Abstract
Determining the fitness of drug-resistant human immunodeficiency virus type 1 (HIV-1) strains is necessary for the development of population-based studies of resistance patterns. For this purpose, we have developed a reproducible, systematic assay to determine the competitive fitness of HIV-1 drug-resistant mutants. To demonstrate the applicability of this assay, we tested the fitness of the five most common nevirapine-resistant mutants (103N, 106A, 181C, 188C, and 190A), with mutations in HIV-1 reverse transcriptase (RT), singly and in combination (for a total of 31 variants) in a defined HIV-1 background. For these experiments, the 27 RT variants that produced viable virus were cocultured with wild-type virus without nevirapine. The ratios of the viral species were determined over time by utilization of a quantitative real-time RT-PCR-based assay. These experiments revealed that all of the viable variants were less fit than the wild type and demonstrated that the order of relative fitness of the single mutants tested was as follows: 103N > 181C > 190A > 188C > 106A. This order correlated with the commonality of these mutants as a result of nevirapine monotherapy. These investigations also revealed that, on average, the double mutants were less fit than the single mutants and the triple mutants were less fit than the double mutants. However, the fitness of the single and double mutants was often not predictive of the fitness of the derivative triple mutants, suggesting the presence of complex interactions between the closely aligned residues that confer nevirapine resistance. This complexity was also evident from the observation that all three of the replication-competent quadruple mutants were fitter than most of the triple mutants, and in some cases, even the double mutants. Our data suggest that, in many cases, viral fitness is the determining factor in the evolution of nevirapine-resistant mutants in vivo, that interactions between the residues that confer nevirapine resistance are complex, and that these interactions substantially affect reverse transcriptase structure and/or function.
Long-term control of human immunodeficiency virus (HIV) infection has been made possible by the use of antiretroviral therapy. Additionally, pharmacological interventions can dramatically reduce the rates of mother-to-child transmission of HIV type 1 (HIV-1) (10, 17). Central to the above advances is the availability of drug-susceptible virus. With the administration of only one or two antiretroviral drugs, treatment failure occurs relatively quickly due to the emergence of resistant virus (37, 43, 46). Following such failure, viral loads may be partially suppressed despite the emergence of high-level drug resistance (2, 34, 50). In addition, it has been noted that after cessation of treatment following failure, resistant virus is often replaced by wild-type virus (5, 10, 12-15, 22, 32, 51). These observations have been attributed to decreased fitness of the resistant virus compared to that of the virus present before therapy (21). The impairment of viral fitness due to resistance to antiretroviral drugs could have clinical relevance in terms of selecting optimal therapies and reducing the rate of progression, as well as on the modeling of the epidemiology of transmission of resistant HIV-1 strains (4, 28, 38, 42, 48). Hence, in the past few years there has been growing interest in the area of HIV-1 fitness in the context of drug resistance.
Research to date on the fitness of drug-resistant HIV-1 has been characterized by methodological inconsistency, which has made the comparison of studies difficult. For example, investigations of the fitness of drug-resistant HIV-1 clinical isolates have been hampered by the inability of researchers to control for the effect of sequences outside of the mutation of interest on viral fitness, i.e., the lack of an isogenic background (16, 26, 34). Thus, the utilization of genotypic resistance assays to determine the fitness of resistant mutants in an infecting viral population has been limited by the ability of the virus to exist as a quasi-species (7-9). A second issue with regards to methodology is the low level of sensitivity of certain assays (p24Gag enzyme-linked immunosorbent assay and plaque reduction assay) for the detection of small differences in fitness between viral variants in cell culture (25), which could be relevant in vivo. In addition, the use of parallel culture designs leads to assessment of replicative efficiency without the condition of competition and, consequently, does not resemble in vivo conditions in which mutant and wild-type strains are maintained in the same environment (18, 29, 31, 35, 39, 40). Furthermore, experiments that utilize probes that anneal to marker gene sequences and not directly to mutant reverse transcriptase (RT) sequences (29) are unable to assess the stability of variants containing multiple drug resistance mutations. In this report, we describe a reproducible, systematic assay to determine the competitive fitness of drug-resistant HIV-1 that addresses these limitations.
MATERIALS AND METHODS
Site-directed mutagenesis.
The mutations K103N, V106A, Y181C, Y188C, and G190A (27) were created individually and in all possible permutations (32 clones, including the wild type) by site-directed mutagenesis with the Quickchange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) in the plasmid p83-2, which contained 5′ NL4-3 sequences (1). Resultant clones were subjected to fluorescence sequencing (ABI, Foster City, Calif.) to verify the presence of the desired mutations and the absence of extraneous mutations.
Generation of virus stocks.
To generate infectious pNL4-3 or derivative HIV-1, DNA from each of the mutant clones was digested with EcoRI (New England Biolabs, Beverly, Mass.), column purified (Qiagen, Valencia, Calif.), and combined with an equal amount of similarly prepared EcoRI-digested p83-10 plasmid DNA (containing 3′ NL4-3 sequences). The two plasmids were fused by use of T4 DNA ligase (New England Biolabs). This mixture was then introduced into the permissive cell line CEMx174 by DEAE-dextran transfection (23, 33). The transfected cells were cultured in RPMI 1640 (Gibco BRL, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (Gibco BRL). For the generation of virus stocks, culture supernatants containing virus were harvested 48 h after the first observation of cytopathic effect (CPE).
Growth curve analyses.
In order to assess the growth kinetics of NL4-3 and the derivative strains, 106 CEMx174 cells were infected with virus diluted to contain 1.0 ng of p24Gag, using stocks derived from transfected CEMx174 cells. Culture supernatants were collected starting at day 5 postinfection. The rate of virus growth was assayed by the concentration of p24Gag in the culture supernatants, as determined by enzyme-linked immunosorbent assay (Coulter, Hialeah, Fla.).
In vitro transcription.
In order to quantify viral RNA, a template was engineered that consisted of a 1,510-bp fragment of the HIV-1 gag gene. For these experiments, the sequences were amplified by PCR and the resultant fragment was cloned into the pSP72 vector (Promega, Madison, Wis.) immediately downstream of the T7 promoter by use of EcoRI and BamHI restriction enzymes (New England Biolabs). Fluorescence sequencing of a specific clone ensured that the exact desired sequence was used in subsequent experiments. The gag sequences were then transcribed in vitro with T7 RNA polymerase and a BD RiboQuant in vitro transcription kit (BD Biosciences Pharmingen, San Diego, Calif.). The resultant RNA was purified by use of a QIAmp viral RNA kit (Qiagen) and quantified by use of a RiboGreen RNA quantification kit (Molecular Probes, Eugene, Oreg.). To establish a standard curve for HIV-1 gag RNA, we used serial dilutions, starting at a determined concentration, in a one-step real-time RT-PCR using an Applied Biosystems Prism 7000 sequence detection system (ABI). Primers 5′ GGCCAGGGAATTTTCTTCAGA 3′ and 5′ TCCCCAAACCTGAAGCTC TCT 3′ and probe 5′ 6FAMA GACCAGAGCCAACAGCCCCACC TAMRA 3′, which annealed to conserved sequences located near the 3′ end of gag and satisfied the manufacturer's design specifications, were used. The cycling parameters used for these experiments were as follows: 48°C for 30 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The number of cycles required to reach threshold fluorescence (Ct) was determined, and the quantity of sequences initially present was calculated by extrapolation onto the standard curve. The number of dilutions required for the extinction of signal was also determined.
Real-time RT-PCR.
Viral RNA from wild-type and mutant stocks was quantified 48 h after infection of 5 × 105 CEMx174 cells. The supernatant was harvested at 48 h postinfection, and HIV-1 RNA was purified by use of the QIAmp viral RNA kit. Relative quantities of viral RNAs were determined by quantitative real-time RT-PCR. For these experiments, primers 4455F (5′ GCAGTTCATGTAGCCAGTGGAT 3′) and 4596R (5′ TGGTGAAATTGCTGCCATTG 3′), which annealed to and amplified a 141-bp fragment in a conserved region of HIV-1 RT, were used. TaqMan SYBR Green PCR amplification of HIV-1 RT was carried out in MicroAmp optical 96-well reaction plates (ABI) in a volume of 50 μl. The reactions contained 3 μl each of the two primers (concentration of primer and probe stock solutions, 100 μM), 25 μl of 2× SYBR Green master mix, 0.25 μl of Multiscribe RT (50 U/μl), 1 μl of RNase inhibitor mix, and 5 μl of viral RNA extract. Cycling parameters were as described above.
Calculation of input virus for competition assay.
The Ct values, obtained from serial dilutions of wild-type virus, were plotted to create a standard curve (data not shown). The results of the regression analyses were used to calculate the ratio of the mutant and wild-type HIV-1 RNAs that were used for the competition experiments. An infectivity assay (using HeLa CD4-β-Gal cells) (24) ensured that a very low multiplicity of infection (MOI) was used for these experiments.
Competition assays.
The fitness of HIV-1 strains containing nevirapine-resistant mutants relative to that of wild-type NL4-3 was determined by simultaneous infection of both viral species in duplicate single wells of a 24-well plate. These wells contained 5 × 105 CEMx174 cells in 2 ml of medium and were infected at an MOI of 0.001. After overnight incubation with virus, the cells were washed once in phosphate-buffered saline (Gibco BRL) to remove residual input virus that remained in the supernatant. After 48 h, the supernatant was harvested and 100 μl of supernatant was transferred to a fresh culture of 5 × 105 CEMx174 cells, in 2 ml of medium, in a 24-well plate. This time point was used as the starting point for the competition experiment. Viral RNA was purified from the supernatant obtained at this time point to determine the ratio of wild-type and mutant viruses. Since we did not know in advance the relative replication capacity of the mutant and wild-type stocks, the infection experiments were repeated until the Ct value for a mutant was one to seven cycles lower than that for the wild type. This ensured a 2- to 100-fold excess of mutant virus at the start of the competition experiment. To determine the change in this ratio over time, 140 μl of supernatant was harvested every 48 h. HIV-1 primers, designed for the two regions of the RT where the mutations resided, were used to amplify 80-bp regions of the HIV-1 RT gene. One set, 2798F (5′ AACTCAAGATTTCGGGAACTTAA 3′) and 2878R (5′ AAAAATATGCATCGCCCACAT 3′), amplified the region that contained amino acids 103 and 106 of RT. The other set, 3051F (5′ AATCCGAAAACATTAGAGCCTTTTCAA 3′) and 3131R (5′ TCTATTTTTGTTCTATGCTGCCCTATT 3′), amplified the region that contained amino acids 181, 188, and 190 of RT. A one-step RT-PCR (performed in duplicate) was used to amplify these templates. For detection of the HIV-1 PCR products, TaqMan MGB probes specific for wild-type and mutant sequences (one mutant and one wild-type probe was included in each reaction) were designed to span regions that were specific for each mutation: positions 2847 to 2866 for residue 103, 2853 to 2875 for residue 106, 3080 to 4000 for residue 181, 3103 to 3121 for residue 188, and 3114 to 3132 for residue 190. All mutant MGB probes were labeled with the reporter dye FAM (6-carboxyfluorescein) at their 5′ ends, all wild-type MGB probes were labeled with the reporter dye VIC (6-carboxyrhodamine 6G, succinimidyl ester) at their 5′ ends, and all probes had a nonfluorescent quencher at their 3′ ends. In addition, both HIV-1 mutant and wild-type probes were phosphorylated at their 3′ ends to prevent elongation during PCR (synthesis and labeling of probes was performed by ABI). Postrun manipulations of data were performed according to the manufacturer's instructions and as described above to generate Ct values for the two viral species in each sample.
Calculation of competing virus ratio.
The Ct values were utilized to determine the ratio of wild-type to mutant virus contained in each sample. The formula used to calculate this ratio was 2(CtWT − CtMut), where CtWT is the Ct value for the wild type and CtMut is the Ct value for the mutant for each coinfection. The ratio of wild-type and mutant virus was plotted on a logarithmic scale, and linear regression for each series of time points was determined with Excel 2000 (Microsoft, Seattle, Wash.). The formula y = b(emx) was used to determine the slope of each line (y is the ratio of mutant to wild-type virus, b is the y intercept, x is the time point, and m is the value of the slope). The term “m” was used for the fitness coefficient in this analysis.
RESULTS
Viability of HIV-1 mutants.
Wild-type HIV-1, along with mutant HIV-1 containing the mutations 103N, 106A, 181C, 188C, and 190A, which confer resistance to nevirapine, either singly or in combination, were transfected into CEMx174 cells. Of these 32 possible permutations, 28 produced CPE in the transfected cultures, which was indicative of replicating HIV-1. The first appearance of CPE (days 3 to 5 posttransfection) (data not shown) and the time of peak virus production (days 7 to 9 posttransfection) (data not shown) were similar for all the replication-competent strains produced in these cultures. Recombinants containing the combination of 103N/106A/188C (one triple mutant, two quadruple mutants, and the quintuple mutant) failed to produce CPE, indicating a lack of virus production. This was confirmed by assaying p24Gag and viral RNA in cultures maintained for an excess of 8 weeks (data not shown).
Growth of triple and quadruple HIV-1 RT mutants.
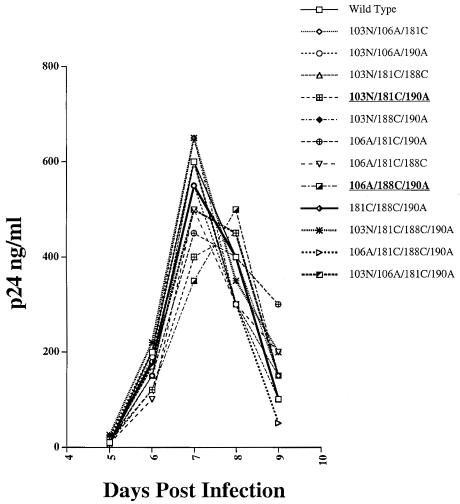
We investigated whether a simple replication assay would be sufficiently sensitive to assess differences in the replicative capacities of wild-type NL4-3 and viable derivative strains containing a maximum number of nevirapine-resistant mutations. For these experiments, equivalent p24Gag concentrations of the wild type as well as the nine viable triple and three viable quadruple mutants were used to infect CEMx174 cells. Under these conditions, we observed that seven of the nine triple mutants and all three quadruple mutants reached peak virus production on day 7 postinfection and two triple mutants reached peak virus production on day 8 postinfection (Fig. 1). The peak level of virus production was similar for all the strains tested (Fig. 1). These experiments revealed that this assay could not detect differences in the replicative capacities of the great majority of these mutants and, consequently, would not be useful for determining relative strain fitness.
FIG. 1.
Growth of the wild type (NL4-3) and of viable derivative triple and quadruple mutants that confer resistance to nevirapine. All of the strains reached peak production on day 7 postinfection, except the 103N/181C/190A and 106A/188C/190A mutants (in bold), which reached peak production on day 8 postinfection.
Parameters of the competitive fitness assay.
Since we could not use a replication-based assay for our studies, we tested the relative fitness of the 27 viable nevirapine-resistant mutants by using a competitive fitness assay. Several parameters were critical to the utility of this assay for our studies. (i) Coinfection of mutant and wild-type HIV-1 was done at a total MOI of 0.001. This low level of viral input ensured that the measurement of viral RNA production would always be based on logarithmic growth since the number of cells would not be limiting through the course of the experiment (18). It also ensured that the likelihood of recombination events between wild-type and mutant strains would be extremely low (41). (ii) The use of an excess of mutant virus was necessary since it allowed for the detection of the relatively unfit mutant strains for multiple time points in our assay. (iii) The use of the 48-h postinfection time point as the starting point of the competition experiment was important, as it served as an internal control for the ratio of wild-type to mutant virus. By measuring the amount of the wild-type and mutant viruses existing at this time, we accounted for errors in the measurement of the inoculum. This allowed for a true measure of the change in the ratio of competing strains and ensured that a 2- to 100-fold excess of mutant virus was present at the start of the competition experiment (Table 1; see Fig. 3 to 6). (iv) A lack of cross-reactivity between wild-type and mutant templates and probes was necessary to allow for reliable quantification of wild-type-to-mutant virus ratios over a wide dynamic linear range. We tested cross-reactivity in two ways, as follows. (a) We assessed whether mutant template-wild-type probe and wild-type template-mutant probe combinations produced detectable fluorescence at all five resistance loci that we studied and did not observe detectable cross-reactivity (data not shown). (b) We ascertained the linear dynamic range of our assay system. For these experiments, we calculated the number of wild-type and mutant RNA molecules, with in vitro-transcribed gag as the standard (see Materials and Methods). The RNAs were then subjected to 10-fold serial dilutions and mixed in various ratios, in the range of 106 (generally the quantity found in undiluted samples) to 102 RNA molecules for either species. The RNAs were then subjected to real-time RT-PCR. These studies revealed that the amount of template was accurately quantified across the entire range used for this assay (Fig. 2 and data not shown). Thus, the competition assay had a linear range of at least 4 logs which meant that the ratio of input virus (2- to 100-fold excess of mutant) was well within this range and therefore did not affect the competition assay.
TABLE 1.
Profile of the competition assay with 181C versus wild-type HIV-1
| Time point |
Ct
|
Difference in Ct | Fold excess of 181C mutant (2Ct) | |
|---|---|---|---|---|
| 181C mutant | Wild type | |||
| 0 | 25.91 | 31.12 | 5.21 | 37.01 |
| 1 | 30.99 | 35.02 | 4.03 | 16.34 |
| 2 | 19.6 | 22.74 | 3.14 | 8.82 |
| 3 | 14.69 | 16.51 | 1.82 | 3.53 |
FIG. 3.
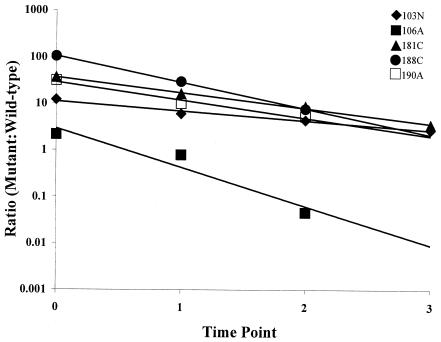
The ratios of nevirapine-resistant single mutants to wild-type HIV-1 plotted over time. The linear regression for each data set was determined with Excel 2000 software. The correlation coefficients (R2) for these data were as follows: for 103N, 0.9766; for 106A, 0.9296; for 181C, 0.9953; for 188C, 0.9995; for 190A, 0.9721. These data represent the averages of two independent experiments.
FIG. 6.
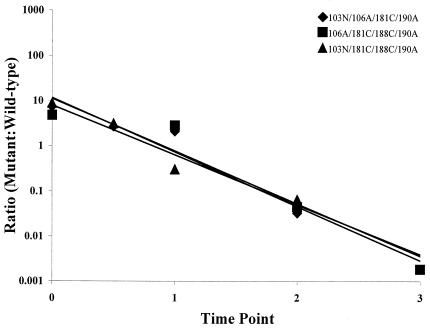
Ratios of viable nevirapine-resistant quadruple mutants to wild-type HIV-1 plotted over time. The linear regression for each data set was determined with Excel 2000 software. The correlation coefficients (R2) for these data were as follows: for 103N/106A/181C/190A, 0.9179; for 103N/181C/188C/190A, 0.9509; for 106A/181C/188C/190A, 0.9319. These data represent the averages of two independent experiments.
FIG. 2.
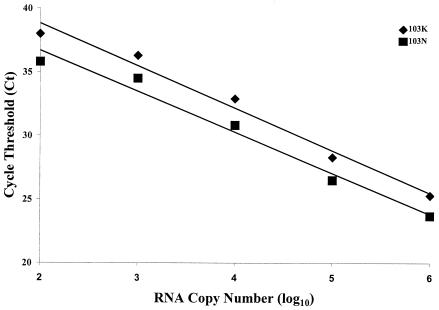
The linear dynamic range of the competition assay. Samples spiked with 103N mutant or wild-type (103K) RNA were assessed for the quantity of each species by RT-PCR, using probes that were specific for either sequence. In one set of reactions, 103N was fixed at 106 copies of RNA and mixed with serial 10-fold dilutions of 103K (diamonds). In the other set, 103K was fixed at 106 copies of RNA and mixed with serial 10-fold dilutions of 103N (squares). In either case, the Ct was unaltered for the species with a fixed copy number in the sample, regardless of the quantity of the diluted species (data not shown). The linear regression for each data set was determined with Excel 2000 software, which revealed that the slopes of the two lines were similar (−3.34 for 103K and −3.22 for 103N) and that the correlation coefficients (R2) for these data were 0.9807 and 0.9769, respectively. These data represent the averages of two independent experiments. Similar data were observed for samples containing mutants at the other four loci, as studied by use of locus-specific probes.
Competitive fitness of the single mutants.
The use of the competitive fitness assay revealed that the variants containing nevirapine-resistant mutants with single mutations were all less fit than the wild type, as a negative slope (fitness coefficient) was observed in every case (Fig. 3). These measurements demonstrated the order of fitness of these variants, which was as follows: 103N > 181C > 190A > 188C > 106A. Our studies also revealed that the fitness coefficients of these variants ranged widely (−0.48 to −1.93) (Table 2) around the mean of −1.07.
TABLE 2.
Summary of fitness data for all viable mutants in this study containing nevirapine resistance mutations
| Mutant | Fitness coefficienta | Mean | Range |
|---|---|---|---|
| Single mutants | |||
| 103N | −0.48 | −1.07 | −0.48, −1.93 |
| 106A | −1.93 | ||
| 181C | −0.77 | ||
| 188C | −1.30 | ||
| 190A | −0.89 | ||
| Double mutants | |||
| 103N/106A | −2.21 | −1.94 | −0.78, −3.46 |
| 103N/181C | −2.28 | ||
| 103N/188C | −1.32 | ||
| 103N/190A | −0.78 | ||
| 106A/181C | −1.07 | ||
| 106A/188C | −3.31 | ||
| 106A/190A | −3.46 | ||
| 181C/188C | −1.65 | ||
| 181C/190A | −1.74 | ||
| 188C/190A | −1.57 | ||
| Triple mutants | |||
| 103N/106A/181C | −2.23 | −3.03 | −2.04, −5.10 |
| 103N/106A/190A | −3.37 | ||
| 103N/181C/188C | −2.04 | ||
| 103N/181C/190A | −5.10 | ||
| 103N/188C/190A | −2.19 | ||
| 106A/181C/190A | −2.99 | ||
| 106A/181C/188C | −2.38 | ||
| 106A/188C/190A | −3.88 | ||
| 181C/188C/190A | −3.09 | ||
| Quadruple mutants | |||
| 103N/106A/181C/190A | −2.68 | −2.66 | −2.54, −2.77 |
| 103N/181C/188C/190A | −2.54 | ||
| 106A/181C/188C/190A | −2.77 |
See Materials and Methods for equation.
Competitive fitness of the double mutants.
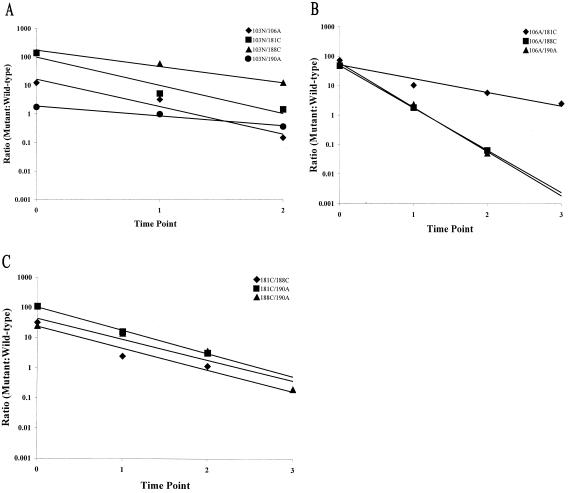
We observed that the nevirapine-resistant double mutants displayed a decreased level of fitness relative to the single mutants, with a mean fitness coefficient of −1.94 (Table 2). As was seen for the single mutants, the fitness coefficients of double mutants ranged widely (−0.78 to −3.46) around the mean (Table 2). Our experiments revealed that the fitness of double mutants was not necessarily a reflection of the additive fitness of the single mutants. For example, the strains that contained 188C in combination with 103N or 190A (Fig. 4; Table 3) were fitter than strains that contained 181C in combination with 103N or 190A, even though 181C was fitter than 188C as a single mutant (Fig. 3; Table 3). Furthermore, one double mutant (103N/190A) was fitter than one of the single mutants (190A) of which it was comprised (Table 3).
FIG. 4.
The ratios of nevirapine-resistant double mutants to wild-type HIV-1 plotted over time. (A) Double mutants in which the N-terminal mutation was 103N. (B) Double mutants in which the N-terminal mutation was 106A. (C) Double mutants in which the N-terminal mutation was 181C or 188C. The linear regression for each data set was determined with Excel 2000 software. The correlation coefficients (R2) for these data were as follows: for 103N/106A, 0.9531; for 103N/181C, 0.9394; for 103N/188C, 0.9901; for 103N/190A, 0.9766; for 106A/181C, 0.9300; for 106A/188C, 0.9999; for 106A/190A, 0.9963; for 181C/188C, 0.9105; for 181C/190A, 0.9976; for 188C/190A, 0.8963. These data represent the averages of two independent experiments.
TABLE 3.
The relative fitness of all viable mutants in this study containing nevirapine resistance mutations
| Mutanta | Fitness coefficientb |
|---|---|
| 103N | −0.48 |
| 181C | −0.77 |
| 103N/190A | −0.78 |
| 190A | −0.89 |
| 106A/181C | −1.07 |
| 188C | −1.30 |
| 103N/188C | −1.32 |
| 188C/190A | −1.57 |
| 181C/188C | −1.65 |
| 181C/190A | −1.74 |
| 106A | −1.93 |
| 103N/181C/188C | −2.04 |
| 103N/188C/190A | −2.19 |
| 103N/106A | −2.21 |
| 103N/106A/181C | −2.23 |
| 103N/181C | −2.28 |
| 106A/181C/188C | −2.38 |
| 103N/181C/188C/190A | −2.54 |
| 103N/106A/181C/190A | −2.68 |
| 106A/181C/188C/190A | −2.77 |
| 106A/181C/190A | −2.99 |
| 181C/188C/190A | −3.09 |
| 106A/188C | −3.31 |
| 103N/106A/190A | −3.37 |
| 106A/190A | −3.46 |
| 106A/188C/190A | −3.88 |
| 103N/181C/190A | −5.10 |
Listed in decreasing order of fitness.
See Materials and Methods for equation.
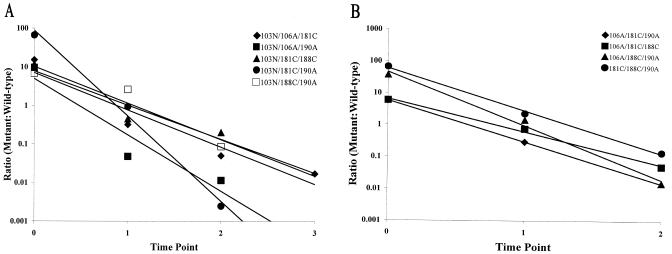
Competitive fitness of the triple mutants.
In transfection experiments, 9 of the 10 possible triple mutants were viable, the exception being the 103N/106A/188C mutant. In the competition assay, the viable triple mutants displayed an overall trend of decreased fitness relative to the single and double mutants, with a mean fitness coefficient of −3.03 (Fig. 5; Table 2). As with the single and double mutants, the fitness coefficients of these mutants ranged widely (−2.04 to −5.10) around the mean (Table 2). The two mutants that displayed the lowest fitness coefficients (106A/188C/190A and 103N/181C/190A) (Table 3) also showed a detectable delay in growth in the replication assay (Fig. 1). One triple mutant (103N/181C/188C) demonstrated a high level of fitness (fitness coefficient of −2.04) (Table 2) which was comparable to the mean fitness coefficient observed for the double mutants (−1.94) (Table 2). Our study also revealed that the two fittest triple mutants (103N/181C/188C and 103N/188C/190A) (Table 3) contained both 103N and 188C. These results were surprising given that the one triple mutant that did not produce a viable virus (103N/106A/188C) also contained these two mutations. We also observed that the four least fit viable triple mutants all contained 190A. Stratifying the viable mutants above and below the mean fitness coefficient for this group (−3.03) revealed that all of the combinations that fell below the mean contained 190A (Table 3).
FIG. 5.
Ratios of viable nevirapine-resistant triple mutants to wild-type HIV-1 plotted over time. (A) Triple mutants in which the N-terminal mutation was 103N. (B) Triple mutants in which the N-terminal mutation was 106A or 181C. The linear regression for each data set was determined with Excel 2000 software. The correlation coefficients (R2) for these data were as follows: for 103N/106A/181C, 0.9250; for 103N/106A/190A, 0.9005; for 103N/181C/188C, 0.8926; for 103N/181C/190A, 0.9910; for 103N/188C/190A, 0.9052; for 106A/181C/190A, 1.0; for 106A/181C/188C, 0.9942; for 106A/188C/190A, 0.9910; for 181C/188C/190A, 0.9971. These data represent the averages of two independent experiments.
Competitive fitness of the quadruple mutants.
Transfection experiments revealed that the two quadruple mutants that contained the nonviable triple combination 103N/106A/188C (103N/106A/181C/188C and 103N/106A/188C/190A) did not produce replication-competent HIV-1. The three quadruple mutants that were viable displayed a mean fitness coefficient of −2.66 (Fig. 6; Table 2). Unlike the groups described above, the viable quadruple mutants had fitness coefficients that clustered very closely (−2.54 to −2.77) around the mean (Table 2). Surprisingly, the mean fitness of this group was higher than that observed for the triple mutants (Table 2). In fact, the 103N/106A/181C/190A and 106A/181C/188C/190A mutants were fitter than five of the eight triple mutants that contained three of these mutations (Table 3). These experiments also revealed that the 103N/106A/181C/190A variant was fitter than one of the double mutants that contained two of these mutations and that the 106A/181C/188C/190A variant was fitter than two of the double mutants that contained two of these mutations (Table 3). In fact, the fitness of these two quadruple mutants was comparable to the fitness of the 106A single mutant (Table 3). The third viable quadruple mutant (103N/181C/188C/190A) was also surprisingly fit, being fitter than two of the four triple mutants that contained three of these mutations (Table 3).
Genetic stability of the quadruple mutants.
Since the fitness coefficients of the quadruple mutants were surprisingly high, we addressed the possibility that reversion events had taken place at the mutant loci during the course of stock generation and/or the competition experiment. For these experiments, we probed RNA obtained from competition experiments containing the quadruple mutants with probes specific for all four mutant loci. Table 4 reveals that the ratio of mutant to wild-type sequence for the competition experiment containing the 103N/181C/188C/190A mutant was decreased to a similar extent at all four loci at the two last time points. Similar results were observed for the other quadruple mutants (data not shown). These data demonstrate that mutant sequences were maintained throughout the course of the experiment, indicating that all of the mutations were equally stable through short-term passage in tissue culture. They also revealed that the mutant and wild-type probes at the loci investigated here (103, 106, 181, 188, and 190) had equivalent affinities for their target sequences, since the difference in the ratio of mutant to wild-type virus between the two time points was consistent for all probe sets (Table 4 and data not shown). These experiments demonstrated that the fitness coefficient derived for a particular variant was not influenced by the particular probe set used in the experiments.
TABLE 4.
Stability of the 103N/106A/181C/190N mutant
| Time point | Probe locusa |
Ct
|
Difference in Ct | |
|---|---|---|---|---|
| WT | Mutant | |||
| 1 | 103 | 20.35 | 21.45 | 1.10 |
| 106 | 21.21 | 22.39 | 1.18 | |
| 181 | 21.55 | 22.76 | 1.21 | |
| 190 | 18.00 | 19.20 | 1.20 | |
| 2 | 103 | 14.20 | 19.01 | −4.81 |
| 106 | 15.78 | 20.79 | −5.01 | |
| 181 | 16.86 | 21.56 | −4.70 | |
| 190 | 13.20 | 17.60 | −4.40 | |
Residue in RT that was the probe target.
DISCUSSION
In this study, we demonstrated the utility of a quantitative real-time RT-PCR-based competition assay for evaluating the relative fitness of drug-resistant HIV-1 strains. This system addressed many of the issues that have limited previous studies pertaining to this topic, as follows. (i) By utilizing isogenic strains differing by as little as a single nucleotide, we eliminated the potential confounding effects on viral fitness of sequence variables outside the mutation of interest, i.e., in other genes, such as env. However, the use of a single genetic background does not allow for the quantification of the effect of nonresistance RT polymorphisms on the fitness of nevirapine-resistant mutants. (ii) This method provided a level of sensitivity that allowed for measurable differences in the relative fitness of variant strains to be determined (Fig. 5 and 6), which could not be done with the growth assay (Fig. 1). (iii) This assay more closely resembled in vivo conditions in which mutant and wild-type strains are maintained in the same environment. (iv) This approach allowed for the determination of the stabilities of variants containing multiple drug resistance mutations as well as for the assessment of the affinities of probes for wild-type and mutant sequences (Table 4).
The elucidation of the crystal structure of HIV-1 RT revealed that the mutations that confer resistance to nevirapine reside in a pocket that is adjacent to the active site of the enzyme (49). Our observation that the fitness of single and double mutants was often not predictive of the fitness of derivative triple mutants indicates that the interactions between these residues are complex. The widely ranging fitness coefficients of the double and triple mutant groups (Table 2) are also evidence of the intricacy of the interactions between these residues. This relationship was underscored by the high degree of fitness of the viable quadruple mutants (Table 3). Since RT is highly conserved among HIV-1 isolates (27), we expected that increasing the number of mutations would increasingly compromise RT structure and/or function. However, we observed that the three replication-competent quadruple mutants were fitter than most of the triple mutants and even some of the double mutants that contain mutations common to the quadruple mutants (Table 3). Such fitness patterns could underlie the puzzling observation that, in some cases, mutations that confer resistance to a drug such as nevirapine can accumulate over time even though a single mutation is sufficient to confer a high level of resistance (3, 6, 11, 20). Although increased viral fitness associated with an increased number of mutations has not been described for HIV-1 RT, it has been described for mutations that decrease sensitivity to HIV-1 protease inhibitors (30, 35, 44). Furthermore, an association of increased fitness with an increased number of mutations, even to the degree of greater replicative capacity than the wild type, has been demonstrated in vitro for other RNA viruses and bacteria (36, 45). Such observations are consistent with the adaptive landscape model of evolution (52). In this model, a population is driven by natural selection to a local optimum but not necessarily to a global optimum. Therefore, a population can become stagnant by developing a suboptimal solution to its environment (in this case, certain double mutants). In such a circumstance, natural selection would not select for fitter variants (in this case, certain quadruple mutants) because it would necessitate passage through a basin of maladapted intermediate variants (in this case, certain triple mutants) (35, 52). Such a mechanism could explain why quadruple mutants are rarely observed in the clinic (47).
The decrease in fitness conferred by the 181C mutation conflicted with a previous observation (19). Although it was among the fittest of the mutants that we tested (Table 3), it was clear that the ratio of this mutant to the wild type decreased at every time point in the competition assay (Fig. 2; Table 1). In the 181C mutant-wild type competition experiment, the amount of virus produced at time point 1 was lower than that produced at time point 0 (representing the start of the competition experiment) (Table 1). These data suggested that the MOI at the start of the competition experiment was likely lower than the 0.001 that we used for the initial coinfection, since it took >2 days for input virus to be completely replaced by newly synthesized virus. We also observed that the peak of virus production occurred between time points 1 and 2 and that less virus production occurred between time points 2 and 3 (Table 1). Therefore, it could be construed that the cell number was limiting at this late time point. However, the fitness coefficient was altered only slightly if time point 3 was not utilized in the calculation (−0.77 [R2 = 0.9953] versus −0.72 [R2 = 0.9942]). This suggested that the decrease in viral production was a reflection of the kinetics of virus replication, that most of the virus obtained at time point 2 was generated just prior to virus harvesting, and that the number of cells was not limiting. It also suggested that there was insufficient time for this virus to completely replace itself prior to the subsequent time point in the experiment. Based on these observations, we believe that the negative fitness coefficient obtained for 181C was accurate and was not a reflection of any bias inherent in our assay. Similar profiles were observed for the additional competition experiments (data not shown).
The mutations with the highest prevalence among HIV-infected individuals, 103N and 181C (3, 6, 11, 20), also exhibited the highest level of fitness in our assay (Table 3). This would suggest that, in most cases, viral fitness has a greater impact on the evolution of nevirapine-resistant HIV-1 than the level of drug resistance. Interestingly, it has been observed that among mothers treated with nevirapine to prevent vertical transmission of HIV-1, there was a higher frequency of the 103N mutation in the mothers and a higher frequency of the 181C mutation in their infants (15). The authors of that study suggested that this may have been due to higher blood concentrations of nevirapine in the infants and, therefore, more exposure to nevirapine than their mothers encountered. Such a situation may have favored the emergence of a fitter mutant, with a lower level of resistance, in the mothers (103N) (Table 3) while favoring the emergence of a less fit mutant, with a higher level of resistance, in the infants (181C) (Table 3). This hypothesis could be tested by the competitive fitness assay in the presence and absence of drug.
Acknowledgments
This work was supported by Elizabeth Glaser Pediatric AIDS Foundation Basic Research grant PG-51179 (L.A.). E.P. was supported by training grant 5T32CA09159-27.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert, J., J. Wahlberg, J. Lundeberg, S. Cox, E. Sandstrom, B. Wahren, and M. Uhlen. 1992. Persistence of azidothymidine-resistant human immunodeficiency virus type 1 RNA genotypes in posttreatment sera. J. Virol. 66:5627-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blower, S. M., A. N. Aschenbach, H. B. Gershengorn, and J. O. Kahn. 2001. Predicting the unpredictable: transmission of drug-resistant HIV. Nat. Med. 7:1016-1020. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, C. A., R. van Leeuwen, P. Kellam, P. Schipper, J. Tijnagel, J. M. Lange, and B. A. Larder. 1993. Effects of discontinuation of zidovudine treatment on zidovudine sensitivity of human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casado, J. L., K. Hertogs, L. Ruiz, F. Dronda, A. Van Cauwenberge, A. Arno, I. Garcia-Arata, S. Bloor, A. Bonjoch, J. Blazquez, B. Clotet, and B. Larder. 2000. Non-nucleoside reverse transcriptase inhibitor resistance among patients failing a nevirapine plus protease inhibitor-containing regimen. AIDS 14:F1-F7. [DOI] [PubMed] [Google Scholar]
- 7.Coffin, J. M. 1992. Genetic diversity and evolution of retroviruses. Curr. Top. Microbiol. Immunol. 176:143-164. [DOI] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1986. Genetic variation in AIDS viruses. Cell 46:1-4. [DOI] [PubMed] [Google Scholar]
- 9.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 10.Connor, E. M., R. S. Sperling, R. Gelber, P. Kiselev, G. Scott, M. J. O'Sullivan, R. VanDyke, M. Bey, W. Shearer, R. L. Jacobson, et al. 1994. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. Pediatric AIDS Clinical Trials Group Protocol 076 Study Group. N. Engl. J. Med. 331:1173-1180. [DOI] [PubMed] [Google Scholar]
- 11.Conway, B., M. A. Wainberg, D. Hall, M. Harris, P. Reiss, D. Cooper, S. Vella, R. Curry, P. Robinson, J. M. Lange, and J. S. Montaner. 2001. Development of drug resistance in patients receiving combinations of zidovudine, didanosine and nevirapine. AIDS 15:1269-1274. [DOI] [PubMed] [Google Scholar]
- 12.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 13.Devereux, H. L., V. C. Emery, M. A. Johnson, and C. Loveday. 2001. Replicative fitness in vivo of HIV-1 variants with multiple drug resistance-associated mutations. J. Med. Virol. 65:218-224. [DOI] [PubMed] [Google Scholar]
- 14.Devereux, H. L., M. Youle, M. A. Johnson, and C. Loveday. 1999. Rapid decline in detectability of HIV-1 drug resistance mutations after stopping therapy. AIDS 13:F123-F127. [PubMed] [Google Scholar]
- 15.Eshleman, S. H., M. Mracna, L. A. Guay, M. Deseyve, S. Cunningham, M. Mirochnick, P. Musoke, T. Fleming, M. Glenn Fowler, L. M. Mofenson, F. Mmiro, and J. B. Jackson. 2001. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). AIDS 15:1951-1957. [DOI] [PubMed] [Google Scholar]
- 16.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guay, L. A., P. Musoke, T. Fleming, D. Bagenda, M. Allen, C. Nakabiito, J. Sherman, P. Bakaki, C. Ducar, M. Deseyve, L. Emel, M. Mirochnick, M. G. Fowler, L. Mofenson, P. Miotti, K. Dransfield, D. Bray, F. Mmiro, and J. B. Jackson. 1999. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet 354:795-802. [DOI] [PubMed] [Google Scholar]
- 18.Holland, J. J., J. C. de la Torre, D. K. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iglesias-Ussel, M. D., C. Casado, E. Yuste, I. Olivares, and C. Lopez-Galindez. 2002. In vitro analysis of human immunodeficiency virus type 1 resistance to nevirapine and fitness determination of resistant variants. J. Gen. Virol. 83:93-101. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, J. B., G. Becker-Pergola, L. A. Guay, P. Musoke, M. Mracna, M. G. Fowler, L. M. Mofenson, M. Mirochnick, F. Mmiro, and S. H. Eshleman. 2000. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS 14:F111-F115. [DOI] [PubMed] [Google Scholar]
- 21.Kaufmann, D., M. Munoz, G. Bleiber, S. Fleury, B. Lotti, R. Martinez, W. Pichler, P. Meylan, and A. Telenti. 2000. Virological and immunological characteristics of HIV treatment failure. AIDS 14:1767-1774. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann, D., G. Pantaleo, P. Sudre, and A. Telenti. 1998. CD4-cell count in HIV-1-infected individuals remaining viraemic with highly active antiretroviral therapy (HAART). Swiss HIV Cohort Study. Lancet 351:723-724. [DOI] [PubMed] [Google Scholar]
- 23.Kestler, H. W., III, Y. N. Naidu, T. Kodama, N. W. King, M. D. Daniel, Y. Li, and R. C. Desrosiers. 1989. Use of infectious molecular clones of simian immunodeficiency virus for pathogenesis studies. J. Med. Primatol. 18:305-309. [PubMed] [Google Scholar]
- 24.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, S. A., S. Karsten, B. Ruster, C. Klebba, M. Pape, O. G. Ottmann, D. Hoelzer, and W. K. Roth. 2003. Comparison of TaqMan real-time PCR and p24 ELISA for quantification of in vitro HIV-1 replication. J. Virol. Methods 107:169-175. [DOI] [PubMed] [Google Scholar]
- 26.Kosalaraksa, P., M. F. Kavlick, V. Maroun, R. Le, and H. Mitsuya. 1999. Comparative fitness of multi-dideoxynucleoside-resistant human immunodeficiency virus type 1 (HIV-1) in an in vitro competitive HIV-1 replication assay. J. Virol. 73:5356-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2001. HIV sequence compendium 2001. Theoretical Biology and Biophysics, Los Alamos, N.Mex.
- 28.Leigh Brown, A. J., S. D. Frost, W. C. Mathews, K. Dawson, N. S. Hellmann, E. S. Daar, D. D. Richman, and S. J. Little. 2003. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J. Infect. Dis. 187:683-686. [DOI] [PubMed] [Google Scholar]
- 29.Lu, J., and D. R. Kuritzkes. 2001. A novel recombinant marker virus assay for comparing the relative fitness of HIV-1 reverse transcriptase variants. J. Acquir. Immune Defic. Syndr. 27:7-13. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Picado, J., A. V. Savara, L. Shi, L. Sutton, and R. T. D'Aquila. 2000. Fitness of human immunodeficiency virus type 1 protease inhibitor-selected single mutants. Virology 275:318-322. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, V., C. Sabin, K. Hertogs, S. Bloor, J. Martinez-Picado, R. D'Aquila, B. Larder, T. Lutz, P. Gute, E. Weidmann, H. Rabenau, A. Phillips, and S. Staszewski. 2000. Virological and immunological effects of treatment interruptions in HIV-1 infected patients with treatment failure. AIDS 14:2857-2867. [DOI] [PubMed] [Google Scholar]
- 33.Naidu, Y. M., H. W. Kestler III, Y. Li, C. V. Butler, D. P. Silva, D. K. Schmidt, C. D. Troup, P. K. Sehgal, P. Sonigo, M. D. Daniel, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nijhuis, M., S. Deeks, and C. Boucher. 2001. Implications of antiretroviral resistance on viral fitness. Curr. Opin. Infect. Dis. 14:23-28. [DOI] [PubMed] [Google Scholar]
- 35.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 36.Novella, I. S., E. A. Duarte, S. F. Elena, A. Moya, E. Domingo, and J. J. Holland. 1995. Exponential increases of RNA virus fitness during large population transmissions. Proc. Natl. Acad. Sci. USA 92:5841-5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nunberg, J. H., W. A. Schleif, E. J. Boots, J. A. O'Brien, J. C. Quintero, J. M. Hoffman, E. A. Emini, and M. E. Goldman. 1991. Viral resistance to human immunodeficiency virus type 1-specific pyridinone reverse transcriptase inhibitors. J. Virol. 65:4887-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips, A. N., M. Youle, M. Johnson, and C. Loveday. 2001. Use of a stochastic model to develop understanding of the impact of different patterns of antiretroviral drug use on resistance development. AIDS 15:2211-2220. [DOI] [PubMed] [Google Scholar]
- 39.Prado, J. G., T. Wrin, J. Beauchaine, L. Ruiz, C. J. Petropoulos, S. D. Frost, B. Clotet, R. T. D'Aquila, and J. Martinez-Picado. 2002. Amprenavir-resistant HIV-1 exhibits lopinavir cross-resistance and reduced replication capacity. AIDS 16:1009-1017. [DOI] [PubMed] [Google Scholar]
- 40.Quiñones-Mateu, M. E., and E. J Arts (ed.). 2001. HIV-1 fitness: implications for drug resistance, disease progression, and global epidemic evolution, p. 134-170. HIV sequence compendium 2001. Theoretical Biology and Biophysics, Los Alamos, N.Mex.
- 41.Quiñones-Mateu, M. E., Y. Gao, S. C. Ball, A. J. Marozsan, A. Abraha, and E. J. Arts. 2002. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J. Virol. 76:9600-9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ribeiro, R. M., S. Bonhoeffer, and M. A. Nowak. 1998. The frequency of resistant mutant virus before antiviral therapy. AIDS 12:461-465. [DOI] [PubMed] [Google Scholar]
- 43.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, et al. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose, J. R., L. M. Babe, and C. S. Craik. 1995. Defining the level of human immunodeficiency virus type 1 (HIV-1) protease activity required for HIV-1 particle maturation and infectivity. J. Virol. 69:2751-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrag, S. J., V. Perrot, and B. R. Levin. 1997. Adaptation to the fitness costs of antibiotic resistance in Escherichia coli. Proc. R. Soc. Lond. B 264:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuurman, R., M. Nijhuis, R. van Leeuwen, P. Schipper, D. de Jong, P. Collis, S. A. Danner, J. Mulder, C. Loveday, C. Christopherson, et al. 1995. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC). J. Infect. Dis. 171:1411-1419. [DOI] [PubMed] [Google Scholar]
- 47.Shafer, R. W., D. Stevenson, and B. Chan. 1999. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 27:348-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simon, V., J. Vanderhoeven, A. Hurley, B. Ramratnam, M. Louie, K. Dawson, N. Parkin, D. Boden, and M. Markowitz. 2002. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS 16:1511-1519. [DOI] [PubMed] [Google Scholar]
- 49.Smerdon, S. J., J. Jager, J. Wang, L. A. Kohlstaedt, A. J. Chirino, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1994. Structure of the binding site for nonnucleoside inhibitors of the reverse transcriptase of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 91:3911-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, M. S., K. L. Koerber, and J. S. Pagano. 1994. Long-term persistence of zidovudine resistance mutations in plasma isolates of human immunodeficiency virus type 1 of dideoxyinosine-treated patients removed from zidovudine therapy. J. Infect. Dis. 169:184-188. [DOI] [PubMed] [Google Scholar]
- 51.Verhofstede, C., F. V. Wanzeele, B. Van Der Gucht, N. De Cabooter, and J. Plum. 1999. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS 13:2541-2546. [DOI] [PubMed] [Google Scholar]
- 52.Wright, S. 1931. Evolution in Mendelian populations. Genetics 16:97-159. [DOI] [PMC free article] [PubMed] [Google Scholar]