Abstract
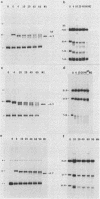
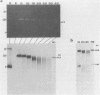
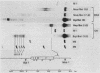
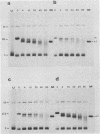
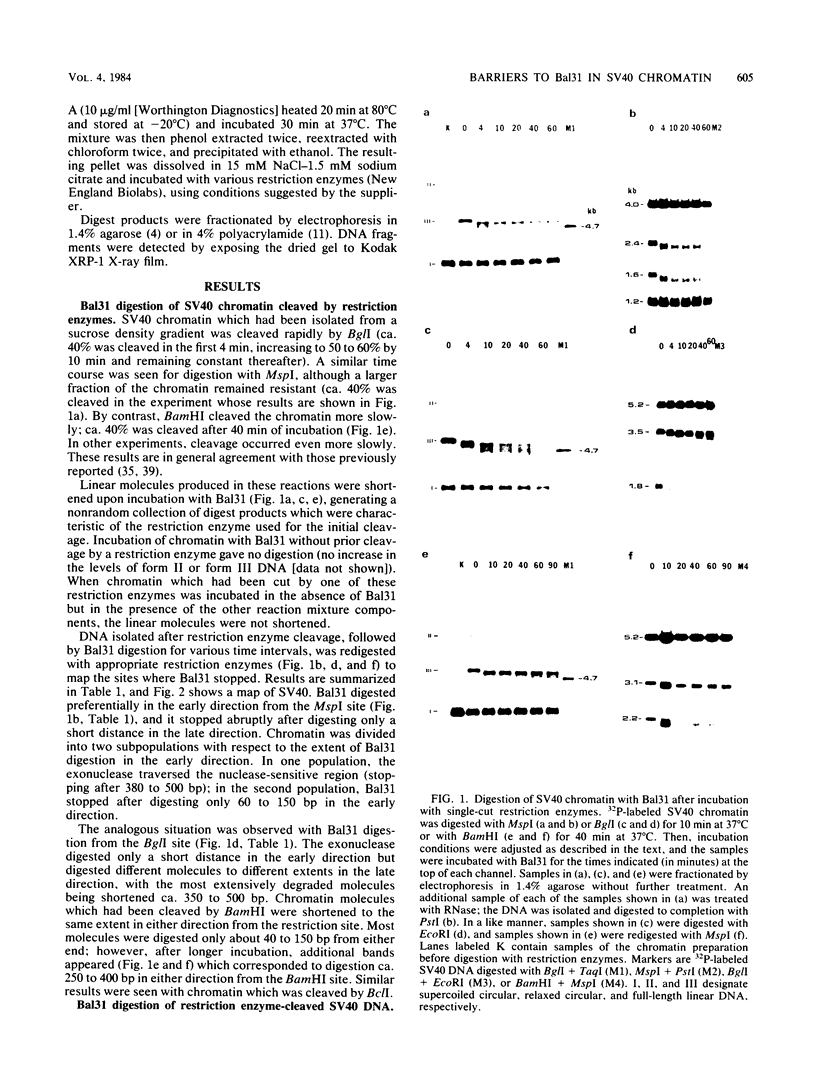
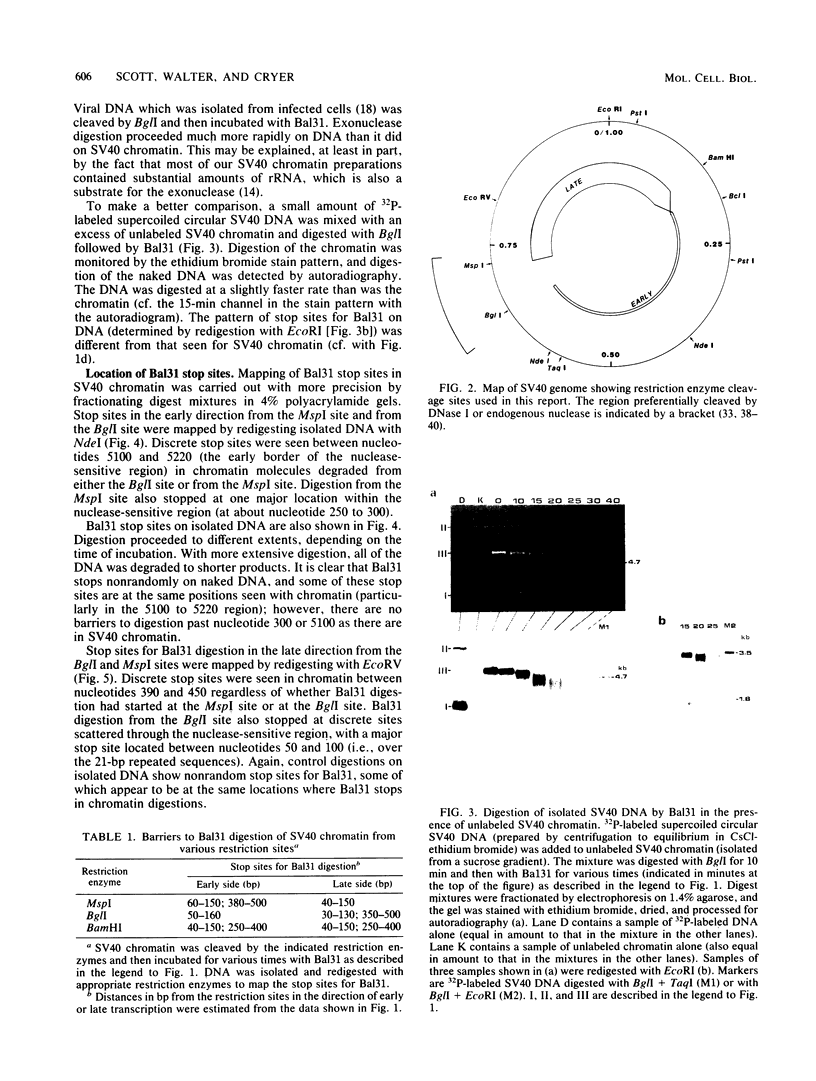
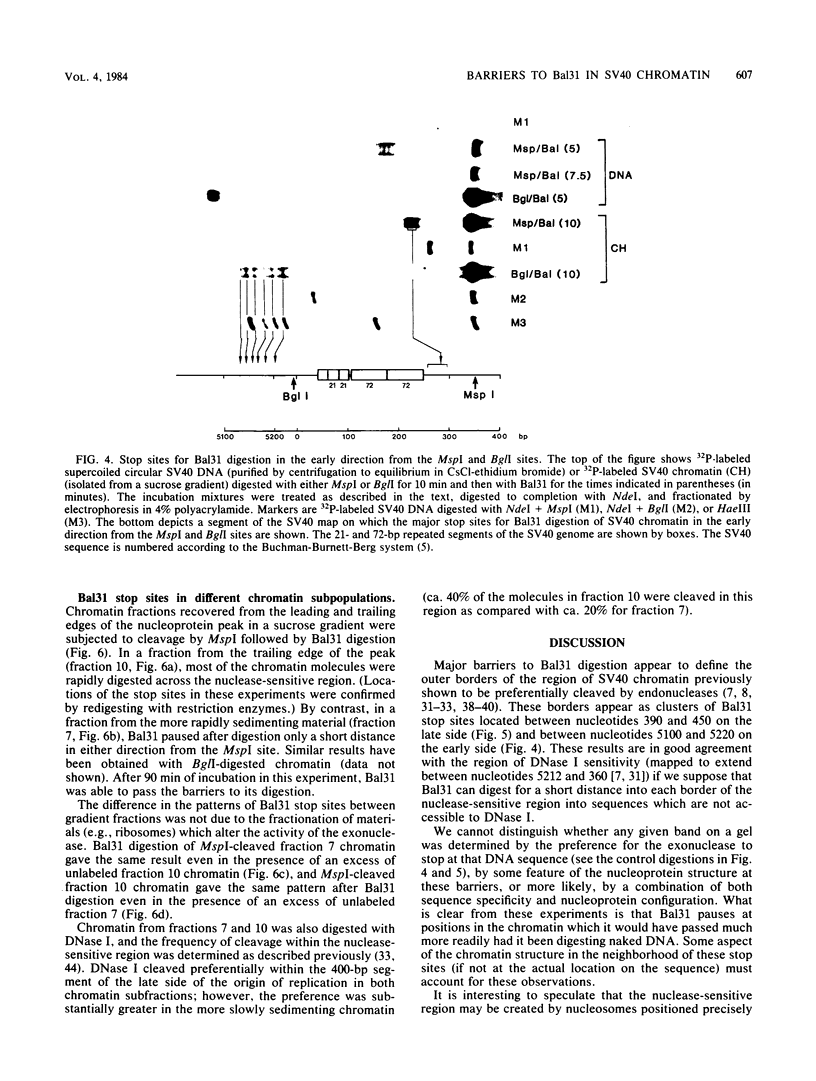
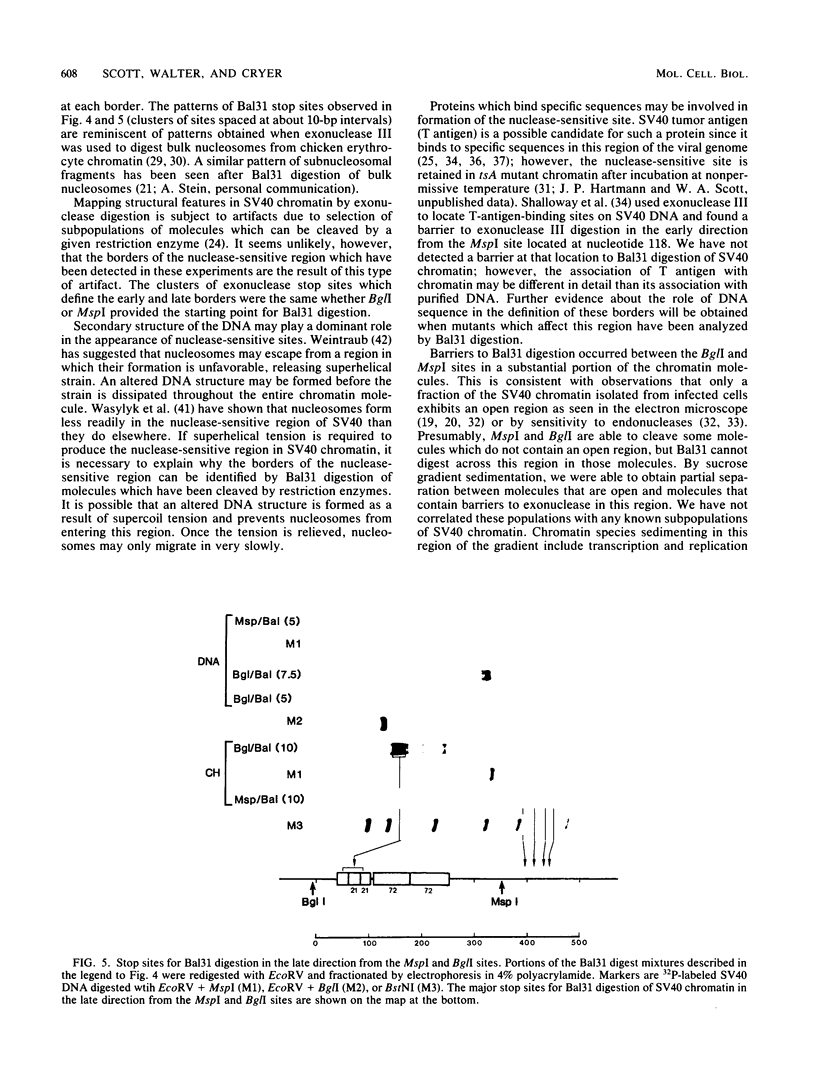
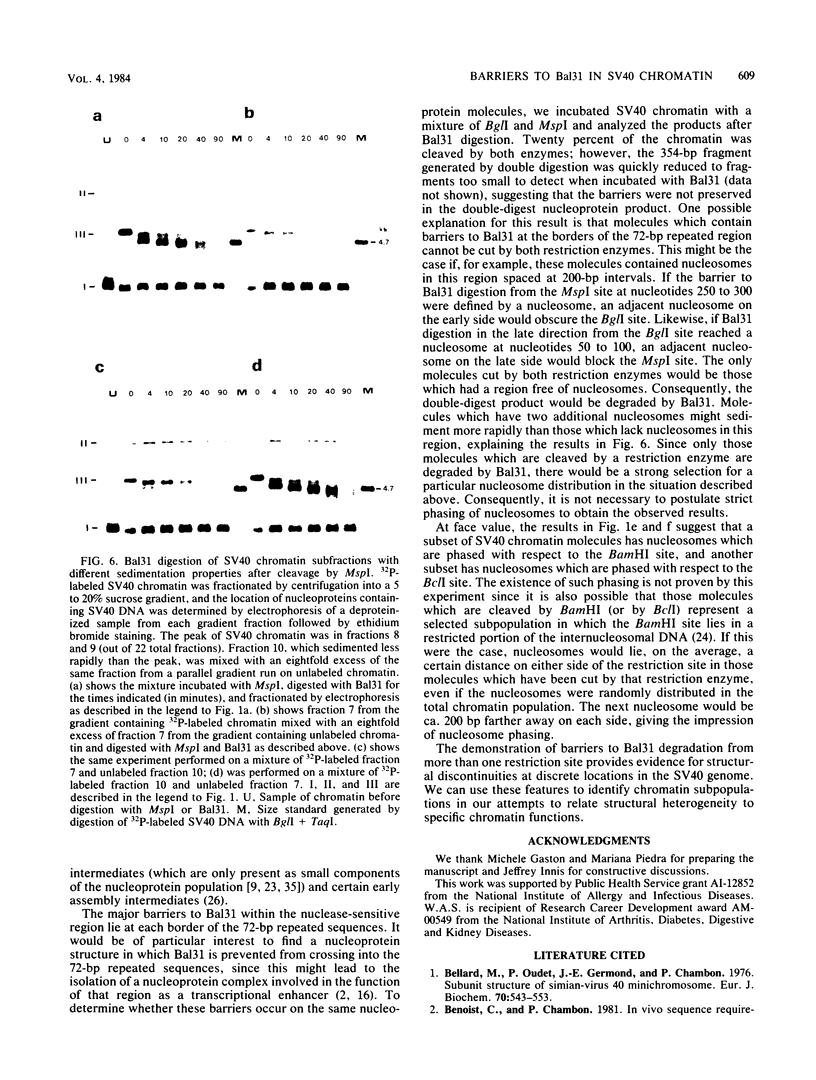
A portion of the nucleoprotein containing viral DNA extracted from cells infected by simian virus (SV40) is preferentially cleaved by endonucleases in a region of the genome encompassing the origin of replication and early and late promoters. To explore this nuclease-sensitive structure, we cleaved SV40 chromatin molecules with restriction enzymes and digested the exposed termini with nuclease Bal31. Digestion proceeded only a short distance in the late direction from the MspI site, but some molecules were degraded 400 to 500 base pairs in the early direction. By comparison, BglI-cleaved chromatin was digested for only a short distance in the early direction, but some molecules were degraded 400 to 450 base pairs in the late direction. These barriers to Bal31 digestion (bracketing the BglI and the MspI sites) define the borders of the same open region in SV40 chromatin that is preferentially digested by DNase I and other endonucleases. In a portion of the SV40 chromatin, Bal31 could not digest through the nuclease-sensitive region and reached barriers after digesting only 50 to 100 base pairs from one end or the other. Chromatin molecules that contain barriers in the BglI to MspI region are physically distinct from molecules that are open in this region as evidenced by partial separation of the two populations on sucrose density gradients.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellard M., Oudet P., Germond J. E., Chambon P. Subunit structure of simian-virus-40 minichromosome. Eur J Biochem. 1976 Nov 15;70(2):543–553. doi: 10.1111/j.1432-1033.1976.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Brockman W. W., Nathans D. The isolation of simian virus 40 variants with specifically altered genomes. Proc Natl Acad Sci U S A. 1974 Mar;71(3):942–946. doi: 10.1073/pnas.71.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright I. L., Abmayr S. M., Fleischmann G., Lowenhaupt K., Elgin S. C., Keene M. A., Howard G. C. Chromatin structure and gene activity: the role of nonhistone chromosomal proteins. CRC Crit Rev Biochem. 1982;13(1):1–86. doi: 10.3109/10409238209108709. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Herbomel P., Jouanneau J., Saragosti S., Katinka M., Bourachot B., de Crombrugghe B., Yaniv M. Structure and function of the promoter-enhancer region of polyoma and SV40. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):935–944. doi: 10.1101/sqb.1983.047.01.107. [DOI] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crémisi C., Chestier A., Yaniv M. Preferential association of newly synthesized histones with replicating SV40 DNA. Cell. 1977 Dec;12(4):947–951. doi: 10.1016/0092-8674(77)90159-3. [DOI] [PubMed] [Google Scholar]
- Crémisi C., Pignatti P. F., Yaniv M. Random location and absence of movement of the nucleosomes on SV 40 nucleoprotein complex isolated from infected cells. Biochem Biophys Res Commun. 1976 Dec 6;73(3):548–554. doi: 10.1016/0006-291x(76)90845-7. [DOI] [PubMed] [Google Scholar]
- Danna K. J., Sack G. H., Jr, Nathans D. Studies of simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol. 1973 Aug 5;78(2):363–376. doi: 10.1016/0022-2836(73)90122-8. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Fernandez-Munoz R., Coca-Prados M., Hsu M. T. Intracellular forms of simian virus 40 nucleoprotein complexes. I. Methods of isolation and characterization in CV-1 cells. J Virol. 1979 Feb;29(2):612–623. doi: 10.1128/jvi.29.2.612-623.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray H. B., Jr, Winston T. P., Hodnett J. L., Legerski R. J., Nees D. W., Wei C. F., Robberson D. L. The extracellular nuclease from Alteromonas espejiana: an enzyme highly specific for nonduplex structure in nominally duplex DNA. Gene Amplif Anal. 1981;2:169–203. [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann J. P., Scott W. A. Nuclease-sensitive sites in the two major intracellular simian virus 40 nucleoproteins. J Virol. 1983 Jun;46(3):1034–1038. doi: 10.1128/jvi.46.3.1034-1038.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. A nucleosome-free region in SV40 minichromosomes. Nature. 1980 May 22;285(5762):263–265. doi: 10.1038/285263a0. [DOI] [PubMed] [Google Scholar]
- Jakobovits E. B., Bratosin S., Aloni Y. Formation of a nucleosome-free region in SV40 minichromosomes is dependent upon a restricted segment of DNA. Virology. 1982 Jul 30;120(2):340–348. doi: 10.1016/0042-6822(82)90035-6. [DOI] [PubMed] [Google Scholar]
- Künzler P., Stein A. Histone H5 can increase the internucleosome spacing in dinucleosomes to nativelike values. Biochemistry. 1983 Apr 12;22(8):1783–1789. doi: 10.1021/bi00277a007. [DOI] [PubMed] [Google Scholar]
- Livingston D. M. A sequence of the yeast 2 micron DNA plasmid chromosome near the origin of replication is exposed to restriction endonuclease digestion. J Mol Biol. 1982 Sep 25;160(3):397–410. doi: 10.1016/0022-2836(82)90304-7. [DOI] [PubMed] [Google Scholar]
- Llopis R., Perrin F., Bellard F., Gariglio P. Quantitation of transcribing native simian virus 40 minichromosomes extracted from CV1 cells late in infection. J Virol. 1981 Apr;38(1):82–90. doi: 10.1128/jvi.38.1.82-90.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Another potential artifact in the study of nucleosome phasing by chromatin digestion with micrococcal nuclease. Cell. 1983 Apr;32(4):1205–1215. doi: 10.1016/0092-8674(83)90303-3. [DOI] [PubMed] [Google Scholar]
- McKay R. D. Binding of a simian virus 40 T antigen-related protein to DNA. J Mol Biol. 1981 Jan 25;145(3):471–488. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- Milavetz B., Hopkins T. Simian virus 40 encapsidation: characterization of early intermediates. J Virol. 1982 Sep;43(3):830–839. doi: 10.1128/jvi.43.3.830-839.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Riley D. E. Deoxyribonuclease I generates single-stranded gaps in chromatin deoxyribonucleic acid. Biochemistry. 1980 Jun 24;19(13):2977–2992. doi: 10.1021/bi00554a024. [DOI] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Cereghini S., Yaniv M. Fine structure of the regulatory region of simian virus 40 minichromosomes revealed by DNAase I digestion. J Mol Biol. 1982 Sep 15;160(2):133–146. doi: 10.1016/0022-2836(82)90171-1. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Scott W. A., Wigmore D. J. Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell. 1978 Dec;15(4):1511–1518. doi: 10.1016/0092-8674(78)90073-9. [DOI] [PubMed] [Google Scholar]
- Shalloway D., Kleinberger T., Livingston D. M. Mapping of SV40 DNA replication origin region binding sites for the SV40 T antigen by protection against exonuclease III digestion. Cell. 1980 Jun;20(2):411–422. doi: 10.1016/0092-8674(80)90627-3. [DOI] [PubMed] [Google Scholar]
- Tack L. C., Wassarman P. M., DePamphilis M. L. Chromatin assembly. Relationship of chromatin structure to DNA sequence during simian virus 40 replication. J Biol Chem. 1981 Aug 25;256(16):8821–8828. [PubMed] [Google Scholar]
- Tegtmeyer P., Andersen B., Shaw S. B., Wilson V. G. Alternative interactions of the SV40 A protein with DNA. Virology. 1981 Nov;115(1):75–87. doi: 10.1016/0042-6822(81)90090-8. [DOI] [PubMed] [Google Scholar]
- Tjian R. The binding site on SV40 DNA for a T antigen-related protein. Cell. 1978 Jan;13(1):165–179. doi: 10.1016/0092-8674(78)90147-2. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O. H., Bohn M. J. SV40 viral minichromosome: preferential exposure of the origin of replication as probed by restriction endonucleases. Nucleic Acids Res. 1978 Oct;5(10):3469–3477. doi: 10.1093/nar/5.10.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Waldeck W., Föhring B., Chowdhury K., Gruss P., Sauer G. Origin of DNA replication in papovavirus chromatin is recognized by endogenous endonuclease. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5964–5968. doi: 10.1073/pnas.75.12.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Oudet P., Chambon P. Preferential in vitro assembly of nucleosome cores on some AT-rich regions of SV40 DNA. Nucleic Acids Res. 1979 Oct 10;7(3):705–713. doi: 10.1093/nar/7.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wigmore D. J., Eaton R. W., Scott W. A. Endonuclease-sensitive regions in SV40 chromatin from cells infected with duplicated mutants. Virology. 1980 Jul 30;104(2):462–473. doi: 10.1016/0042-6822(80)90348-7. [DOI] [PubMed] [Google Scholar]