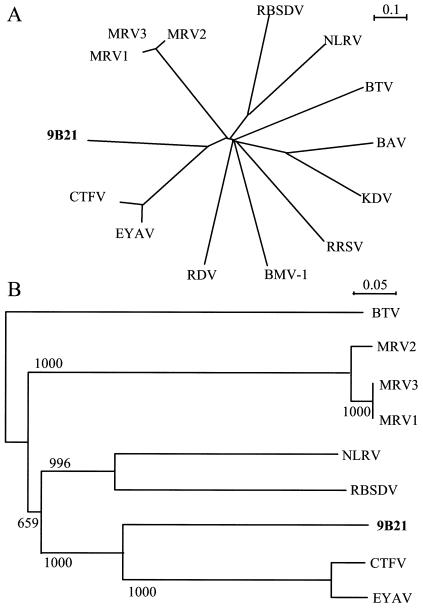
A strongly supported clade that included the C. parasitica 9B21 virus, CTFV, and EYAV was observed in tree diagrams drawn from alignments of amino acid sequences. In complete alignments of segments 1 and 2, the grouping of the 9B21 virus with CTFV and EYAV was supported in 1,000 of 1,000 bootstrap replicates. The topology of the tree drawn for alignments of the entire segment 1 amino acid sequences from a subset of reoviruses (Fig. 4A) was very similar to that presented for a more complete set by Attoui et al. (3). A robust alignment could be generated from sequences around the GDD motif with the reoviruses most closely related to 9B21. The resulting bootstrapped neighbor-joining tree drawn from these alignments, using bluetongue virus (BTV) as an outgroup, is presented in Fig. 4B. Sequences for the R. necatrix W370 virus segments 1 and 3 are not available at this writing and are not included in the comparison.
FIG. 4.
Phylogenetic relationship of 9B21 reovirus VP1 sequence with sequences of selected Reoviridae family members. (A) Unrooted tree from complete alignments. (B) Bootstrapped neighbor-joining tree from alignments of the core polymerase domain of the most closely related viruses from panel A, using the BTV sequence to root the tree, as described in the text. Abbreviations and accession numbers are as follows: MRV1, Mammalian reovirus 1 (NC004271); MRV2, Mammalian reovirus 2 (NC004272); MRV3, Mammalian reovirus 3 (NC004282); RBSDV, Rice black streak dwarf virus (AY144568); NLRV, Nilaparvata ligulans reovirus (D49693); BTV, Bluetongue virus (P13840); BAV, Banna virus (NC004211); KAV, Kadipiro virus (NC004210); RRSV, Rice ragged stunt virus (NC003749); BMV-1, Bombyx mori virus 1 (NC004138); RDV, Rice dwarf virus (Q02119); EYAV, Eyach virus (NC003696); CTFV, Colorado tick fever virus (AF004181).
Virus infectivity.
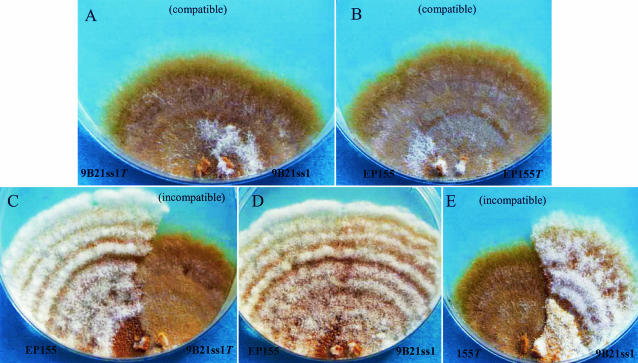
To examine virus infectivity, we transfected two strains of the fungus with particles purified by high-speed centrifugation and sucrose pellets. One of the strains, 9B21ss1, is a virus-free single-spore isolate of the original 9B21 colony. The other strain, EP155 (ATCC 38755) from Connecticut, has become the standard virus-free isolate used in many C. parasitica studies. Importantly, EP155 is not vegetatively compatible with 9B21. If infection of EP155 was observed, it could only be explained reasonably as a transfection event, since plating of the particle preparations alone, as expected, resulted in no growth on PDA. With both 9B21ss1 and EP155 as hosts, the introduction of purified virus into protoplasts, regeneration of colonies, and subculturing onto PDA plates resulted in colonies that were morphologically identical to the original 9B21 colony (Fig. 5). Virus particles containing 11 segments of dsRNA were isolated from different subcultures of both transfected isolates. Anastomosis of the newly transfected 9B21ss1 (9B21ss1T) with its uninfected counterpart resulted in the transfer of virus between these vegetatively compatible, isogenic colonies (Fig. 5A). Similarly, the newly transfected EP155 bearing the 9B21 virus (EP155T) was able to transmit the virus to its uninfected isogenic counterpart (Fig. 5B). However, attempts to transmit virus to the heterologous, incompatible host resulted in visible incompatibility reactions, with no change in colony morphology and no transfer of virus (Fig. 5C and E). These transfectants have proven to be stable upon repeated subculturing.
FIG. 5.
Infectivity of 9B21 reovirus following introduction of particles into two vegetatively incompatible strains of C. parasitica. Virus particles were introduced into protoplasts of either strain 9B21ss1 or EP155 and plated on regeneration medium. After 10 days, subcultures were grown from various regions of the primary transfectant culture. Two such subcultures with phenotypes typical of 9B21, labeled 9B21ss1T and EP155T, were plated beside each of the original uninfected isolates and allowed to grow for 10 days, until the edge of the plate was reached. Virus was transmitted consistently from the transfected to the isogenic compatible isolates, with resulting phenotypic changes (A and B), but in no case did virus move from EP155T into 9B21ss1 or from 9B21ss1T into EP155 (C and E). (D) Virus-free isolates plated adjacent to each other.
DISCUSSION
The characterization of a member of the Reoviridae family of dsRNA viruses that is infectious in a filamentous fungus is important from several perspectives. From the perspective of the study of fungal biological control, yet another viral agent has been described. From the perspective of the study of fungal viruses, new approaches to their study have been described. Perhaps most importantly, the identification of a member of the reovirus family in a haploid host will allow us to use the power of haploid genetics to investigate reovirus biology. Although we have just begun experiments to examine the experimental host range of these viruses, it is not unrealistic to conjecture that they may be able to infect other hosts, as has been done, for example, with brome mosaic virus (17) and flock house virus (32) in the yeast host Saccharomyces cerevisiae. Moreover, the success of transfection of the 9B21 virus may lead to progress in reassortant virus genetics, which is still impeded for plant reoviruses.
Reoviruses may be difficult to purify in their complete forms with all of their virion proteins intact (27). Adding to this difficulty is the fact that there are large differences in the numbers of copies of each of the virion proteins (21). The most intensely staining protein of the 9B21 virus observed on polyacrylamide gels migrated at an Mr of approximately 65,000. Based on results from other reoviruses (21), this likely represents the major capsid protein, which is present in much greater abundance than other structural proteins and is probably encoded by segment 4, 5, or 6. We have just begun the tasks of determining how many virion proteins are associated with the 9B21 virus and assigning them to their cognate segments. The relatively low virus yield from infected fungal tissue has made it somewhat difficult to determine the best purification scheme for the virus. However, the virus is quite stable and retains its infectivity upon storage, so the task should be relatively straightforward once antibodies are available.
A related feature that must be resolved is the difference in the number of segments for the two Cryphonectria viruses, 9B21 and C18, and their closest relatives. Based on preliminary sequence analysis, it has been determined that the C18 virus is also a true reovirus (Hillman, unpublished data). Using a variety of different gel types, we have identified only 11 dsRNA segments from both of these Cryphonectria viruses, whereas the coltiviruses EYAV and CTFV (3) as well as R. necatrix W370 virus (27) have 12 segments. Segment number is a feature that is conserved within genera of other reoviruses: no fewer than 10 segments and no more than 12 segments have been observed. Even when unnecessary segments are removed from the genomes of reoviruses, deletion mutants containing only the termini and little or no coding sequence have been detected (1, 33).
Terminal sequences of different segments of a given reovirus are highly conserved, with segment-specific variation near the extreme termini (21). Extreme terminal sequences tend to be similar or identical within members of a given genus, and large differences may be one of the criteria for removing species from a genus (3). The genome segments S1 to S3 of 9B21 have common terminal sequences (5′GAUCA…CGCAGUCA3′) that are unique among the known reoviruses. It is noteworthy that the sequence GC/UAGU/AC/G is conserved among four related reoviruses (CTFV, EYAV, R. necatrix W370, and 9B21), although the position of the 9B21 segment relative to the 3′ end is different from those of the other three. Together with the amino acid sequence similarities found among viral proteins encoded by the four viruses, the similarity in the 3′ terminal sequences supports evolutionarily close relatedness of these reoviruses.
The most thoroughly studied virus of a filamentous fungus, CHV1-EP713, moderately depresses fungal virulence (30) but dramatically depresses pigmentation and spore production (15). During infection, >400 fungal genes are up- or down-regulated (5). The effects of specific viral coding regions on these and other fungal processes have been examined in detail (reviewed in reference 26). In contrast, the 9B21 virus has quite a different effect on the fungus: virulence is greatly depressed while pigmentation and sporulation are affected little or even increased. Laccase activity, which serves as a natural reporter for virus infection and is associated with multiple genes in C. parasitica, is down-regulated in a complex fashion by infection with CHV1-EP713 (18, 29, 35) and is reduced even more by infection with the 9B21 virus. This system should therefore provide an interesting counterpoint model to further examine virus effects on a filamentous fungal host, and the virus may be interesting to examine as a biological control agent.
The introduction of the 9B21 reovirus into Cryphonectria may be a relatively recent event, possibly by lateral transfer from another fungus. The introduction of C. parasitica into North America in the late 19th century and its spread through the northeast United States in the early 20th century are well documented (see reference 22 for review). During the last 20 to 30 years, thousands of isolates of the fungus from different areas of the natural chestnut tree range have been examined, but the two isolates from West Virginia are the only ones found bearing reoviruses. An interesting connection in this regard is that between the W370 virus from R. necatrix, identified in Japan (28), and the virus from C. parasitica, which is thought to have been introduced to North America from Japan (22). Our preliminary analysis based on partial sequences of several of the smaller segments of the 9B21 reovirus reveals that it is related to the R. necatrix W370 virus. This is expected, as several segments of the R. necatrix virus were shown to be related to coltivirus segments (27). We would expect to find closer relationships between the reoviruses of the two fungal hosts once all segments of both viruses have been sequenced.
If such viruses can move by lateral transfer among fungi, what is the connection to coltiviruses? One intriguing feature here is their arthropod association. Coltiviruses are tick-borne. While Attoui et al. (3) considered the evolution and movement of coltiviruses in lagomorph hosts, somewhat less mention was made of the tick vectors. An interesting connection with Cryphonectria involves its well-documented association with mites (24, 40), which, like ticks, belong to the Acari. Since this fungus has been found in the guts of mites, it is possible that the progenitor to the fungal reoviruses and coltiviruses evolved from a virus native to an Acari host.
Regarding the naming of this virus, the genus name Mycoreovirus was proposed by Enebak (8) and again by Osaki et al. (28). A proposal to the International Committee on Taxonomy of Viruses to determine whether this constitutes a new genus of the family Reoviridae and to describe its nomenclature is under consideration.
Acknowledgments
B.I.H. is grateful for a visiting professorship at RIB, Okayama University, The Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank William MacDonald, Mark Double, and Scott Enebak for isolate 9B21 and Don Nuss for the infectious cDNA clone of CHV1-EP713. We are grateful to K. Maruyama and Bernadette Glasheen for their technical assistance and to Michael Milgroom for valuable comments on an earlier draft of this paper.
REFERENCES
- 1.Anzola, J. A., Z. Xu, T. Asamizu, and D. L. Nuss. 1987. Segment-specific inverted terminal repeats found adjacent to conserved terminal sequences in wound tumor virus genome and defective interfering RNAs. Proc. Natl. Acad. Sci. USA 84:8301-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asamizu, T., D. Summers, M. B. Motika, J. V. Anzola, and D. L. Nuss. 1985. Molecular cloning and characterization of the genome of wound tumor virus: a tumor-inducing plant reovirus. Virology 144:398-409. [DOI] [PubMed] [Google Scholar]
- 3.Attoui, H., F. Mohd Jaafar, P. Biagini, J. F. Cantaloube, P. de Micco, F. A. Murphy, and X. de Lamballerie. 2002. Genus Coltivirus (family Reoviridae): genomic and morphologic characterization of Old World and New World viruses. Arch. Virol. 147:533-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, B., G. H. Choi, and D. L. Nuss. 1994. Attenuation of fungal virulence by synthetic infectious hypovirus transcripts. Science 264:1762-1764. [DOI] [PubMed] [Google Scholar]
- 5.Chen, B., S. Gau, G. H. Choi, and D. L. Nuss. 1996. Extensive alteration of fungal gene transcript accumulation and elevation of G-protein-regulated cAMP levels by a virulence-attenuating hypovirus. Proc. Natl. Acad. Sci. USA 93:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchill, A. C. L., L. M. Ciuffetti, D. R. Hansen, H. D. van Etten, and N. K. van Alfen. 1990. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 17:25-31. [Google Scholar]
- 7.el-Sherbeini, M., and K. A. Bostian. 1987. Viruses in fungi: infection of yeast with the K1 and K2 killer viruses. Proc. Natl. Acad. Sci. USA 84:4293-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enebak, S. A. 1992. Characterization of dsRNA-containing strains of Cryphonectria parasitica recovered from the central Appalachian. Ph.D. thesis. West Virginia University, Morgantown.
- 9.Enebak, S. A., B. I. Hillman, and W. L. MacDonald. 1994. A hypovirulent isolate of Cryphonectria parasitica with multiple genetically unique dsRNA elements. Mol. Plant-Microbe Interact. 7:590-595. [Google Scholar]
- 10.Fukushi, T. 1969. Relationships between propagative rice viruses and their vectors, p. 279-301. In K. Maramorosch (ed.), Viruses, vectors, and vegetation. Wiley Interscience, New York, N.Y.
- 11.Fulbright, D. W. 1984. Effect of eliminating dsRNA in hypovirulent Endothia parasitica. Phytopathology 74:722-724. [Google Scholar]
- 12.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghabrial, S. A., and B. I. Hillman. 1999. Partitiviruses—fungal (Partitiviridae), p. 1147-1151. In A. Granoff and R. Webster (ed.), Encyclopedia of virology, 2nd ed., vol. 2. Academic Press, New York, N.Y.
- 14.Hillman, B. I., and D. L. Nuss. 1999. Phytoreoviruses (Reoviridae), p. 1262-1267. In A. Granoff and R. Webster (ed.), Encyclopedia of virology, 2nd ed., vol. 3. Academic Press, New York, N.Y.
- 15.Hillman, B. I., R. Shapira, and D. L. Nuss. 1990. Hypovirulence-associated suppression of host functions in Cryphonectria parasitica can be partially relieved by high light intensity. Phytopathology 80:950-956. [Google Scholar]
- 16.Hillman, B. I., Y. Tian, P. J. Bedker, and M. P. Brown. 1992. A North American hypovirulent isolate of the chestnut blight fungus with European isolate-related dsRNA. J. Gen. Virol. 73:681-686. [DOI] [PubMed] [Google Scholar]
- 17.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 18.Larson, T. G., G. H. Choi, and D. L. Nuss. 1992. Regulatory pathways governing modulation of fungal gene expression by a virulence-attenuating mycovirus. EMBO J. 11:4539-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maruyama, K., and S. Sugano. 1994. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene 138:171-174. [DOI] [PubMed] [Google Scholar]
- 20.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 21.Mertens, P. P. C., et al. 2000. Family Reoviridae, p. 395-480. In M. H. V. van Regenmortel et al., Virus taxonomy. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 22.Milgroom, M. G., K. Wang, Y. Zhou, S. E. Lipari, and S. Kaneno. 1996. Intercontinental population structure of the chestnut blight fungus, Cryphonectria parasitica. Mycologia 88:179-190. [Google Scholar]
- 23.Morris, T. J., and J. A. Dodds. 1979. Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854-858. [Google Scholar]
- 24.Nannelli, R., T. Turchetti, and G. Maresi. 1999. Corticolous mites (Acari) as potential vectors of Cryphonectria parasitica (Murr.) Barr hypovirulent strains. Int. J. Acarol. 24:237-244. [Google Scholar]
- 25.Nibert, M. L., and L. A. Schiff. 2001. Reoviruses and their replication, p. 1679-1729. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 26.Nuss, D. L. 2000. Hypovirulence and chestnut blight: from the field to the laboratory and back, p. 149-170. In J. W. Kronstad (ed.), Fungal pathology. Kluwer, Amsterdam, The Netherlands.
- 27.Omura, T., and J. Yan. 1999. Role of outer capsid proteins in transmission of Phytoreovirus by insect vectors. Adv. Virus Res. 54:15-43. [DOI] [PubMed] [Google Scholar]
- 28.Osaki, H., C. Z. Wei, M. Arakawa, T. Iwanami, K. Nomura, N. Matsumoto, and Y. Ohtsu. 2002. Nucleotide sequences of double-stranded RNA segments from a hypovirulent strain of the white root rot fungus Rosellinia necatrix: possibility of the first member of the Reoviridae from fungus. Virus Genes 25:101-107. [DOI] [PubMed] [Google Scholar]
- 29.Parsley, T. B., B. Chen, L. M. Geletka, and D. L. Nuss. 2002. Differential modulation of cellular signaling pathways by mild and severe hypovirus strains. Eukaryot. Cell 1:401-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polashock, J. J., and B. I. Hillman. 1994. A small mitochondrial double-stranded (ds) RNA element associated with a hypovirulent strain of the chestnut blight fungus and ancestrally related to yeast cytoplasmic T and W dsRNAs. Proc. Natl. Acad. Sci. USA 91:8680-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad, B. V. V., and M. K. Estes. 2000. Electron cryomicroscopy and computer imaging techniques, p. 9-31. In J. Gray and U. Duesselberger (ed.), Methods in molecular medicine. Humana, Totowa, N.J. [DOI] [PubMed]
- 32.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy, D. V., and L. M. Black. 1974. Deletion mutations of the genome segments of wound tumor virus. Virology 61:458-473. [DOI] [PubMed] [Google Scholar]
- 34.Rigling, D., U. Heiniger, and H. R. Hohl. 1989. Reduction of laccase activity in dsRNA-containing hypovirulent strains of Cryphonectria (Endothia) parasitica. Phytopathology 79:219-223. [Google Scholar]
- 35.Rigling, D., and N. K. Van Alfen. 1993. Extra- and intracellular laccases of the chestnut blight fungus, Cryphonectria parasitica. Appl. Environ. Microbiol. 59:3634-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smart, C. D., W. Yuan, R. Foglia, D. L. Nuss, D. W. Fulbright, and B. I. Hillman. 1999. Cryphonectria hypovirus 3-GH2, a virus species in the family Hypoviridae with a single open reading frame. Virology 265:66-73. [DOI] [PubMed] [Google Scholar]
- 37.Stanway, C., and K. W. Buck. 1984. Infection of protoplasts of the wheat take-all fungus, Gauemannomyces graminis var. tritici, with double-stranded RNA viruses. J. Gen. Virol. 65:2061-2066. [Google Scholar]
- 38.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4867-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei, C. Z., H. Osaki, T. Iwanami, N. Matsumoto, and Y. Ohtsu. 2003. Molecular characterization of dsRNA segments 2 and 5 and electron microscopy of a novel reovirus from a hypovirulent isolate, W370, of the plant pathogen Rosellinia necatrix. J. Gen. Virol. 84:2431-2437. [DOI] [PubMed] [Google Scholar]
- 40.Wendt, R., J. Weidhaas, G. J. Griffin, and J. R. Elkins. 1983. Association of Endothia parasitica with mites isolated from cankers on American chestnut trees. Plant Dis. 67:757-758. [Google Scholar]
- 41.Wickner, R. B. 2001. Viruses of yeasts, fungi, and parasitic microorganisms, p. 629-658. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 42.Xu, Z., J. V. Anzola, C. M. Nalin, and N. D. L. 1989. The 3′-terminal sequence of a wound tumor virus transcript can influence conformational and functional properties associated with the 5′-terminus. Virology 170:511-522. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Y. P., and A. Rowhani. 2000. A strategy for rapid cDNA cloning from double-stranded RNA isolated from plants infected with RNA viruses by using Taq DNA polymerase. J. Virol. Methods 84:59-63. [DOI] [PubMed] [Google Scholar]
- 44.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and bio/technology. Kluwer, Amsterdam, The Netherlands.