Abstract
Inactivating mutations of THRB, which encodes the thyroid hormone receptor β (TRβ), cause resistance to thyroid hormone (RTH; OMIM 190160). To date, more than 100 THRB mutations have been reported among RTH patients. Most mutations substitute a single amino-acid residue in the ligand-binding domain. In this report, we describe clinical and molecular findings of three families with RTH. Three families harbored one novel (p.I431M) and two recurrent (p.R320H and p.R383C) THRB mutations. To examine the pathogenicity of identified mutations, we introduced a novel computational mutation prediction method based on three-dimensional structure data of TRβ-T3 complex. First, to define the accuracy of our prediction system, we evaluated ten previously reported ‘positive control’ mutations, as well as 30 seemingly benign sequence variations observed among vertebral species as ‘negative controls’. We found that our system had a sensitivity of 80% and a specificity of 93%. We then analyzed three mutations detected in the present study and found that all three mutations are predicted to be deleterious. Our data suggest that our structure-based prediction system would be a prompt, inexpensive and feasible method for evaluating the pathogenicity of missense THRB mutations.
Keywords: thyroid hormone resistance syndrome, thyroid hormone receptor beta, mutation, genetics, computational biology
Introduction
Monoallelic inactivating mutations of THRB, which encodes the thyroid hormone receptor β (TRβ), cause an inherited syndrome of reduced end-organ responsiveness to thyroid hormone (resistance to thyroid hormone, RTH; OMIM 190160) (1, 2). RTH is hallmarked by high circulating thyroid hormone levels accompanied by unsuppressive TSH levels. In the majority of cases, thyroid hormone resistance develops systemically (generalized RTH), although the severity of resistance varies tissue to tissue. One major determinant for the tissue specificity is the relative expression levels of two TR isoforms (α and β): in RTH patients, TRβ-dominant tissues, such as bone, become more thyroid hormone resistant than TRα-dominant ones, such as the heart (3). Therefore, RTH patients can manifest hypothyroidism, hyperthyroidism or a mixture of both, depending on the balance between elevation of circulating thyroid hormone and the severity of thyroid hormone resistance of each tissue.
To date, more than 150 THRB mutations have been reported among RTH patients (a partial listing is available from http://www.receptors.org/cgi-bin/nrmd/nrmd.py). Most are missense mutations occurring in the ligand-binding domain, and the mutations have scattered locations throughout the domain. Of clinical genetic importance, a considerable number of mutations are ‘private’ (i.e., a specific mutation is observed only within a single family). Thus, finding a novel mutation is not exceptional when one performs genetic testing, requiring an examiner to distinguish true deleterious mutations from uncommon but benign sequence variations. Conventionally, whether a THRB mutation is deleterious or not has been tested by expression experiments using cultured cell lines. Although such an experimental approach has provided fruitful insights into the molecular pathogenesis of RTH, routine use in a clinical genetics setting is still impractical because it is time-consuming and expensive. Computational mutation prediction based on protein sequence conservation (sequence-based method) is a practical alternative method to evaluate the pathogenicity of a putative mutation. Today, a handful of sequence-based prediction programs are freely available as web servers (reviewed in Ref. 4). In general, the sensitivities of these programs are high (70–90%), while the specificities remain relatively low (70–75%) (4). Another alternative method is mutation prediction based on three-dimensional structure data (structure-based method). In this method, the effect of a putative mutation on protein structure is predicted from one or more rules (e.g., location of the mutation and change in amino acid size). There are several programs with algorithms that use structure data in addition to sequence data (4). At present, however, structure-based methods have only limited prediction power, probably because the programs cannot handle complicated information such as the interaction between residues that are distant in the protein sequence but proximal in its three-dimensional structure.
In the present report, we describe three families with RTH carrying a total of three missense THRB mutations. To evaluate the pathogenicity of identified mutations, we introduced a novel structure-based mutation prediction method that uses protein structure modeling.
Materials and Methods
Mutation detection
Written informed consent for molecular studies was obtained from the patients or parents. This study was approved by the institutional review board of Keio University School of Medicine. We extracted genomic DNA from the three probands and the family member, and sequenced all coding exons and flanking introns of THRB using a standard PCR-based technique. Detected mutations were tested in 100 control Japanese individuals.
Computational mutation prediction
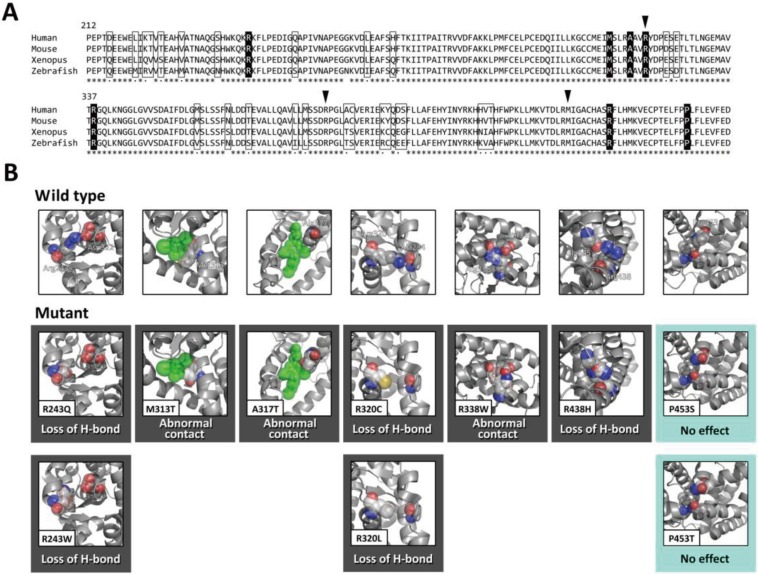
Crystal structure data of TRβ-T3 complex (5) (protein data bank ID 3GWS; http://www.rcsb.org/pdb) were used as a reference WT structure. The data were also used as a template to model each mutation (or ‘polymorphism’). We modeled the structures of mutations (or ‘polymorphisms’) using two modeling softwares: the PyMOL Molecular Graphics System (http://www.pymol.org) and DeepView (http://spdbv.vital-it.ch/). A mutation (or ‘polymorphism’) was predicted as deleterious when the amino acid change causes (i) loss of a hydrogen bond, (ii) loss of a van der Waals contact or (iii) formation of an abnormal residue-residue (or residue-ligand) contact. The pictures in Figs. 2 and 3 were produced with PyMOL.
Fig. 2.
Computational prediction of missense THRB mutations. A, Comparative protein sequence alignment of THRB in the human, mouse, Xenopus and zebrafish. To evaluate the accuracy of the structure-based mutation prediction, we analyzed ten known hotspot mutations affecting seven amino acid residues (shown in solid boxes) and 30 polymorphic changes occurring at 26 amino acid residues (shown in open boxes). Arrowheads indicate the positions of the mutations that were detected in the present study. Asterisks denote invariable sites, and dots indicate conservative substitutions. B, Modeled structures of the ten mutant TRβ (affecting seven amino acid residues) in comparison with the wild-type structure (upper panels). Predicted effects of the mutations are also shown. Modeling of mutants was performed using a built-in mutagenesis function of the PyMOL Molecular Graphics System. A mutation was predicted as deleterious (indicated by black boxes) when the amino acid change causes (i) loss of a hydrogen bond (‘Loss of H-bond’), (ii) loss of a van der Waals contact or (iii) formation of an abnormal residue-residue (or residue-ligand) contact (‘Abnormal contact’). Mutations that were predicted to have no deleterious effect on protein structure (“No effect”) are shown in sky blue boxes. Atom color code: red, oxygen; blue, nitrogen; yellow, sulfur; grey, others. T3 is colored green.
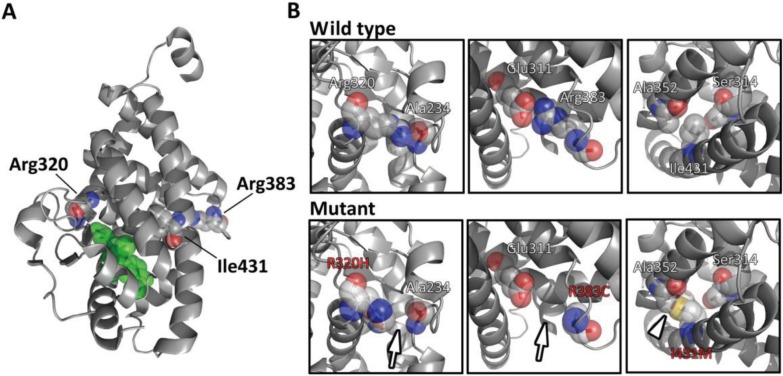
Fig. 3.
Computational mutation prediction of R320H, R383C and I431M. A, The three-dimensional structure of the TRβ (ligand-binding domain)-T3 complex is shown. Residues corresponding to Arg320, Arg383 and Ile431 are shown with their side chains as spheres. B, Modeled structures of R320H, R383C and I431M are shown (lower panels) in comparison with the wild-type structure (upper panels). R320H was predicted to lose the hydrogen bond between Arg320 and Ala234 (arrow). Similarly, the R383C mutation was predicted to lose the hydrogen bond between Arg383 and Glu311 (arrow). As for the I431M mutation, formation of the abnormal residue-residue contact (between mutated Met431 and Ala352) was predicted (arrowhead). Atom color code: red, oxygen; blue, nitrogen; yellow, sulfur; grey, others. T3 is colored green.
PolyPhen (http://genetics.bwh.harvard.edu/pph/), one of the most common sequence-based mutation prediction programs, was used to obtain prediction results based on sequence conservation information.
Results
Case reports
The three probands were born at term after uneventful pregnancies and deliveries from non-consanguineous parents. They each had a negative result in newborn screening for congenital hypothyroidism, in which their blood spot TSH level was measured. None had a family history of hypothyroidism or hyperthyroidism. All three probands were negative for thyroid autoantibodies, including anti-TSH receptor antibody, anti-thyroglobulin antibody and anti-thyroid peroxidase antibody.
Family 1
The proband (a 15-yr-old boy) was evaluated for his thyroid function at age 10 yr due to weight loss (1.5 kg) and fatigue, which were transient. He had high serum thyroid hormone levels (free T4, 2.3 ng/dl; free T3, 5.0 pg/ml; reference ranges of 0.9–1.8 and 2.3–4.3, respectively) with a normal serum TSH level (1.6 mU/l; reference 0.5–5.0). His height and weight were normal (167 cm, +0.4 SD; 52.6 kg, –0.2 SD, at age 14 yr). Although tachycardia was noted (heart rate, 108/min), no other consistent symptoms suggesting hyperthyroidism or hypothyroidism were observed. He had diffuse goiter (+2.7 SD; the size was evaluated by ultrasonography (6)), and a high normal 123I uptake (37.3% at 24 h; reference, 8–40). He had no intellectual problems, verbal disability or behavioral problems. As of his last visit, he had had maintained a good general condition without treatment.
The 42-yr-old mother of the proband has been healthy and had no specific complaint. She had neither goiter nor tachycardia (heart rate, 78/min). She received her first thyroid function tests as a family study of RTH. She had high serum thyroid hormone levels (free T4, 3.2 ng/dl; free T3, 4.1 pg/ml) with a normal serum TSH level (1.6 mU/l).
Family 2
The proband (a 5-yr-old boy) was first evaluated for his thyroid function at age 10 mo due to poor weight gain. He had high serum thyroid hormone levels (free T4, 2.4 ng/dl; free T3, 8.3 pg/ml) accompanied by a slightly elevated serum TSH level (6.2 mU/l). His height and weight were normal (112 cm, +0.4 SD; 17.2 kg, –0.6 SD). He had no symptoms or physical findings suggesting hyperthyroidism or hypothyroidism, and his heart rate was 80/min. He had delayed speech for his toddler age (says words, 1 yr 6 mo), but the delay was normalized by age 3 yr. He had no intellectual or behavioral problems. Ultrasonography revealed a normal-sized gland (+1.0 SD). He has never been treated.
The 37-yr-old father of the proband has been healthy and had no specific complaints; his height and weight were 175 cm and weight 68 kg, respectively. He had neither goiter nor tachycardia. Reportedly, he had delayed speech for his toddler age, but subsequently the delay was normalized. Thyroid function tests, which were conducted as a family study of RTH, showed high serum thyroid hormone levels (free T4, 2.7 ng/dl; free T3, 2.9 pg/ml) with a normal serum TSH level (1.7 mU/l).
Family 3
The proband (an 18-yr-old girl) was first noted to have goiter at age 13 yr. She had no symptoms or physical findings suggesting hyperthyroidism or hypothyroidism, except for mild tachycardia (heart rate, 80–120/min). Thyroid function tests revealed high serum thyroid hormone levels (free T4, 2.7 ng/dl; free T3, 8.2 pg/ml) with a normal serum TSH level (0.9 mU/l). Her height and weight were normal (160.5 cm, 50 kg). She had no intellectual problems, verbal disability or behavioral problems. She had normal 123I uptake (18.9% at 24 h). Her basal metabolic rate measurement was slightly low at –15.6% (reference, –15 to 15). She has never been treated. She has been in good general condition throughout the clinical course.
Mutation detection
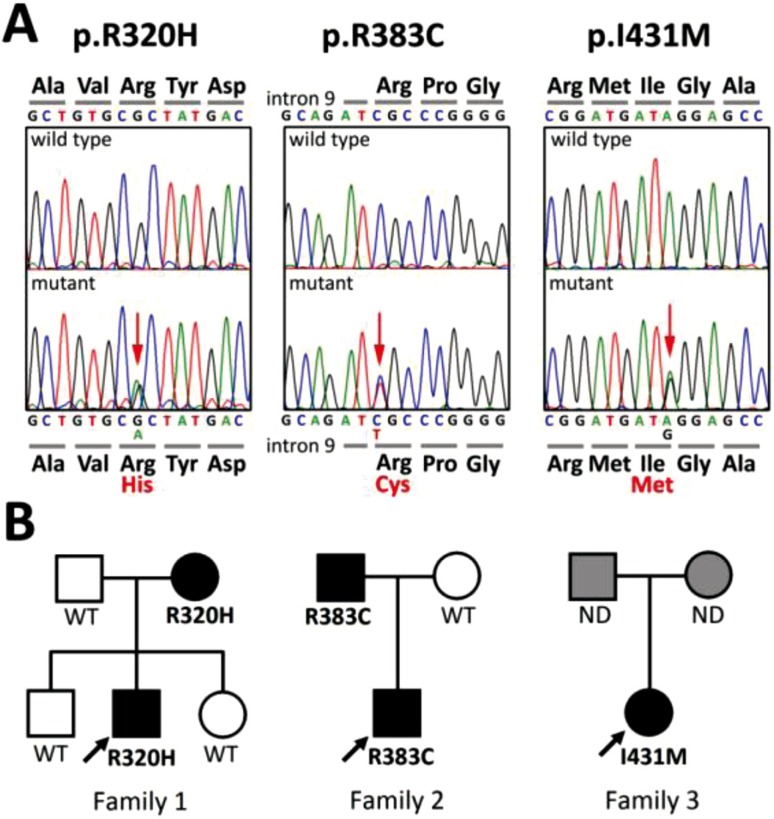
We found three heterozygous THRB mutations in the three families: one was novel (c.1293A>G, p.I431M in family 3), and the other two were recurrent (c.1244G>A, p.R320H (7) in family 1; c.959C>T, p.R383C (8) in family 2) (Fig. 1A). These mutations were absent among the 100 healthy controls. Family studies revealed that the mutation was transmitted by the mother in family 1 and by the father in family 2 (Fig. 1B). As for family 3, the parents declined the family study.
Fig. 1.
Identification of three THRB mutations. A, Partial sequences of PCR products of subjects are shown. The heterozygous substitutions of histidine in place of Arg320 (p.R320H), cysteine in place of Arg383 (p.R383C) and methionine in place of Ile431 (p.I431M) are indicated by the red arrows. B, The pedigrees of the three families with THRB mutations are shown. Thyroid function tests and THRB genotyping of family members showed 100% concordance between the mutant alleles and resistance to thyroid hormone phenotype. Squares, men; circles, women; solid symbols, affected by resistance to thyroid hormone; open symbols, unaffected; grey symbols, unknown affected status. ND denotes genotype not determined.
Computational mutation prediction
To examine whether or not the three mutations are likely to have deleterious effects, we introduced a novel structure-based mutation prediction system. We modeled the structure of each mutation (or ‘polymorphism’) using the WT structure as a template and predicted the effect of the mutation based on the three rules (See Materials and Methods for details).
To verify the accuracy of our prediction system, we evaluated previously reported mutations as ‘positive controls’. We choose ten mutations (R243Q, R243W, M313T, A317T, R320C, R320L, R338W, R438W, P453S and P453T; Fig. 2A and Table 1) that have been identified in five or more unrelated families (9) because they represent the most convincing deleterious mutations. We also evaluated 30 seemingly benign sequence variations (‘polymorphisms’) observed among vertebral species (mouse, Xenopus or zebrafish) as ‘negative controls’ (Fig. 2A). As a result, eight out of ten ‘positive control’ mutations were predicted as deleterious (Fig. 2B and Table 1). As for the 30 ‘polymorphisms’, two were predicted as deleterious (data not shown). The prediction results were not affected by which modeling software (PyMOL or DeepView) we used. Collectively, in our verification study, the sensitivity and specificity of our system were 80% (8/10) and 93% (28/30), respectively. Two ‘positive control’ mutations (P453S and P453T) were incorrectly predicted as benign (Fig. 2B), while two ‘polymorphisms’ (L221M and Y409C) were incorrectly predicted as deleterious (formation of the abnormal residue-residue contact, and loss of the van der Waals contact, respectively; data not shown).
Table 1. T3 binding affinity of the mutant TRβs compared with that of the wild-type TRβ.
| Mutation | T3 binding capacity Mean (SEM) | Ref. | Sequence-based predictiona | Structure-based prediction |
| R243Q | 0.84 (NA) | 14 | Benign | Deleterious |
| R243W | 0.74 (NA) | 14 | Probably damaging | Deleterious |
| M313T | NA | Probably damaging | Deleterious | |
| A317T | 0.12 (0.02) | 13 | Benign | Deleterious |
| R320C | 0.46 (0.17) | 15 | Probably damaging | Deleterious |
| R320L | 0.10 (0.03) | 13 | Probably damaging | Deleterious |
| R338W | 0.10 (0.03) | 13 | Probably damaging | Deleterious |
| R438H | 0.23 (0.09) | 13 | Possibly damaging | Deleterious |
| P453S | 0.36 (0.09) | 13 | Possibly damaging | Not deleterious |
| P453T | 0.20 (0.04) | 13 | Possibly damaging | Not deleterious |
Incorrect prediction results are indicated in bold. NA denotes not available. aPrediction based on PolyPhen (http://genetics.bwh.harvard.edu/pph/).
To compare prediction results between a sequence-based method and our structure-based method, we tested ten ‘positive control’ mutations by PolyPhen, one of the most common sequence-based programs. PolyPhen predicted eight ‘positive control’ mutations to be deleterious, but incorrectly predicted R243Q and A317T as benign (Table 1).
Finally, we analyzed the three mutations detected in the present study (locations of the mutations are shown in Fig. 3A) using our structure-based system. The R320H mutation was predicted to lose the hydrogen bond between Arg320 and Ala234 (Fig. 3B, left panel). The R383C mutation was also predicted to lose the hydrogen bond (between Arg383 and Glu311; Fig. 3B, middle panel). As for I431M, the isoleucine to methionine substitution was predicted to make an abnormal residue-residue contact between the mutated Met431 residue and Ala352 (Fig. 3B, right panel). Collectively, all three mutations were predicted to be deleterious.
Discussion
We have described five subjects (belonging to three families) carrying THRB mutations. These mutation carriers had high serum thyroid hormone levels accompanied by normal to slightly high serum TSH levels. The differential diagnosis for the biochemical phenotype includes RTH, TSH-producing pituitary adenoma and familial dysalbuminemic hyperthyroxinemia (FDH; OMIM 103600). It is unlikely that the three families have TSH-producing pituitary adenoma because i) none of the five subjects had manifestations of hyperthyroidism, except for mild tachycardia, and ii) less than 1% of TSH-producing adenomas shows familial occurrence (10). FDH is an autosomal dominant genetic disorder characterized by the increased thyroxine-binding capacity of mutated albumin and resultant erroneously high serum thyroid hormone measurements (11, 12). Patients with FDH usually have no physical abnormalities. Because the three families had goiter, tachycardia or verbal disability, which are common findings of RTH (3, 9), FDH is also unlikely. Collectively, the clinical diagnosis of generalized RTH seems convincing for the three families.
In the present study, we tried to predict the pathogenicity of each mutation using a novel structure-based computational method. We verified the accuracy of our system with ten ‘positive control’ mutations and 30 ‘polymorphisms’ and produced a reasonable result (sensitivity of 80% and specificity of 93%). The sensitivity was comparable to that of the sequence-based method (80%) evaluated by the same dataset.
Among the ten ‘positive control’ mutations, two mutations (M313T and A317T) seem to involve the T3-TRβ contact directly (Fig. 2). At least for A317T, this prediction was consistent with in vitro data showing very limited T3-binding capacity of the mutation (Table 1) (13). As for M313T, no functional data have been reported. Nonetheless, such very low T3-binding capacity has also been shown in other mutations that do not involve the T3-TRβ contact directly (e.g., R320L and R338W; Table 1). This implies that the T3-binding capacity of TRβ can be affected not only by local changes at the T3 binding site but also by global structural changes.
Our structure-based computational method has clear advantages as compared with experimental methods in its promptness and inexpensiveness. Nevertheless, its limitations should be noted. In our verification study, false negative results were seen in two out of ten ‘positive control’ mutations, making the sensitivity relatively low. The two mutations that were incorrectly predicted as benign (P453S and P453T) have impaired T3 binding in vitro (13). Thus, these two mutations should affect the structure of TRβ. Why does our system fail to predict deleterious effects of P453S and P453T? This is presumably due to the nature of the residue (proline) that was affected by the mutations. It is well known that proline, which has a pentacyclic side chain and more rigid conformation than other amino acids, is commonly used in the formation of a turn structure. This is true for Pro453 of TRβ (5). The P453S and P453T mutations probably affect the turn linked by Pro453 and result in erroneous rearrangement of carboxyl-terminal residues. However, no currently available software can model such a drastic change appropriately.
Of interest, the two 'positive control' mutations missed by our structure-based method could be predicted by the sequence-based method (PolyPhen), and two other mutations that Polyphen failed to predict could be correctly predicted by our system. This observation indicates that the two distinct methods can work complimentarily in predicting mutations. Therefore, using the two methods in combination might improve the overall prediction power of the computational approach. Further studies, including ones dealing with other genes, will be needed to validate the performance of such a combinatory approach.
In summary, we found one novel and two recurrent THRB mutations in three families with RTH. We examined the effects of the mutations using a novel structure-based method and found that all three mutations are likely to be deleterious. Our experience exemplifies the usefulness of the structure-based mutation prediction method, which might strengthen the prediction power of computational approaches.
Acknowledgments
We thank Prof. Takao Takahashi for fruitful discussion. This work was supported by a Grant-in-Aid for Scientific Research (C) (21791006) from the Japan Society for the Promotion of Science and a Grant for Child Health and Development (20C-2) from the Ministry of Health, Labour and Welfare of Japan.
References
- 1.Refetoff S, DeWind LT, DeGroot LJ. Familial syndrome combining deaf-mutism, stuppled epiphyses, goiter and abnormally high PBI: possible target organ refractoriness to thyroid hormone. J Clin Endocrinol Metab 1967;27: 279–94 [DOI] [PubMed] [Google Scholar]
- 2.Sakurai A, Takeda K, Ain K, Ceccarelli P, Nakai A, Seino S, et al. Generalized resistance to thyroid hormone associated with a mutation in the ligand-binding domain of the human thyroid hormone receptor beta. Proc Natl Acad Sci U S A 1989;86: 8977–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brucker-Davis F, Skarulis MC, Grace MB, Benichou J, Hauser P, Wiggs E, et al. Genetic and clinical features of 42 kindreds with resistance to thyroid hormone. The National Institutes of Health Prospective Study. Ann Intern Med 1995;123: 572–83 [DOI] [PubMed] [Google Scholar]
- 4.Laskowski RA, Thornton JM. Understanding the molecular machinery of genetics through 3D structures. Nat Rev Genet 2008;9: 141–51 [DOI] [PubMed] [Google Scholar]
- 5.Nascimento AS, Dias SM, Nunes FM, Aparicio R, Ambrosio AL, Bleicher L, et al. Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function. J Mol Biol 2006;360: 586–98 [DOI] [PubMed] [Google Scholar]
- 6.Yasumoto M, Inoue H, Ohashi I, Shibuya H, Onishi T. Simple new technique for sonographic measurement of the thyroid in neonates and small children. J Clin Ultrasound 2004;32: 82–5 [DOI] [PubMed] [Google Scholar]
- 7.Weiss RE, Weinberg M, Refetoff S. Identical mutations in unrelated families with generalized resistance to thyroid hormone occur in cytosine-guanine-rich areas of the thyroid hormone receptor beta gene. Analysis of 15 families. J Clin Invest 1993;91: 2408–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owen PJ, Chatterjee VK, John R, Halsall D, Lazarus JH. Augmentation index in resistance to thyroid hormone (RTH). Clin Endocrinol (Oxf) 2009;70: 650–4 [DOI] [PubMed] [Google Scholar]
- 9.Weiss RE, Refetoff S. Resistance to thyroid hormone. Rev Endocr Metab Disord 2000;1: 97–108 [DOI] [PubMed] [Google Scholar]
- 10.Refetoff S, Weiss RE, Usala SJ. The syndromes of resistance to thyroid hormone. Endocr Rev 1993;14: 348–99 [DOI] [PubMed] [Google Scholar]
- 11.Ruiz M, Rajatanavin R, Young RA, Taylor C, Brown R, Braverman LE, et al. Familial dysalbuminemic hyperthyroxinemia: a syndrome that can be confused with thyrotoxicosis. N Engl J Med 1982;306: 635–9 [DOI] [PubMed] [Google Scholar]
- 12.Wada N, Chiba H, Shimizu C, Kijima H, Kubo M, Koike T. A novel missense mutation in codon 218 of the albumin gene in a distinct phenotype of familial dysalbuminemic hyperthyroxinemia in a Japanese kindred. J Clin Endocrinol Metab 1997;82: 3246–50 [DOI] [PubMed] [Google Scholar]
- 13.Adams M, Matthews C, Collingwood TN, Tone Y, Beck-Peccoz P, Chatterjee KK. Genetic analysis of 29 kindreds with generalized and pituitary resistance to thyroid hormone. Identification of thirteen novel mutations in the thyroid hormone receptor beta gene. J Clin Invest 1994;94: 506–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagi H, Pohlenz J, Hayashi Y, Sakurai A, Refetoff S. Resistance to thyroid hormone caused by two mutant thyroid hormone receptors beta, R243Q and R243W, with marked impairment of function that cannot be explained by altered in vitro 3,5,3'-triiodothyroinine binding affinity. J Clin Endocrinol Metab 1997;82: 1608–14 [DOI] [PubMed] [Google Scholar]
- 15.Cugini CD , Leidy JW , Chertow BS, Bérard J, Bradley WE, Menke JB, et al. An arginine to histidine mutation in codon 315 of the c-erbA beta thyroid hormone receptor in a kindred with generalized resistance to thyroid hormones results in a receptor with significant 3,5,3’-triiodothyronine binding activity. J Clin Endocrinol Metab 1992;74: 1164–70 [DOI] [PubMed] [Google Scholar]