Summary
While environmental stress likely plays a significant role in promoting aging, the relationship remains poorly understood. In order to characterize this interaction in a more comprehensive manner, we examined the stress response profiles for 46 long-lived yeast mutant strains across four different stress conditions (oxidative, ER, DNA damage, and thermal), grouping genes based on their associated stress response profiles. Unexpectedly, cells lacking the mitochondrial AAA protease gene AFG3 clustered strongly with long-lived strains lacking cytosolic ribosomal proteins of the large subunit. Similar to these ribosomal protein mutants, afg3Δ cells show reduced cytoplasmic mRNA translation, enhanced resistance to tunicamycin that is independent of the ER unfolded protein response, and Sir2-independent but Gcn4-dependent life span extension. These data demonstrate an unexpected link between a mitochondrial protease, cytoplasmic mRNA translation, and aging.
Keywords: aging, stress response, translation, mitochondria, ER stress, replicative lifespan, longevity, yeast, epistasis, phenotype mapping
Introduction
Defining the molecular mechanisms of aging is one of the most challenging problems in modern biology. Hundreds of life span-extending mutations have been discovered in yeast, nematodes, and fruit flies, and a handful of conserved longevity pathways have been identified (Fontana et al. 2010; Kenyon 2010; Sutphin et al. 2012). For the vast majority of these genes, however, the pathways in which they act, and the mechanisms by which they modulate aging, remain poorly understood.
One feature that has been observed to correlate with longevity, both across species and among individuals of the same species, is altered resistance to different forms of stress. In general, long-lived mutants across a variety of species tend to be stress-resistant; however, there is specificity depending on the type(s) of stress applied and the longevity pathway(s) under study. For example, cell lines derived from long-lived species show enhanced resistance to several forms of stress, but also show enhanced sensitivity to other forms of stress (Harper et al. 2007; Salmon et al. 2008; Harper et al. 2011). In yeast, nematodes and fruit flies, many long-lived mutants are resistant to oxidative and thermal stress. For example, in yeast, long-lived cells deleted for the S6 kinase homolog SCH9 or the mammalian target of rapamycin homolog TOR1, up-regulate stress-responsive transcription factors, and also show enhanced stress resistance (Fabrizio et al. 2001; Powers et al. 2006). To date, however, no comprehensive analysis of the relationship between stress resistance and longevity has been performed in any system.
The budding yeast Saccharomyces cerevisiae provides an ideal model for exploring the relationship between stress resistance and longevity. The availability of collections containing individual single gene deletions for a majority of yeast genes has allowed for genome-scale studies of sensitivity and resistance for multiple forms of environmental stress (Thorpe et al. 2004; Postma et al. 2009). Replicative life span (RLS) in yeast is defined as the number of daughter cells produced by a mother cell before cessation of cell division (Mortimer & Johnston 1959). Several types of molecular damage are asymmetrically inherited by the mother cell and are proposed to limit RLS, including nuclear ribosomal DNA circles, cytoplasmic protein aggregates, and damaged mitochondria (Steinkraus et al. 2008; Kaeberlein 2010). Over the past several years, we have been screening strains derived from the yeast ORF deletion collection to identify single-gene deletions that increase RLS (Kaeberlein & Kennedy 2005). This has resulted in the identification of several dozen long-lived mutants chosen for study here.
One well-studied longevity pathway in yeast consists of long-lived mutants with reduced nutrient signaling and impaired mRNA translation. TOR1 and SCH9 are nutrient responsive kinases that regulate ribosome biogenesis and mRNA translation in response to nutrient availability (Longo & Fabrizio 2012). Under conditions of nutrient restriction, such as dietary restriction, reduced signaling through Tor1 and Sch9 along with other factors, coordinate a reduction in mRNA translation, an increase in autophagy, and a metabolic shift from fermentation to respiration (Kennedy et al. 2007). Deletion of either TOR1 or SCH9 is sufficient to increase RLS, and subjecting these mutants to dietary restriction fails to further increase life span (Kaeberlein et al. 2005b). The particular importance of mRNA translation in this pathway was suggested by the finding that deletion of multiple ribosomal protein genes is also sufficient to increase RLS (Steffen et al. 2008). With one exception (Chiocchetti et al. 2007), life span extension from ribosomal protein gene deletions in yeast appears to be specific for large ribosomal subunit (60S) genes that result in a deficiency of mature large ribosomal subunits (Steffen et al. 2008). Like tor1Δ or sch9Δ cells, RLS extension in mutants deficient for large ribosomal subunits is non-additive with dietary restriction and independent of the Sir2 protein deacetylase (Steffen et al. 2008; Delaney et al. 2011b). Although general mRNA translation is impaired, it is thought that RLS extension in these mutants results primarily from increased translation of the Gcn4 transcription factor under conditions where large ribosomal subunits are limiting (Steffen et al. 2008). This increase in Gcn4 translation has been attributed to the presence of inhibitory upstream open reading frames in the Gcn4 mRNA 5’ untranslated region, and Gcn4 is required for life span extension in several of the long-lived ribosomal large subunit gene deletion mutants (Steffen et al. 2008).
In this study, we have performed a systematic analysis of the stress response profiles for 46 long-lived deletion strains across four different stress conditions. The growth properties of each strain were assessed under conditions designed to induce ER stress (tunicamycin), oxidative stress (paraquat), DNA damage stress (methyl methanesulfonate, or MMS), or thermal stress (heat shock). Although long-lived mutants were often resistant to one or more stressors, relative to the parental wild type strain (BY4742), none of the stressors elicited a similar response across the entire panel of long-lived strains. Analysis of the stress response profiles using cluster algorithms grouped long-lived strains into distinct classes. One such class included several large subunit ribosomal protein deletion strains that showed resistance to growth inhibition by tunicamycin and paraquat, as well as resistance to heat shock. Also included in this class was a strain lacking the gene coding for the mitochondrial m-AAA protease Afg3 (also referred to as Yta10). Afg3 is known to play an important role in regulating electron transport chain complexes and is also required for maturation of the mitochondrial ribosomal protein Mrpl32 (Arlt et al. 1998; Nolden et al. 2005)
In this study, deletion of AFG3 was found to cause a profound reduction in cytoplasmic mRNA translation and an increase in resistance to tunicamycin that was independent of the ER unfolded protein response transcription factor Hac1. Similar to deletion of RPL20B, life span extension from deletion of AFG3 is independent of both Hac1 and Sir2, but requires Gcn4. These observations demonstrate an important new role for the mitochondrial protease Afg3 in modulating cytoplasmic mRNA translation and ER stress resistance, and suggest that Afg3 modulates longevity in yeast by a mechanism similar to cytoplasmic ribosomal large subunit proteins.
Results
Quantitative determination of stress induced growth rate changes among long-lived mutants
In order to quantitatively assess the stress response profiles of long-lived yeast deletion strains, we utilized a Bioscreen C MBR machine to determine doubling time in the presence or absence of a stressor for 46 long-lived mutants. The Bioscreen C MBR is a combined shaker, incubator, and plate reader that has been shown to provide high quality growth rate determinations for yeast cells maintained under a variety of conditions (Murakami et al. 2008; Burtner et al. 2009a; Burtner et al. 2009b; Murakami & Kaeberlein 2009; Burtner et al. 2011; Kruegel et al. 2011). Since many long-lived deletion strains have doubling times that differ from the wild type control (Delaney et al. 2011a), we normalized growth rate changes induced by stress (GRCs) by normalizing to the growth rate under untreated conditions using the following equation:
where Ds is the doubling time of the strain in rich growth medium (YPD) in the presence of the stressor and Du is the doubling time of the strain in the same medium in the absence of the stressor. Using this method, we determined the growth inhibition stress responses for each of 46 long lived mutants as well as wild type and control sensitive strains to three different chemical stress conditions: ER stress induced by tunicamycin, oxidative stress induced by paraquat, and DNA damage stress induced by MMS (Supplemental Tables 1–3). It should be noted that each of these chemicals may induce additional forms of stress in addition to those described above. Resistance to thermal stress was also assessed as the percent of cells that maintained viability following a heat shock (Supplemental Table 4).
Quantitative determination of tunicamycin induced growth rate changes among long-lived mutants
Growth inhibition in response to ER stress was determined by monitoring outgrowth kinetics in the presence of 1 ug/mL tunicamycin, an inhibitor of N-linked protein glycosylation that causes an accumulation of proteins in the ER and induces the ER unfolded protein response. This concentration of tunicamycin was sufficient to increase the doubling time of wild type BY4742 cells by nearly 200% (Figure 1a, Supplemental Table 1). In order to validate the method, we included as a control cells lacking HAC1, which encodes a transcription factor required for induction of many proteins involved in the ER unfolded protein response (Travers et al. 2000). As expected, hac1Δ cells showed extreme growth inhibition by 1 µg/mL tunicamycin, such that not even a single doubling in cell density was observed over 24 hours (Figure 1a, Supplemental Table 1).
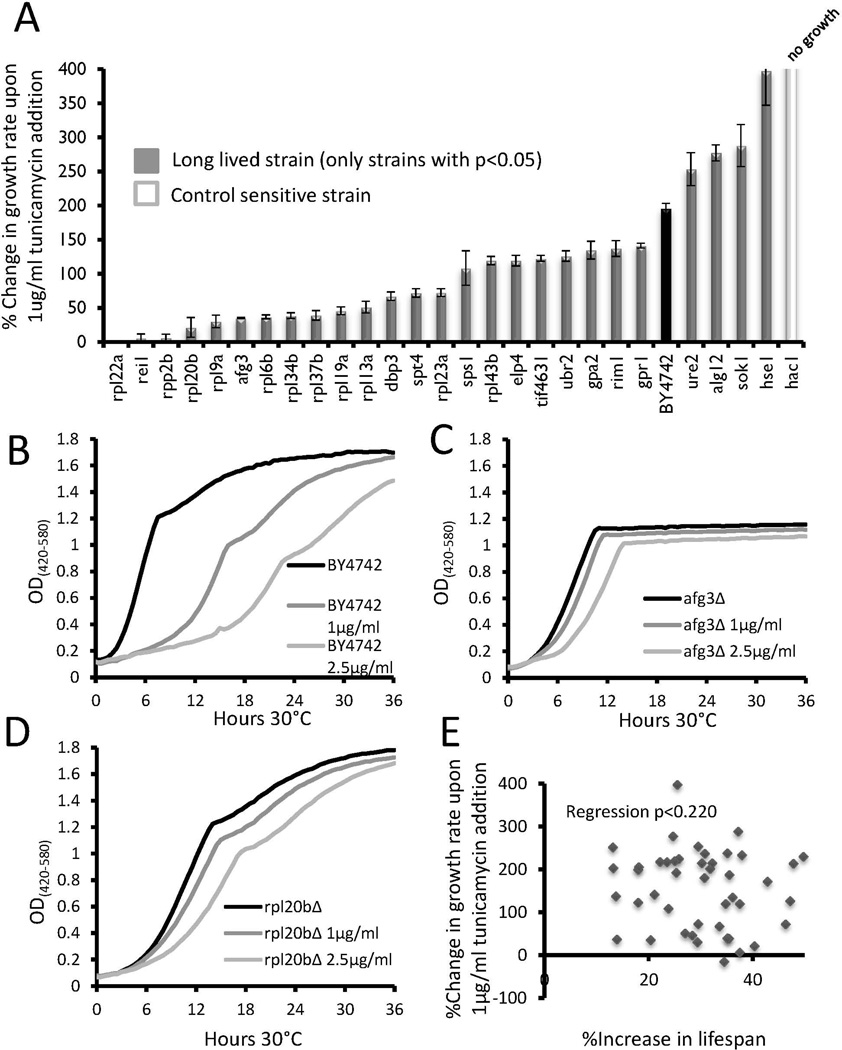
Figure 1. Growth inhibition of long lived strains in response to tunicamycin.
(a) Long lived strains which showed significant (p < 0.05) changes in relative growth inhibition compared to wild type BY4742 are shown. The control sensitive strain hac1Δ did not grow in the media. Error bars are s.e.m. Data for all strains tested can be found in Supplemental Table 1.
(b–d) Representative outgrowth curves for wild type, rpl20bΔ, and afg3Δ strains in 0, 1, or 5 µg/mL tunicamycin.
(e) Scatter plot comparing the percent change in replicative lifespan to percent change in growth rate for each long-lived mutant.
Of the 46 long-lived mutants tested, 22 had a lower GRCs than wild type BY4742 (WT) cells in the presence of tunicamycin, 4 showed greater growth inhibition (higher GRCs), and 20 had no significant change from WT (p < 0.05 in each case, Figure 1a, Supplemental Table 1). Notably, all ten long-lived large ribosomal protein deletion strains had significantly lower GRCs than WT. Three additional mutants with known defects in large ribosomal subunit biogenesis or function (rei1Δ, rpp2bΔ, and dbp3Δ) showed a similar resistance to tunicamycin growth inhibition, while mutations in the signaling kinases that regulate mRNA translation (tor1Δ and sch9Δ) did not.
Afg3 encodes a component of a mitochondrial inner membrane AAA protease that functions to maintain proteostasis in the mitochondria and promote proper assembly of electron transport chain complexes (Guelin et al. 1994; Arlt et al. 1996; Arlt et al. 1998). Interestingly, long-lived afg3Δ cells showed reduced growth inhibition in the presence of tunicamycin that was comparable to the large subunit ribosomal protein deletion mutants, such as rpl20bΔ (Figure 1b–d). This resistance to growth inhibition is particularly evident at higher concentrations of tunicamycin, such as 2.5 µg/mL, where the doubling time of afg3Δ (156 min) and rpl20bΔ (185 min) was faster than that of WT cells (284 min), despite both mutants being slow-growing in unstressed conditions. The absence of respiratory growth in afg3Δ cells is evident from the flat growth curve after glucose has been consumed. The Bioscreen C MBR machine provides a linear estimate of cell density from approximately OD 0.1-1.0, so anything above this range is an underestimate of the cell density.
Across the entire panel of 46 long-lived mutants, there was no significant correlation between tunicamycin growth rate changes and RLS (Figure 1e).
Quantitative determination of paraquat induced growth rate changes among long-lived mutants
The effect of oxidative stress on growth inhibition was determined by culturing cells in the presence of the superoxide-generating chemical paraquat. Addition of 5 mM paraquat was sufficient to increase the doubling time of wild type cells by 69% (Figure 2a, Supplemental Table 2). As controls, we examined growth inhibition for three strains previously annotated as sensitive to oxidative stress: ctt1Δ, cta1Δ, and sod1Δ (Cohen et al. 1985; Slekar et al. 1996). CTT1 and CTA1 encode cytosolic and peroxisomal catalase enzymes necessary for detoxification of hydrogen peroxide, respectively, while SOD1 encodes the cytosolic superoxide dismutase that is primarily responsible for converting superoxide to hydrogen peroxide. Cells deleted for SOD1 are significantly shorter-lived than BY4742 wild type cells, while cells lacking either CTT1 or CTA1 have a RLS that does not differ significantly from wild type in this background (Supplemental Figure 2).
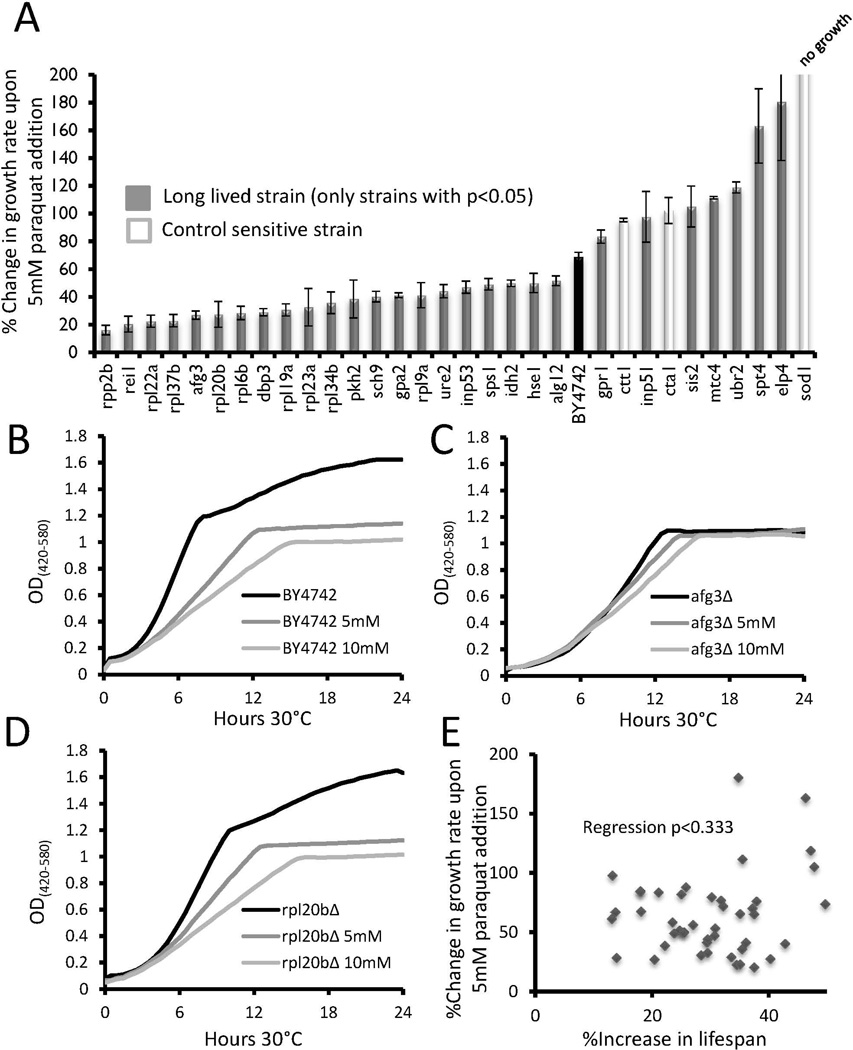
Figure 2. Growth inhibition of long lived strains in response to paraquat.
(a) Long lived strains which showed significant (p < 0.05) changes in relative growth inhibition compared to wild type BY4742 are shown. The control sensitive strains ctt1 Δ,cta1 Δ, and sod1Δ are shown for comparison. Error bars are s.e.m. Data for all strains tested can be found in Supplemental Table 2.
(b–d) Representative outgrowth curves for wild type, rpl20bΔ, and afg3Δ strains in 0, 5, or 10 mM paraquat.
(c) Scatter plot comparing the percent change in replicative lifespan to percent change in growth rate for each long-lived mutant.
Among the 46 long-lived strains examined, eight were significantly more growth inhibited by paraquat, relative to the wild type control, while twenty-one were less inhibited (p<0.05 in each case, Figure 2a, Supplemental Table 2). The remaining 17 long-lived mutants showed changes in doubling time (GRCs values) that did not differ significantly from that of wild type cells. As was observed for tunicamycin, afg3Δ and rpl20bΔ cells were significantly less growth inhibited by paraquat compared to WT (Figures 2b–d). No significant correlation was observed between the effect of paraquat on growth and magnitude of life span extension across the 43 long-lived strains (Figure 2e).
Quantitative determination of MMS induced growth rate changes among long-lived mutants
The DNA alkylating agent methyl methanesulfonate (MMS) was utilized to assay for the effect of DNA damage stress on growth inhibition. At 0.01%, MMS increased the doubling time of WT cells by 66% (Figure 3a, Supplemental Table S3). As a control, we included rad52Δ cells defective for homologous recombination (Herzberg et al., 2006). The same concentration of MMS increased the doubling time of rad52Δ by 506%.
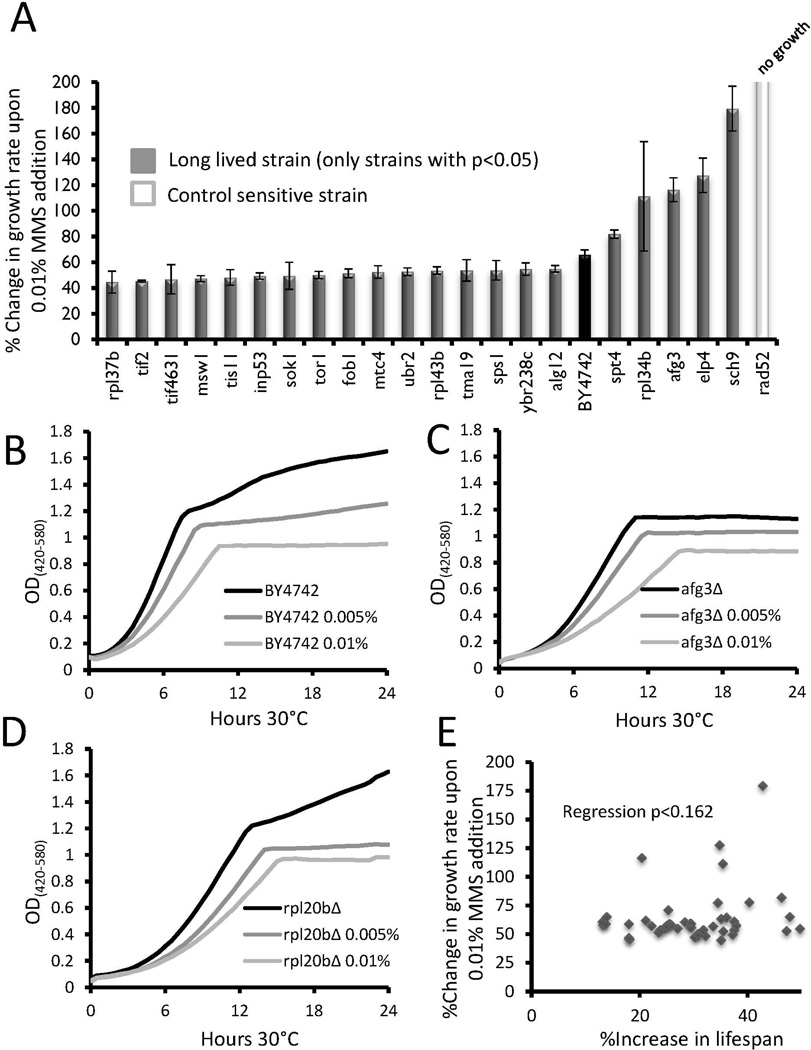
Figure 3. Growth inhibition of long lived strains in response to MMS.
(a) Long lived strains which showed significant (p < 0.05) changes in relative growth inhibition compared to wild type BY4742 are shown. The control sensitive rad52Δ strain is shown for comparison. Error bars are s.e.m. Data for all strains tested can be found in Supplemental Table 3.
(b–d) Representative outgrowth curves for wild type, rpl20bΔ, and afg3Δ strains in 0, 0.005%, or 0.01% MMS.
(c) Scatter plot comparing the percent change in replicative lifespan to percent change in growth rate for each long-lived mutant.
Of the 46 long-lived mutants, 15 showed a significant reduction in growth inhibition compared to WT, while 6 had a higher GRCs (p<0.05 in each case, Figure 3a, Supplemental Table S3). The long-lived fob1Δ strain showed less growth inhibition by MMS (Figure 3a), perhaps related to the enhanced genomic stability of this strain at the ribosomal DNA. Once again, afg3Δ and rpl20bΔ showed a similar response to MMS, although in this case both strains trended toward enhanced growth inhibition rather than reduced growth inhibition, relative to WT (Figures 3b–d). As with tunicamycin and paraquat, no significant correlation between growth inhibition in the presence of MMS and longevity was observed (Figure 3e).
Quantitative determination of heat shock stress profiles among long-lived mutants
Resistance to thermal stress was assessed by quantifying viability following a 10 minute transient 55°C heat shock. After thermal stress, 15.8% of wild type control cells from an overnight culture retained viability. Within the set of 46 long-lived mutants, 23 had a higher survival than wild type whereas only 4 showed increased death (p < 0.05, Figure S1a, Supplemental Table S4). No significant difference in survival was detected for 19 long-lived mutants. As expected, the control sensitive mutants, heat shock response factor double mutant msn2Δmsn4Δ and ER stress response factor deletion hac1Δ, were sensitive to heat shock.
As with growth inhibition from tunicamycin, many long-lived ribosomal protein deletion mutants showed resistance to heat shock, relative to WT. Also, once again, afg3Δ cells showed a response similar to ribosomal protein deletion mutants, such as rpl20bΔ (Figure S1b); however, no significant correlation between heat shock resistance and RLS was detected across all 46 long-lived strains (Figure S1c).
Stress response profiling places afg3Δ cells into the mRNA translation longevity pathway
We next asked whether the stress response profiles of long-lived mutants could be informative regarding potential mechanisms of lifespan extension. To assess this, we applied the Array Track™ clustering algorithm to the stress response profiles for each strain subjected to the four stress inducers (Figure 4). We utilized the “complete linkage analysis” option to maximize distances between clusters in order to identify distinct pathways. Although the power of this approach is likely to be limited be the relatively small number of conditions examined, we noted that many of the mutants with known defects in mRNA translation clustered together (Cluster I). These included several strains lacking ribosomal proteins of the large subunit or rRNA processing factors (dbp3Δ and rei1Δ) that were characterized by significantly reduced growth inhibition from paraquat, tunicamycin, and heat shock. Cells lacking AFG3 were also contained in this Cluster.
Figure 4. Clustering of long lived mutants based on their stress response profiles.
Clusters were formed using Euclidean distance calculations of vectors constructed from the percent difference in sensitivity of a mutant strain compared to wild type, as described in Methods.
The remaining genes could be classified into three additional clusters (II–IV). Unlike Cluster I, the functional and mechanistic relationships among the grouped genes were less clear. Cluster II differed from Cluster I primarily in the response to paraquat, with strains in this group showing stronger growth inhibition than wild type following oxidative stress. Cluster II contained the recently characterized ubr2Δ mutant, which extends life span by up-regulation of proteasome gene expression and activity (Kruegel et al. 2011), as well as mtc4Δ, spt4Δ, and elp4Δ. Cluster III comprises mutants largely unaffected (less growth rate changes) by paraquat but growth-inhibited by tunicamycin. This cluster contains the tor1Δ strain, as well as translation initiation factor deletion strains, tif1Δ and tif2Δ. The remaining Group IV is more likely an unspecific group rather than a bona fide cluster, since it contains a variety of mutants that generally showed similar growth inhibition as WT across the different stress conditions.
Deletion of AFG3 promotes resistance to tunicamycin by a HAC1-independent mechanism
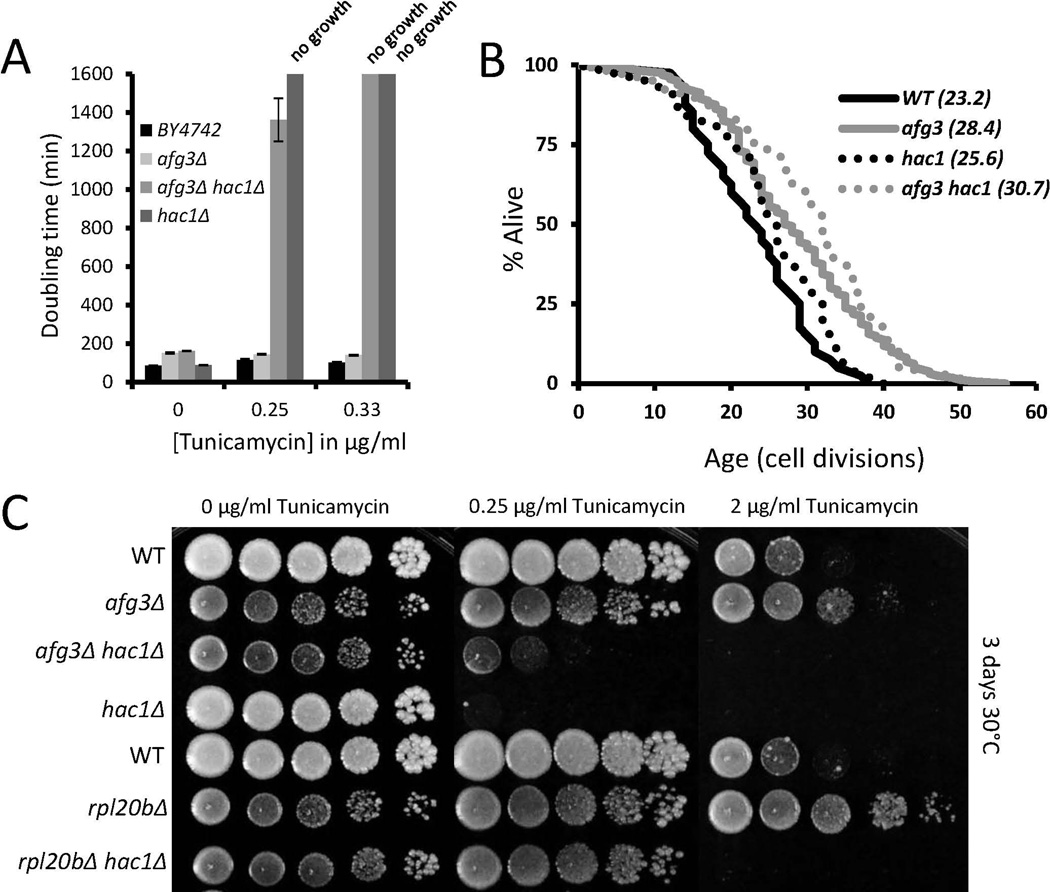
Based on the significant resistance to growth inhibition from tunicamycin observed for afg3Δ cells in liquid culture, we asked whether this phenotype could be recapitulated using traditional spot assays on solid agar medium. Cells deleted for AFG3 were also found to be resistant to tunicamycin in agar plates, although rpl20bΔ cells appeared to be even more resistant (Figure 5a,c). We reasoned that resistance to tunicamycin in afg3Δ cells could be caused by constitutive induction of the ER unfolded protein response. To test this possibility we deleted HAC1, which is required for induction of the ER unfolded protein response, in afg3Δ cells. Interestingly, afg3Δhac1Δ double mutants still showed enhanced resistance to tunicamycin, relative to hac1 Δ cells, demonstrating that loss of AFG3 promotes resistance to ER stress by a mechanism that is at least partially independent of the canonical unfolded protein response. Consistent with these observations, life span extension from deletion of AFG3 did not require HAC1 (Figure 5b). These results are similar to our recent studies of cytoplasmic ribosomal protein deletions, where deletion of RPL20B showed a similar HAC1-independent resistance to tunicamycin (as in Figure 5c) and life span extension (Steffen et al. 2012).
Figure 5. Hac1-independent resistance to tunicamycin in afg3Δ and rpl20bΔ cells.
(a) Average doubling times for wild type BY4742, afg3Δ, hac1Δ, and afg3Δhac1Δ strains at the indicated concentrations of tunicamcycin. Note how afg3Δhac1Δ double mutant cells can grow, albeit very slowly, at 0.25µg/ml tunicamycin, while hac1Δ cells cannot, indicating Hac1-independent tunicamycin resistance.
(b) Replicative lifespan curves of the indicated strains. Parentheses denote mean lifespan. afg3Δ does not require Hac1 for lifespan extension.
(c) Spot assays of the indicated strains across tunicamycin concentrations. Spots were diluted 1:10. Panels were grown for 3 days at 30°C.
Deletion of AFG3 reduces cytoplasmic translation and extends RLS by a Gcn4-dependent mechanism
Based on the stress response studies described above, we speculated that deletion of AFG3 might extend life span by a similar mechanism to that of cells deficient for ribosomal large subunits. We have previously shown that life span extending mutations resulting in large subunit deficiency, such as rpl20bΔ, decrease overall mRNA translation as measured by polysome analysis (Steffen et al. 2008). Similar to rpl20bΔ cells, afg3Δ cells showed a significant decrease in mRNAs with 2 or more ribosomes bound, consistent with a dramatic decrease in overall cytoplasmic mRNA translation (Figure 6a, b). In contrast, however, no significant change in the relative abundance of free large ribosomal subunits (or small subunits) was detected. This suggests that, while overall mRNA translation is reduced, the mechanism of translation inhibition in afg3Δ cells does not involve specific depletion of large ribosomal subunits.
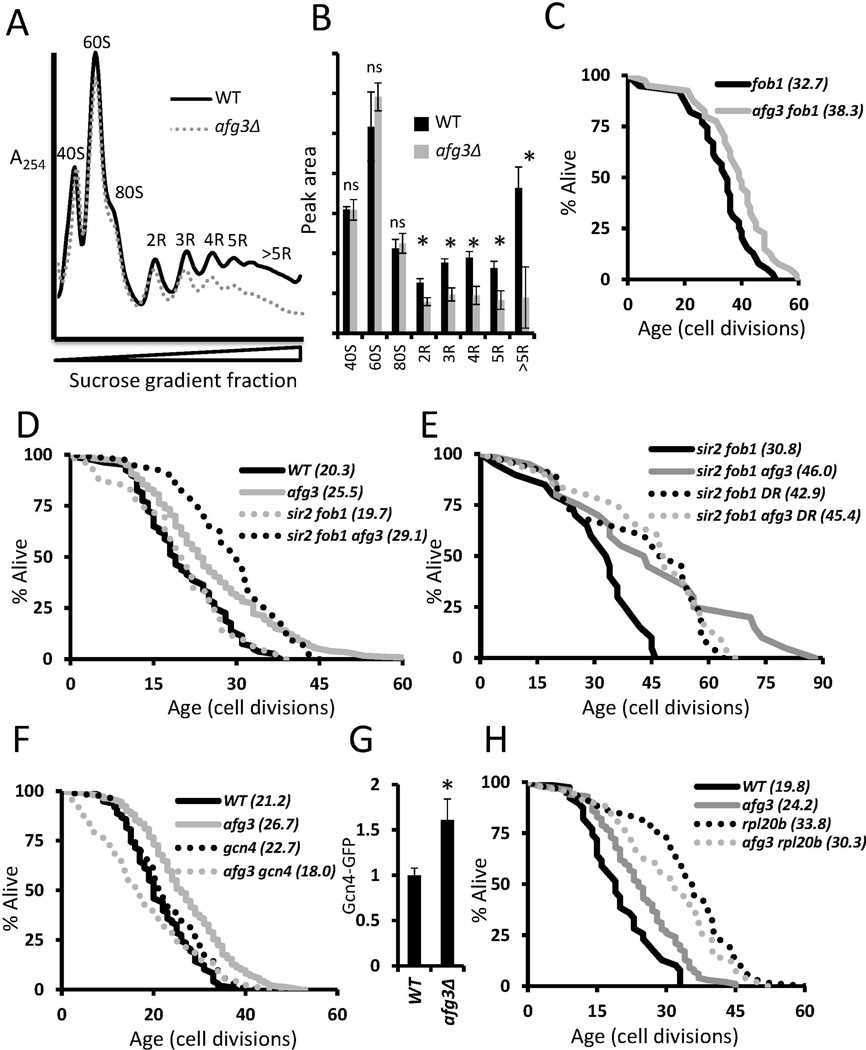
Figure 6. Deletion of AFG3 reduces cytoplasmic translation and extends life span by a Sir2-independent, Gcn4-dependent mechanism.
(a) Representative polysome profiles of log phase wild type BY4742 yeast and afg3Δ mutant yeast grown in YPD at 30°C. Deletion of AFG3 causes a shift of ribosomes from highly translated mRNA to unbound forms. Curves are normalized by the minima between 80S free subunits and 2R disomes.
(b) Quantitation of peak areas from triplicate polysomes. *p<0.05, ns= not significant, error bars are s.e.m.
(c–f) Replicative lifespan curves of the indicated strains. Parentheses denote mean lifespan. afg3Δ requires Gcn4 for lifespan extension but does not require Sir2 or Fob1. afg3Δ is not additive with rpl20bΔ nor DR (0.05% glucose) for lifespan extension.
(g) Flow cytometric analysis of log phase genomically tagged Gcn4-GFP strains.
Cold sensitive growth is associated with defects in cytoplasmic mRNA translation, as well as some mitochondrial proteases (Francis & Thorsness 2011). We therefore examined the effect of reduced temperature on afg3Δ and rpl20bΔ cells. Interestingly, neither mutant significantly extended RLS at 15°C. We were also unable to detect enhanced resistance to tunicamycin at 15°C (Figure S5). These data further support the model that AFG3 and RPL20B influence stress resistance and RLS by similar mechanisms.
We have previously shown that life span extension from deletion of RPL20B or other large ribosomal subunit genes occurs by a mechanism that is independent of both the Sir2 protein deacetylase, the fork blocking protein Fob1, but involves activation of the Gcn4 transcription factor (Steffen et al. 2008; Delaney et al. 2011b). In order to assess whether AFG3 is modulating aging by a similar mechanism, we determined the RLS of afg3Δfob1Δ double mutant cells, afg3Δsir2Δfob1Δ triple mutant cells, and afg3Δgcn4Δ double mutant cells. As is the case for cells lacking large ribosomal protein subunits (Delaney et al. 2011b), deletion of AFG3 further extends the RLS of cells lacking both SIR2 and FOB1 (Figure 6c,d), but fails to extend RLS of dietary restricted cells (Figure 6e) or in cells lacking GCN4 (Figure 6f). Additionally, we observed an increase in Gcn4 protein levels in the afg3Δ mutant via flow cytometric analysis of a Gcn4-GFP strain (Figure 6g). Combining deletion of AFG3 with deletion of RPL20B also fails to result in an additive increase in RLS (Figure 6h). Along with the stress response, temperature response, and polysome data, these longevity epistasis data support the model that deletion of the mitochondrial protease gene AFG3 unexpectedly reduces cytoplasmic mRNA translation and increases life span by a mechanism similar to deletion of large subunit ribosomal protein genes.
Discussion
The relationship between stress resistance and longevity is complex. A systematic analysis of stress response for 43 long-lived yeast deletion strains across 4 different stress conditions revealed that a majority of mutants show altered response to one or more types of stress. More often than not, the change in stress response was toward enhanced resistance and decreased growth inhibition, although there were many observed cases of increased sensitivity or growth inhibition in the long-lived mutants, and no significant correlation between absolute RLS and altered stress resistance was detected for any of the stress conditions tested. Despite the complexity of the data, this analysis allowed for successful classification of long-lived mutants by their stress response profiles. In particular, the striking similarly in stress response of afg3Δ cells with ribosomal protein gene deletions led us to uncover a previously unsuspected role for this mitochondrial protease in cytoplasmic mRNA translation, ER stress resistance, and Gcn4-dependent longevity control. Thus, successful classification of Afg3 represents proof-of-principle that this type of stress response profiling approach can be used to predict the longevity pathway for unknown factors, as well as uncover novel cellular functions.
It should be noted that our method for quantitatively assessing growth inhibition in responses to paraquat, tunicamycin, and MMS differs from traditional methods such as spot assays or plating for colony forming units. Unlike these traditional methods, which identify changes in sensitivity of about 2-fold, we were able to reproducibly quantify changes in doubling time of less than 10% using this method (Tables S1-S3). Two possible interpretations can explain a difference in growth rate in these assays. One is simply that a different percentage of cells are dying and cannot give rise to the next generation, thereby influencing the doubling time of the population as a whole. This interpretation is supported by the fact that all of our sensitive mutant controls displayed longer doubling times. Another possibility is that the drug is inducing a slowed cell cycle and that while most cells are indeed viable, the population doubling time increases due to more cells reaching a damage level that activates a cellular response or checkpoint. Either case is physiologically relevant. It is possible that differing stress response profiles, and the resulting clusters, would be obtained if alternative methods that directly measure cell death rather than growth inhibition were used. In the case of tunicamycin, however, we observed similar resistance in both growth inhibition assays and less sensitive spot assays for rpl20bΔ and afg3Δ.
The finding that loss of Afg3 results in reduced cytoplasmic mRNA translation and enhanced resistance to tunicamycin was unexpected, as prior studies had indicated purely mitochondrial functions for Afg3 in yeast, as well as its orthologs in mammals (Juhola et al. 2000; Andreou & Tavernarakis 2010). Although it is possible that Afg3 may also function in the cytoplasm to modulate mRNA translation, we know of no evidence to support this model. Instead, we speculate that the mitochondrial function of Afg3 indirectly promotes cytoplasmic mRNA translation, perhaps by ensuring appropriate regulation of mitochondrial mRNA translation. In support of this idea, it has been shown that cleavage of the mitochondrial ribosomal protein Mrpl32 by Afg3 is required for assembly of ribosome particles in the mitochondria (Nolden et al. 2005). It may be that failure to properly assemble mitochondrial ribosomes or other mitochondria complexes induces a signal from the mitochondria to the cytoplasm to inhibit cytoplasmic mRNA translation. Further support for this idea is provided by the observation that mrpl32Δ cells are also slow-growing and long-lived (Figure S4). Such a response may be important for preventing an imbalance between nuclear encoded mitochondrial proteins and mitochondrially encoded proteins under conditions where mitochondrial translation is impaired. Interestingly several other mitochondrial ribosomal protein deletion strains have been found to have normal or reduced RLS (Kaeberlein et al. 2005b; Managbanag et al. 2008), indicating that there is at least some specificity with respect to reduced mitochondrial translation and RLS.
A possible alternative explanation for our data is that the inability to carry out mitochondrial respiration directly causes a deficit in mRNA translation due to depletion of ATP needed to synthesize ribosomes and new proteins. We do not favor this model, however, since not all respiratory deficient mutants show growth defects or RLS extension similar to afg3Δ cells (Figure S3). Also, deletion of AFG3 results in slow growth and respiratory deficiency in the W303AR strain background, but fails to extend RLS in this strain (Figure S4d). This is consistent with our prior data that DR also fails to extend RLS in this strain background (Kaeberlein et al. 2006). A second alternative explanation is the possibility that loss of AFG3 induces yeast cells to lose their mitochondrial DNA (become rho0), which can reduce growth rate and has previously been shown to extend RLS in some strain backgrounds (Kirchman et al. 1999). However, rho0 mutants in the BY background do not have an extended RLS (Kaeberlein et al. 2005a) (Fig. S4b), but afg3Δ rho0 cells are still long-lived (Fig S4b). Furthermore, we crossed our long-lived afg3Δ haploid to a rho0 haploid of the opposite mating type and observed that the resulting diploid is able to carry out mitochondrial respiration as evidenced by growth on the non-fermentable carbon source, glycerol (Figure S4c). Thus, cells lacking AFG3 are long-lived independent of the status of their mitochondrial DNA.
One prior study reported resistance to oxidative stress and RLS extension in cells lacking the mitochondrial ribosomal protein Afo1 (Heeren et al. 2009). It is tempting to speculate that the mechanism of RLS extension and stress resistance is similar between deletion of AFG3 and AFO1. Unlike the case for afg3Δ cells, however, afo1Δ cells were reported to have a normal growth rate (and presumably normal mRNA translation). Further studies will be needed to establish the precise relationship between Afo1 and Afg3 with respect to longevity, stress resistance, and cytoplasmic mRNA translation.
In addition to RLS, a second type of aging (referred to as chronological aging) is studied in yeast by culturing the cells into stationary phase and monitoring viability over time. Many chronologically long-lived mutants are resistant to multiple forms of stress; however, unlike RLS, the vast majority of respiratory-deficient mutants have reduced chronological life span, likely as a result of an inability to properly enter stationary phase. Consistent with this, afg3Δ cells also have reduced chronological lifespan (Burtner et al. 2011).
The Hac1-independent tunicamycin resistance displayed by afg3Δ cells can likely be attributed to the profound reduction in cytoplasmic mRNA translation, since rpl20bΔ cells showed an even more pronounced tunicamycin-resistance. The mechanism accounting for this phenotype in rpl20bΔ and afg3Δ cells is not known, but may simply reflect reduced flux of proteins through the ER (Steffen et al. 2012). Since tunicamycin induces ER stress by inhibiting glycosylation of ER proteins, it may be that simply reducing translation of proteins destined for the ER partially alleviates this stress. Such a model would also explain why the enhanced resistance to tunicamycin does not fully require Hac1 and the ER unfolded protein response. As noted above, however, we cannot rule out the possibility that tunicamycin is inhibiting growth by affecting cellular processes other than ER stress and that reduced mRNA translation is interacting with these additional processes to alleviate the growth-inhibitory effects of tunicamycin.
It is of particular interest that the extension of RLS in afg3Δ cells requires Gcn4, similar to ribosomal large subunit deletion mutants such as rpl20bΔ. While deletion of either AFG3 or RPL20B results in a similar reduction in overall mRNA translation, the polysome profile of afg3Δ cells does not show the profound depletion of 60S ribosomal subunits seen in long-lived ribosomal protein gene deletion strains (Steffen et al. 2008). Instead, the polysome profile of afg3Δ cells more resembles the general decrease in translation without an imbalance of free 40S and 60S subunits seen in sch9Δ cells; however, in the case of SCH9 deletion the RLS extension is only partially dependent on Gcn4 (Steffen et al. 2008). The situation is made more complex by the observation that reduced mRNA translation is not always sufficient to extend RLS; for example, small ribosomal subunit protein gene deletions or treatment with the translation inhibitor cycloheximide do not extend RLS (Steffen et al. 2008). This may suggest that loss of AFG3 extends life span via a reduction in mRNA translation that mimics the Gcn4-dependent component of RLS extension in sch9Δ cells, but not the Gcn4-independent component.
This study demonstrates the utility of stress response profiling as a method for classifying longevity mutants in yeast. The observation that loss of the highly conserved m-AAA protease Afg3 phenocopies the longevity extension, enhanced tunicamycin resistance, and mRNA translation deficit of ribosomal protein deletion mutants suggests a previously unknown mechanism for regulating these processes in response to a mitochondrial signal. This is of particular interest in light of recent work in C. elegans showing that mitochondrial defects can modulate cytoplasmic translation through phosphorylation of eIF2α, although in that system the mitochondrial signal appears to be growth promoting rather than growth inhibitory (Baker et al. 2012). It will be of interest to further define the detailed mechanisms linking Afg3 function and cytoplasmic mRNA translation, as well as the specificity of translation-inhibition in afg3Δ mother cells, which allows for enhanced longevity. These future studies are likely to provide key insights into the complex nature by which cells sense and respond to mitochondrial stress, and how such responses impact longevity and health.
Experimental Procedures
Yeast strains
Yeast strains were derived from the MAT α ORF deletion collection and are isogenic to the parental BY4742 strain, with the following exceptions. The SIR2OX strain is a tandem two copy SIR2 strain with a URA3 marker generated by integration of a second copy of SIR2 in BY4742 as previously described (Kaeberlein et al. 1999). With concern for spontaneous suppressors, all ribosomal gene deletions were generated by sporulation from the heterozygous diploid deletion collection and are isogenic to the MAT α ORF deletion collection. The afg3Δ and mrpl32Δ mutants were remade through homologous recombination of a URA3 marker at the endogenous locus and then backcrossed to BY4741 and sporulated. The sir2Δfob1Δafg3Δ mutant was generated by homologous recombination of URA3 into a previously described (Kaeberlein et al. 2004) sir2Δfob1Δ strain. The afg3Δgcn4Δ and afg3Δhac1Δ strains were made from standard sporulation and tetrad dissection procedures from the remade afg3Δ strain and deletion collection strains. The rpl20bΔhac1Δ strain was a gift from K. Steffen, and was made similarly from an rpl20b Δ::HIS3 marked strain. Full genotypes are included in the strain list in Table S5.
Growth rate and viability analysis
Cultures were typically grown at 30°C overnight in a 96 well plate in YPD and then 2.5µl of the overnight culture was inoculated into 147.5µl of culture media (YPD+diluent or YPD+drug) in 100 well Bioscreen plates. Unless otherwise indicated, paraquat was used at a 5mM concentration, tunicamycin at a 1µg/ml concentration, and MMS at a 0.01% concentration.
Yeast growth rates were analyzed in the OD420–580 range in the Bioscreen C MBR machine (Growth Curves USA) as previously described using the Yeast Outgrowth Data Analyzer (YODA)(Olsen B 2010). The Bioscreen C MBR machine provides a linear estimate of cell density from approximately OD 0.1–1.0, so anything above this range is an underestimate of the cell density. Reported doubling times in 30°C YPD are taken from interval readings from the OD420–580 0.2–0.5 range of maximum growth rate. Inhibition of growth in response to stress was calculated as described above using the formula:
where Ds is the doubling time of the strain in rich growth medium (YPD) in the presence of the stressor and Du is the doubling time of the strain in the same medium in the absence of the stressor.
Heat shock survival experiments were performed with 10 minutes of 55°C incubation of overnight cultures grown in 96 well plates and then placed into the Bioscreen analyzer and survival integrals calculated using YODA. While the Bioscreen C MBR machine only measures culture density, we have shown in the past that growth curves can quantitatively assay for cell viability. We utilized the chronological aging features of YODA to calculate the viability of cultures that had undergone heat shock, which we have previously verified as an equally quantitative measure of viability as counts of colony forming units. Cell death is not significant during outgrowth, as cells show full viability after the growth phase ends, as assayed at 24 hours of incubation (Figure S6).
Statistical significance was determined by applying a two tailed student t-test of GRCs in YPD+drug media compared to wild type in YPD+drug media. Error bars are standard error of the mean (s.e.m.). A minimum of four biological replicate cultures were examined for each condition. Trendlines comparing change in RLS to stress phenotypes utilized Excel’s Analysis ToolPak add-in using the P value generated in the ToolPak’s regression analysis.
Cluster analysis
Clustering was performed using the Array Track™ clustering algorithms. Complete linkage analysis using Euclidean distances was used in the settings. Vectors are shown in Supplemental Tables S1–4 as the log2(Vs) value, where the percent difference between the long lived mutant and wild type was calculated (Vs= (GRCs BY4742-GRCs mutant)/(GRCs BY4742)) and then log2 was taken of this value to mimic microarray style data. Heat maps are shown by taking the minimum values and assigning them the brightest green color and the maximum values as the brightest red color, and black represents a log2(Vs)=0.
Replicative lifespan analysis
Yeast RLS assays were performed as previously described (Kaeberlein et al. 2004; Steffen et al. 2009). In short, virgin daughter cells were isolated from each strain and then allowed to grow into mother cells while their corresponding daughters were microdissected and counted until the mother cell could no longer divide. YEP agar plates (1% yeast extract, 2% bacto-peptone, 2% agar) containing 2% glucose were utilized and strains were grown at 30°C. Statistical significance was determined using the Wilcoxon Rank-Sum test. Dietary restriction was accomplished by reducing the glucose present in the medium from 2% to 0.05%.
Flow cytometry
Flow cytometry was performed on a BD Biosciences Influx Cell Sorter. For GFP experiments, cells were grown overnight with dilute streaks on YPD or YEP + 0.05% glucose plates to maintain log phase growth across different conditions. Cells were directly taken from the plates and resuspended in iced 50mM Na Citrate. GFP intensity values were calculated as mean raw GFP intensity subtracted by GFP auto fluorescence of isogenic cells grown under identical conditions. All values were also normalized to relative cell size, as measured by forward scatter. GFP fold changes (Fig. 6g) were normalized to normal 2% glucose growth conditions of WT (but GCN4-GFP tagged) cells as an intensity of “1”. Values were compared by student’s t-test for statistical analysis. FCS Express was used for flow cytometry data analysis. For cell viability (Fig. S6), SYTOX® Green (Invitrogen, S7020) was utilized at a 2µM final concentration after washing the cells and resuspending cells in 50mM Na Citrate.
Polysome Analysis
Polysome analysis was carried out as described previously (MacKay et al. 2004; Steffen et al. 2008). Briefly, 125ml of log phase (0.4–0.6 OD, as close to 0.5 OD as possible) yeast were quickly cooled by addition of 60ml frozen YPD containing 133µg/ml cycloheximide to halt translation immediately and prevent ribosomes from dissociating from RNA. Yeast were spun down and then lysed by glass beads and protein-containing fractions were isolated. 20 OD260 units were used for each polysome run by adding the lysate to 7–47% sucrose gradients. Gradients were spun for 2 hours at 39k rpm in a Beckmann ultracentrifuge. The resulting gradients were then fractionated and the A254 read, resulting in the polysome graphs in Figure 6a.
Supplementary Material
Acknowledgements
We thank K. Steffen for remade ribosomal gene deletion strains and V. MacKay for technical assistance with polysome analysis. This work was supported by NIH Grant R01AG039390 to MK. JRD and GLS were supported by NIH Training Grant T32AG000057. JS was supported by NIH Training Grant T32ES007032. MK is an Ellison Medical Foundation New Scholar in Aging.
Footnotes
Author contributions:
J.D. and M.K. jointly conceived this study. All except B. K. K. and M.K. performed experiments. J.D., B. K. K. and M.K. wrote the manuscript.
References
- Andreou AM, Tavernarakis N. Protein metabolism and homeostasis in aging. Preface. Adv Exp Med Biol. 2010;694:vii–viii. [PubMed] [Google Scholar]
- Arlt H, Steglich G, Perryman R, Guiard B, Neupert W, Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. Embo J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012;8:e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kaeberlein M. A genomic approach to yeast chronological aging. Methods in molecular biology. 2009a;548:101–114. doi: 10.1007/978-1-59745-540-4_6. [DOI] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Kennedy BK, Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009b;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner CR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. A genomic analysis of chronological longevity factors in budding yeast. Cell Cycle. 2011;10:1385–1396. doi: 10.4161/cc.10.9.15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, Breitenbach M, Breitenbach-Koller L. Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Exp Gerontol. 2007;42:275–286. doi: 10.1016/j.exger.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Cohen G, Fessl F, Traczyk A, Rytka J, Ruis H. Isolation of the catalase A gene of Saccharomyces cerevisiae by complementation of the cta1 mutation. Mol Gen Genet. 1985;200:74–79. doi: 10.1007/BF00383315. [DOI] [PubMed] [Google Scholar]
- Delaney JR, Murakami CJ, Olsen B, Kennedy BK, Kaeberlein M. Quantitative evidence for early life fitness defects from 32 longevity-associated alleles in yeast. Cell Cycle. 2011a;10:156–165. doi: 10.4161/cc.10.1.14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney JR, Sutphin GL, Dulken B, Sim S, Kim JR, Robison B, Schleit J, Murakami CJ, Carr D, An EH, Choi E, Chou A, Fletcher M, Jelic M, Liu B, Lockshon D, Moller RM, Pak DN, Peng Q, Peng ZJ, Pham KM, Sage M, Solanky A, Steffen KK, Tsuchiya M, Tsuchiyama S, Johnson S, Raabe C, Suh Y, Zhou Z, Liu X, Kennedy BK, Kaeberlein M. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell. 2011b doi: 10.1111/j.1474-9726.2011.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis BR, Thorsness PE. Hsp90 and mitochondrial proteases Yme1 and Yta10/12 participate in ATP synthase assembly in Saccharomyces cerevisiae. Mitochondrion. 2011;11:587–600. doi: 10.1016/j.mito.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelin E, Rep M, Grivell LA. Sequence of the AFG3 gene encoding a new member of the FtsH/Yme1/Tma subfamily of the AAA-protein family. Yeast. 1994;10:1389–1394. doi: 10.1002/yea.320101016. [DOI] [PubMed] [Google Scholar]
- Harper JM, Salmon AB, Leiser SF, Galecki AT, Miller RA. Skin-derived fibroblasts from long-lived species are resistant to some, but not all lethal stresses and to the mitochondrial inhibitor rotenone. Aging Cell. 2007;6:1–13. doi: 10.1111/j.1474-9726.2006.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JM, Wang M, Galecki AT, Ro J, Williams JB, Miller RA. Fibroblasts from long-lived bird species are resistant to multiple forms of stress. J Exp Biol. 2011;214:1902–1910. doi: 10.1242/jeb.054643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeren G, Rinnerthaler M, Laun P, von Seyerl P, Kossler S, Klinger H, Hager M, Bogengruber E, Jarolim S, Simon-Nobbe B, Schuller C, Carmona-Gutierrez D, Breitenbach-Koller L, Muck C, Jansen-Durr P, Criollo A, Kroemer G, Madeo F, Breitenbach M. The mitochondrial ribosomal protein of the large subunit, Afo1p, determines cellular longevity through mitochondrial back-signaling via TOR1. Aging (Albany NY) 2009;1:622–636. doi: 10.18632/aging.100065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhola MK, Shah ZH, Grivell LA, Jacobs HT. The mitochondrial inner membrane AAA metalloprotease family in metazoans. FEBS Lett. 2000;481:91–95. doi: 10.1016/s0014-5793(00)01989-x. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kennedy BK. Large-scale identification in yeast of conserved aging genes. Mech Ageing Dev. 2005;126:17–21. doi: 10.1016/j.mad.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005a;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005b;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Steffen KK, Hu D, Dang N, Kerr EO, Tsuchiya M, Fields S, Kennedy BK. Comment on "HST2 mediates SIR2-independent life-span extension by calorie restriction". Science. 2006;312:1312. doi: 10.1126/science.1124608. author reply-1312. [DOI] [PubMed] [Google Scholar]
- Kennedy BK, Steffen KK, Kaeberlein M. Ruminations on dietary restriction and aging. Cell Mol Life Sci. 2007;64:1323–1328. doi: 10.1007/s00018-007-6470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy BK, Schmidt M. Elevated Proteasome Capacity Extends Replicative Lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo VD, Fabrizio P. Chronological Aging in Saccharomyces cerevisiae. Subcell Biochem. 2012;57:101–121. doi: 10.1007/978-94-007-2561-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay VL, Li X, Flory MR, Turcott E, Law GL, Serikawa KA, Xu XL, Lee H, Goodlett DR, Aebersold R, Zhao LP, Morris DR. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol Cell Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- Managbanag JR, Witten TM, Bonchev D, Fox LA, Tsuchiya M, Kennedy BK, Kaeberlein M. Shortest-path network analysis is a useful approach toward identifying genetic determinants of longevity. PLoS One. 2008;3:e3802. doi: 10.1371/journal.pone.0003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Murakami C, Kaeberlein M. Quantifying yeast chronological life span by outgrowth of aged cells. Journal of visualized experiments : JoVE. 2009 doi: 10.3791/1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami CJ, Burtner CR, Kennedy BK, Kaeberlein M. A method for high-throughput quantitative analysis of yeast chronological life span. J Gerontol A Biol Sci Med Sci. 2008;63:113–121. doi: 10.1093/gerona/63.2.113. [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Olsen BMC, Kaeberlein M. YODA: Software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma L, Lehrach H, Ralser M. Surviving in the cold: yeast mutants with extended hibernating lifespan are oxidant sensitive. Aging (Albany NY) 2009;1:957–960. doi: 10.18632/aging.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–184. doi: 10.1101/gad.1381406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Sadighi Akha AA, Buffenstein R, Miller RA. Fibroblasts from naked mole-rats are resistant to multiple forms of cell injury, but sensitive to peroxide, ultraviolet light, and endoplasmic reticulum stress. J Gerontol A Biol Sci Med Sci. 2008;63:232–241. doi: 10.1093/gerona/63.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slekar KH, Kosman DJ, Culotta VC. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- Steffen KK, Kennedy BK, Kaeberlein M. Measuring replicative life span in the budding yeast. Journal of visualized experiments : JoVE. 2009 doi: 10.3791/1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, McCormick MA, Pham KM, Mackay VL, Delaney JR, Murakami CJ, Kaeberlein M, Kennedy BK. Ribosome Deficiency Protects Against ER Stress in Saccharomyces cerevisiae. Genetics. 2012 doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annual review of cell and developmental biology. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutphin GL, Olsen BA, Kennedy BK, Kaeberlein M. Genome-wide analysis of yeast aging. Subcell Biochem. 2012;57:251–289. doi: 10.1007/978-94-007-2561-4_12. [DOI] [PubMed] [Google Scholar]
- Thorpe GW, Fong CS, Alic N, Higgins VJ, Dawes IW. Cells have distinct mechanisms to maintain protection against different reactive oxygen species: oxidative-stress-response genes. Proc Natl Acad Sci U S A. 2004;101:6564–6569. doi: 10.1073/pnas.0305888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.