Summary
Skeletal muscle possesses a strong ability to regenerate following injury, a fact that has been largely attributed to satellite cells. Satellite cells are skeletal muscle stem cells located beneath the basal lamina of the myofiber, and are the principal cellular source of growth and regeneration in skeletal muscle. MicroRNAs (miRNAs) play key roles in modulating several cellular processes by targeting multiple mRNAs that comprise a single or multiple signaling pathway. Several miRNAs have been shown to regulate satellite cell activity, such as miRNA-489, which functions to maintain satellite cells in a quiescent state. Although muscle-specific miRNAs have been identified, many of the molecular mechanisms that regulate myogenesis that are regulated by miRNAs still remain unknown. In this study, we have shown that miR-128a is highly expressed in brain and skeletal muscle, and increases during myoblast differentiation. MiR-128a was found to regulate the target genes involved in insulin signaling, which include Insr (insulin receptor), Irs1 (insulin receptor substrate 1) and Pik3r1 (phosphatidylinositol 3-kinases regulatory 1) at both the mRNA and protein level. Overexpression of miR-128a in myoblasts inhibited cell proliferation by targeting IRS1. By contrast, inhibition of miR-128a induced myotube maturation and myofiber hypertrophy in vitro and in vivo. Moreover, our results demonstrate that miR-128a expression levels are negatively controlled by tumor necrosis factor α (TNF-α). TNF-α promoted myoblast proliferation and myotube hypertrophy by facilitating IRS1/Akt signaling via a direct decrease of miR-128a expression in both myoblasts and myotubes. In summary, we demonstrate that miR-128a regulates myoblast proliferation and myotube hypertrophy, and provides a novel mechanism through which IRS1-dependent insulin signaling is regulated in skeletal muscle.
Key words: Muscle satellite cell, microRNA, Cell proliferation, Insulin signalling, Muscular dystrophy
Introduction
Skeletal muscle has the ability to regenerate following injury or disease. Muscle regeneration is mediated primarily by mononucleated myogenic stem cells, referred to as satellite cells (Collins et al., 2005; Mauro, 1961). Satellite cells are found beneath the basal lamina of myofibers and remain in a quiescent state. Satellite cells are activated by muscle injury, exercise or disease in which they are induced to proliferate, differentiate and fuse to form multinucleated myofibers. Duchenne muscular dystrophy (DMD) is caused by lack of dystrophin protein at the sarcolemma of muscle fibers and is characterized by progressive muscle weakness associated with continuous degeneration and regeneration of skeletal myofibers (Hoffman et al., 1987; Hoffman et al., 1988; Monaco et al., 1986). It has been reported that loss of satellite cell regenerative capacity due to continual needs for regeneration may contribute to disease progression in DMD (Luz et al., 2002; Mouly et al., 2005; Wright, 1985).
Skeletal muscle also exhibits tremendous plasticity depending on their activity, which is tightly regulated via a balance of protein synthesis and degradation. Increased protein synthesis results in muscle hypertrophy, whereas elevated protein degradation causes muscle atrophy. Additionally, skeletal muscle function is regulated by the nervous system. Muscle atrophy is caused by the prolonged electrical inactivity including immobilization, denervation or muscle unloading. These observations indicate that the stimulation of physiological activity of muscles is an important regulator to maintain skeletal muscle mass (Jackman and Kandarian, 2004).
Recently, a number of studies have proposed novel aspects of muscle gene regulation in which small non-coding RNAs, known as microRNAs (miRNAs), play fundamental roles in various biological processes including development, cell proliferation and differentiation (Bartel, 2004; Krol et al., 2010; Stefani and Slack, 2008). MiRNAs are from 17 to 24 nucleotides in length and are known to interact with 3′-untranslated regions (3′ UTRs) of target mRNAs, resulting in the repression of mRNA translation and/or stability (Bartel, 2004). Since miRNAs can repress multiple target mRNAs, miRNA expression patterns and their predicted target mRNAs might represent a novel regulatory network (Eisenberg et al., 2007; Greco et al., 2009; Williams et al., 2009). MiRNAs are involved in the process of regeneration and hypertrophy/atrophy in skeletal muscle. In particular, miR-1, -133 and -206, which have been identified as muscle-enriched mRNAs, are known to play central regulatory roles in myogenesis (Chen et al., 2006; Kim et al., 2006; Rao et al., 2006). Additional miRNAs have been reported to regulate myoblast proliferation or differentiation which include miR-24, -26a, -27b, -125b, -148a, -181 and -214 (Crist et al., 2009; Ge et al., 2011; Liu et al., 2010; Naguibneva et al., 2006; Sun et al., 2008; Wong and Tellam, 2008; Zhang et al., 2012). Another microRNA, miR-489, is highly expressed in quiescent satellite cells and maintains satellite cell quiescence by suppressing expression of the oncogene Dek (Cheung et al., 2012). Another class of muscle-enriched microRNAs or myomiRs, which consist of miR-23b, -195 and -208 are involved in muscle hypertrophy/atrophy (van Rooij et al., 2006; van Rooij et al., 2007; Wada et al., 2011). These observations suggest that miRNAs regulate several stages of myogenesis by repressing many target genes and critical time points during development and disease.
Here, we investigated the function of miR-128a in skeletal muscle during several myogenic processes. MiR-128a is expressed in hematopoietic stem cells and has been shown to play an important role in cell lineage determination in neuronal stem cell, as well as muscle side population cells (Georgantas et al., 2007; Motohashi et al., 2012; Smirnova et al., 2005). In addition, it has been reported that cell proliferation in several types of tumor cells is regulated by miR-128a expression (Godlewski et al., 2008; Venkataraman et al., 2010). However, despite its enriched expression in skeletal muscle (Lee et al., 2008) the function of miR-128a in this tissue has not been fully elucidated. Using biological pathway analysis programs, we identified several predicted miR-128a targets in the insulin signaling pathway including insulin receptor (Insr), insulin receptor substrate 1 (Irs1) and phosphatidylinositol 3-kinases regulatory 1 (Pik3r1; also referred to as p85α). We show that miR-128a directly targets the 3′ UTR region of these insulin signaling genes to negatively regulate their expression levels. We also demonstrate that miR-128a modulates myoblast proliferation and myotube formation in vitro and regulates muscle hypertrophy in vivo. Furthermore, we identify the proinflammatory cytokine tumor necrosis factor α (TNF-α), as an enhancer of activation of IRS1/Akt signaling pathway via a decrease of miR-128a expression in myoblasts and myotubes. Our observations highlight the role of miR-128a in skeletal muscle and suggest that regulation of muscle mass may be possible through modulation of miR-128a activity.
Results
Expression of miR-128a in skeletal muscle and myogenic cells
To identify the expression profile of miR-128a in adult tissues, we first performed qPCR analysis of miR-128a levels in various adult mouse tissues. MiR-128a expression was the highest in brain and skeletal muscle, which is consistent with previously published findings (Fig. 1A) (Lee et al., 2008). Previously, it was shown that miR-128a was increased in expression during myoblast differentiation (Chen et al., 2010; Sun et al., 2010). To confirm this, miR-128a expression was analyzed in quiescent satellite cells analyzed immediately after isolation by fluorescence-activated cell sorting (FACS), in myoblasts derived from satellite cells cultured in growth medium for 4 days, and in myotubes, which were derived from myoblasts maintained in differentiation medium for 5 days. Mononuclear cells from uninjured fore- and hind-limb muscle of adult mice were purified by FACS as CD45-negative, CD31-negative and Vcam1-positive cells (supplementary material Fig. S1A). Expression of Pax7 and MyoD were assessed to confirm cell purity and further validate the isolated myogenic population as satellite cells (supplementary material Fig. S1B) (Fukada et al., 2007). The level of miR-128a expression increased during myogenic differentiation from quiescent satellite cells to myotubes (Fig. 1B) which is in agreement with previous reports (Chen et al., 2010; Sun et al., 2010). We then investigated miR-128a expression during skeletal muscle regeneration in mouse tibialis anterior (TA) muscle following a cardiotoxin (CTX)-induced injury. MiR-128a expression levels were analyzed by qPCR during a time course of 0 (uninjured), 1, 3, 5, 7 and 14 days post cardiotoxin-induced muscle injuries. MiR-128a expression decreased immediately following CTX-injury (Fig. 1C). Interestingly, during the myoblast proliferative phase 3 days following CTX muscle injury, miR-128a levels were slightly increased (Fig. 1C). MiR-128a expression remained at low levels until day 14 when the levels began to increase (Fig. 1C).
Fig. 1.

Expression of miR-128a in tissues and myogenic cells. (A) Expression of miR-128a in adult mouse tissues. Quantitative PCR results show that miR-128a is highly expressed in brain and skeletal muscle. (B) MiR-128a expression during myogenesis. Total RNA enriched for miRNAs was extracted from mouse quiescent (Q) satellite cells, myoblasts (cultured satellite cells in growth medium for 3 days) and myotube (fused myoblasts in differentiation medium for 5 days), and analyzed by qPCR to assess miR-128 expression. The levels of miR-128a increased during myogenesis. The expression values were normalized with U6 snRNA expression, and are represented as mean±s.e. (n = 3). (C) MiR-128a expression during skeletal muscle regeneration. Mouse TA muscles were injured with cardiotoxin (CTX), and isolated 1, 3, 5, 7 and 14 days after CTX injection. After extraction of total RNAs, miR-128a expression was subjected to analysis by qPCR. The levels of miR-128a declined immediately after CTX injection. Values are represented as mean±s.e. (n = 3).
KEGG pathway analysis of miR-128a predicted target genes
To identify potential targets of miR-128a in muscle, we utilized the computational prediction program TargetScan (http://www.targetscan.org/) which yielded >500 predicted target genes for miR-128a. The molecular functional clustering of the predicted target genes of miR-128a was determined by the use of Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation analyses in the Database for Annotation, Visualization and Integrated Discovery tool (DAVID: http://david.abcc.ncifcrf.gov) (Table 1 and supplementary material Table S1) (Dennis et al., 2003; Huang et al., 2009). GO term annotation analysis of miR-128a target genes showed that over 60% of predicted target genes were classified directly as “cellular process”, while other GO terms (biological regulation and developmental process) were enriched as well (supplementary material Table S1). KEGG pathway annotation analysis was then performed on miR-128a predicted target genes within the “Cellular process” GO category. This analysis revealed that members of the insulin signaling pathway, which includes Pde3a, Pde3b, Insr, Irs1, Pik3r1, Pdpk1, Tsc1 and Rps6kb1, was the most enriched target pathway (Table 1) that might be directly regulated by miR-128a.
Table 1. The top ten pathways that are enriched with predicted target genes of miR-128a in GO:0009987 cellular processes.

MiR-128a targets several genes involved in the insulin signaling pathway
To determine if miR-128a could be directly involved in suppressing the predicted target genes in the insulin signaling pathway, the 3′ UTR of each gene was cloned into a luciferase reporter construct and transfected into HEK293T cells along with plasmid constructs that overexpressed either mouse miR-128a or scrambled miR controls (CON). The 3′ UTR in genes related to insulin signaling (IRS1, INSR, PIK3r1) was predicted to have a single miR-128a target site with complete complementary sequence in the seed region that is comprised of 6–8 nucleotides essential for miRNA/mRNA binding (Fig. 2A). MiR-128a overexpression dramatically inhibited the reporter activity of the Irs1, Insr, and Pik3r1 luciferase reporter constructs (Fig. 2B). Importantly, after miR-128a predicted target site in the 3′ UTR was mutated, the mutated reporter construct failed to activate the luciferase reporter following miR-128a overexpression (Fig. 2B). These findings suggest that predicted target site of each gene is directly affected by miR-128a expression levels and that miR-128a targeted the 3′ UTR of these three insulin pathway-related genes.
Fig. 2.

Predicted miR-128a targets related to insulin signaling. (A) Alignment of the predicted binding sequence of miR-128a given below the binding site in the 3′ UTR of insulin signaling pathway-related genes, and the mutated 3′ UTR binding sequence below. The miR-128a seed match region is highlighted in bold and mutated region is underlined. (B) Luciferase reporter gene linked to the 3′ UTR of the individual genes (WT) or the 3′ UTR with a mutated miR-128a binding site (Mut) was co-transfected with miR-128a (black bar) or a scrambled miRNA control (CON) expression plasmid (white bar) into 293T cells. Forty-eight hours after transfection, luciferase assay was performed. The transfection of miR-128a overexpression vector significantly inhibited the levels of luciferase activity in cells that were transfected with 3′ UTR-pGL2 construct of individual genes, whereas it had no effect when cells were transfected with mutated 3′ UTR-pGL2 and miR-128a.
MiR-128a regulates myoblast cell proliferation by targeting IRS1
To address the potential role of miR-128a during myoblast proliferation, we induced or inhibited miR-128a expression in primary mouse myoblasts using lentiviral vectors containing green fluorescent protein (GFP) fluorescent reporters. Three days after culture of primary myoblasts, the viruses were added to the culture medium. Seventy-two hours after infection, infected myoblasts were purified by FACS based on GFP expression and subjected to cell proliferation assays. MiR-128a expression was approximately 40-fold higher in cells overexpressing miR-128a compared with control cells after lentivirus infection, whereas in miR-128a silenced cells, miR-128a expression was not at detectable levels by qPCR (data not shown). As shown in Fig. 3A, overexpression of miR-128a attenuated myoblast proliferation, whereas miR-128a inhibition in myoblasts promoted cellular proliferation. Additionally, when the cells were stained with phospho-histone-H3 antibody, a marker for mitotic cells, overexpression of miR-128a reduced myoblast proliferation which is indicated by significantly decreased numbers of phospho-histone-H3-positive cells in the miR-128a overexpressing myoblasts as compared with scrambled miR control (CON) cells (Fig. 3B). Conversely, we found that inhibition of miR-128a promoted myoblast proliferation, as shown by the increase in the number of phospho-histone H3 cells as compared with scrambled miR control cells (Fig. 3B).
Fig. 3.
miR-128a negatively regulates myoblast proliferation through targeting of IRS1. (A) Myoblasts, where miR-128a is overexpressed or inhibited, were seeded at the same number (10,000 cells per well in a 48-well culture plate) and then counted every 2 days for 8 days. Overexpression of miR-128a repressed myoblast proliferation compared with scrambled control (CON), whereas inhibition of miR-128 promoted proliferation of myoblasts, indicating that miR-128a is the regulator of cell proliferation in myoblasts. Values are represented as mean±s.e. (n = 3). (B) Immunostaining with phospho-histone-H3 antibody (H3-P), which is a mitosis marker. Myoblasts cultured in growth medium were stained with H3-P antibody and the number of H3-P-positive cells was counted. The ratio of H3-P-positive cells to total nuclei was significantly lower in myoblasts where miR-128a is overexpressed than in control myoblasts, whereas anti miR-128-containing myoblasts had more H3-P-positive cells in comparison with control or miR-128-overexpressed myoblasts. This would indicate that myoblast proliferation is regulated by miR-128a expression levels. Values are represented as mean±s.e. (n = 4). Scale bar: 50 µm. (C,D) Protein levels in genes related to insulin signaling during myoblast proliferation indicated that miR-128a induced the decline of IRS1 and phospho-Akt levels, and the elevation of p21 protein levels. All quantitative results are normalized by GAPDH, and are shown as mean±s.e. from three independent experiments. (E) Effect of miR-128a on the expression of myogenic transcriptional factors (Pax7, Myf5, MyoD and Myogenin) during myoblast proliferation. The levels of myogenic factors were slightly elevated by expression of the anti miR-128, although not significantly. These transcription factors were not affected by miR-128a overexpression in primary myoblasts. The expression values were normalized with 18s rRNA expression, and are represented as mean±s.e. (n = 3).
Insulin and insulin-like growth factors (IGFs) are well known to stimulate proliferation and differentiation of myoblasts (Engert et al., 1996), in addition to increase protein synthesis (Fryburg, 1994). IRS1/PI3K/Akt signaling is the major pathway controlled by IGF1 (Walker et al., 2010). Binding of IGF1 with INSR induces tyrosine phosphorylation of IRS1, which in turn activates phosphatidylinositol 3-kinase (PI3K) and its downstream target, Akt. Our observations that miR-128a targets genes related to insulin signaling and represses myoblast proliferation led us to hypothesize that miR-128a directly restricted INSR or IRS1 expression which would result in a subsequent activation of Akt in myoblasts. To test this hypothesis, miR-128a was overexpressed in primary mouse myoblasts cultured in growth medium, and followed by analysis of the protein levels of genes related to insulin signaling and Akt phosphorylation. IRS1 protein was reduced in proliferating myoblasts that overexpressed miR-128a, while protein levels of INSR and PIK3r1 did not show significant changes in expression levels (Fig. 3C,D). Phosphorylated Akt levels were significantly decreased in miR-128a overexpressing myoblasts (Fig. 3C,D). These results indicated that overexpression of miR-128a in myoblasts attenuated cell proliferation by suppressing the levels of IRS1 and phosphorylated Akt. At the same time, mRNA expression of myogenic transcriptional factors (Myf5, Pax7, MyoD and Myogenin) did not significantly change by overexpression or suppression of miR-128a (Fig. 3E). As previous studies indicated, the Akt pathway regulates cell cycle and cellular proliferation by modulating the expression levels of critical cell cycle regulatory genes, such as p21, independent of MRF gene expression (Lawlor and Rotwein, 2000). Collectively, these studies suggest that miR-128a might regulate cell proliferation via expression of Akt signaling pathway components, not via MyoD or Myogenin expression.
MiR-128 inhibition does not affect myoblast differentiation, but promotes IRS1/Akt signaling and myotube hypertrophy
To study the function of miR-128a during myoblasts differentiation, we overexpressed or inhibited miR-128a in primary mouse myoblasts and then induced them to differentiate for 3 days. Differentiated myotubes were stained with myosin heavy chain (MyHC) antibody (supplementary material Fig. S2), and the percentage of nuclei in MyHC-positive cells (fusion index) were quantified. We observed no significant differences in the overall fusion indexes among each group (Fig. 4A). Interestingly, the number of myotubes with 10 or more nuclei was significantly increased when miR-128a was reduced as compared with cultures where miR-128a was overexpressed (Fig. 4B). To determine what effect miR-128a expression levels had on myogenic factors during differentiation, the expression levels of myogenic genes (Myf5, Pax7, MyoD and Myogenin) were analyzed by qPCR. Both Pax7 and MyoD, which are expressed during myoblast proliferation and early phase of myoblast differentiation and which are abolished in late stage of myoblast differentiation, were significantly decreased in anti miR-128 myotubes compared with control myotubes. No change in Myogenin expression was detected in the miR-128a inhibited myotubes (Fig. 4C). These results indicate that inhibition of miR-128a may promote the progression of myotube maturation instead of myoblast fusion.
Fig. 4.
miR-128a does not affect myoblast differentiation, but inhibition of miR-128a promotes myotube hypertrophy. (A) Primary myoblasts where miR-128a is overexpressed or inhibited were induced to differentiate with DMEM medium containing 5% horse serum for 3 days. The differentiated cells were stained with myosin heavy chain (MyHC) antibody and DAPI. The ratio of MyHC-positive cells to total nuclei (DAPI-positive cells) was quantified. There were no changes among each group during myoblast differentiation. Values are represented as mean±s.e. (n = 3). (B) Quantification of myotube formation. Three days after differentiation, the number of nuclei per myotube was calculated and it was found that overexpression of miR-128a induced smaller myotubes with fewer nuclei relative to anti miR-128a-transduced myotubes. Values are represented as mean±s.e. (n = 3). (C) Evaluation of the mRNA levels of myogenic transcriptional factors (Myf5, Pax7, MyoD and Myogenin). Five days after differentiation, total RNA was extracted from myotubes, and the RNAs were subjected to analysis with qPCR. The expression of Pax7 and MyoD in myotubes where miR-128 was suppressed was significantly reduced compared with scrambled control (CON) myotubes. The expression values were normalized with 18s rRNA expression, and are represented as mean±s.e. (n = 3). (D,E) C2C12 myoblasts transduced by anti miR-128 or scrambled miR control lentivirus vectors were induced to differentiate with DMEM medium containing 5% horse serum for 1 and 3 days, and starved with serum-free medium 24 hours before extraction of protein. The protein levels were explored by western blot analysis. Protein levels of IRS1 and Akt phosphorylation were higher in anti miR-128-transduced myotubes compared with that of control myotubes 1 day after differentiation. Three days after differentiation, the level of IRS1 was still higher in anti miR-128 myotubes than in control myotubes, although phosphorylated Akt level was slightly lower in anti miR-128 myotubes relative to that of control myotubes. All quantitative results were normalized by GAPDH, and are shown as mean±s.e. from four independent experiments. (F) C2C12 myoblasts transduced by anti miR-128 or scrambled control lentivirus vectors were induced to differentiate for 5 days, and starved with serum-free medium 24 hours before extraction of protein. The protein levels of IRS1, INSR, PIK3r1, and Akt phosphorylation were analyzed and quantified, indicating that IRS1 and phospho-Akt levels were reduced whereas INSR and PIK3r1 levels were not altered in anti miR-128-expressing myotubes. Quantitative data were normalized by GAPDH, and are shown as mean±s.e. (n = 3). (G) Inhibition of miR-128 induced myotube hypertrophy. C2C12 myoblasts transduced by anti miR-128 or scrambled control lentivirus vectors were induced to differentiate for 5 days, and then stained with myosin heavy chain (MyHC) antibody (red) and DAPI (blue). Myofiber diameter was measured and quantified. Representative data show that anti miR-128-expressing myotubes were thicker than control-expressing myotubes. Values are shown as mean±s.e. Scale bar: 50 µm.
To assess the protein levels of miR-128a targets and Akt signaling components during myoblast differentiation, C2C12 myoblasts transfected with either miR-128a, anti miR-128a or control viruses were induced to differentiate in differentiation medium. One day after differentiation, the protein levels of IRS1 and phosphorylated Akt were significantly higher in anti miR-128 myotubes than in control myotubes (Fig. 4D,E). Three days after differentiation, the level of IRS1 was still higher in anti miR-128 myotubes than in control myotubes, while the phosphorylated Akt level was slightly lower in anti miR-128 myotubes relative to control myotubes (Fig. 4D,E). Five days after the start of differentiation, IRS1 and phospho-Akt levels could no longer be detected in anti miR-128 myotubes, even though INSR and PIK3r1 expression levels could still be detected in both control and anti miR-128 myotubes (Fig. 4F). Moreover, to determine if anti miR-128 induces myotube hypertrophy, we measured C2C12 myotube size, since IGF1-mediated activated Akt has been proposed to increase the size of myotubes (Rommel et al., 2001). The diameter of myotubes where miR-128 was inhibited was measured 5 days after differentiation, and quantified as shown in Fig. 4G and supplementary material Fig. S3. Inhibition of miR-128a resulted in significantly larger myotubes (36.1±0.75 µm; n = 3) when compared with controls (29.2±0.65 µm; n = 3) (Fig. 4G and supplementary material Fig. S3). Collectively, these studies suggested that inhibition of miR-128a may promote myotube maturation or hypertrophy.
Inhibition of miR-128 by minicircle vectors induced muscle hypertrophy in vivo
To explore the function of miR-128a in skeletal muscle in vivo, non-viral minicircle vectors containing antisense miR-128 (anti miR-128) or scrambled miRNA control (CON) were introduced into wild-type mice via tail vein (supplementary material Fig. S4). Minicircle vectors lack a bacterial backbone sequence and the antibiotic selection marker, which can provide higher transfection efficiency, less immunological rejection and longer transgene expression (Chen et al., 2003). We performed intravenous injection with 50 µg of minicircle vectors carrying anti miR-128 or scrambled miRNA into 4-week-old C57BL/6 wild-type mice twice a week for 4 weeks. Total body and individual mass of muscles was examined following treatment [gastrocnemius (GA), quadriceps (QC) and TA]. Anti miR-128 treatment resulted in significant gain of entire body weight and muscle volume, compared with control miR-treated mice (Fig. 5A,B). At the same time, the transcriptional levels of miR-128a target-containing genes, Irs1, Insr and Pik3r1, were slightly elevated by anti miR-128 compared with scrambled control miR, but there were no significant differences between control and anti miR-128 injected muscle (Fig. 5C). Consistently, anti miR-128 inhibition significantly induced myofiber hypertrophy, as shown by the increased distribution of myofiber diameter (Fig. 5D). To examine a potential relationship between muscle hypertrophy induced by anti miR-128 and muscle fiber type, we compared myosin heavy chain (MyHC) isoform expression by qPCR in GA muscles. Although there were no significant differences, Myh7 (MyHC type I) expression was slightly decreased for the mice treated with anti miR-128, while the expression levels of Myh2 (MyHC type IIa), Myh1 (MyHC type IIX) and Myh4 (MyHC type IIb) were slightly elevated in muscle where miR-128a was inhibited as compared with control injections. However, there were no significant differences between control and anti miR-128 injected muscles (Fig. 5E).
Fig. 5.
Inhibition of miR-128 with minicircle vector increases skeletal muscle mass and fiber size. (A) C57BL/6 wild type mice were intravenously injected with 50 µg of minicircle vectors, which have the H1 promoter with anti miR-128 or scrambled miRNA, every 4 days, and each animal was analyzed for body weight. The results indicated that the body weight of anti miR-128-transduced mice was heavier than control (CON) mice. Values are shown as mean±s.e. (n = 4). (B) Thirty-five days after first injection, muscles from mice where anti miR-128 or scrambled miRNA was transduced via minicircle were isolated and each muscle was weighed. The results indicate that the volume of muscles was increased in anti miR-128-transduced mice in comparison with control mice. Values are represented as mean±s.e. (n = 3). (C) The expression levels of miR-128a target genes in the GA muscle from mice where anti miR-128 or scrambled miRNA was transduced were investigated by qPCR. The expression levels of miR-128a targets were elevated in anti miR-128-transduced muscle relative to control muscle, indicating that the expression of miR-128a targets was effectively promoted by anti miR-128 minicircle vector injection in vivo. Values are shown as mean±s.e. (n = 4). (D) QC muscles were isolated 35 days after first injection, sectioned and then stained with H&E. Images show representative H&E staining of QC muscle from mice where anti miR-128 (right) or scrambled miRNA (left) was transduced. The graph shows the frequency distribution of muscle fiber diameter, revealing that transduction of anti miR-128 via minicircles promoted muscle hypertrophy in QC muscles. Values are shown as mean±s.e. (n = 3). Scale bar: 100 µm. (E) Expression levels of Myh7 (MyHC type I slow), Myh2 (MyHC type IIa), Myh1 (MyHC type IIx) and Myh4 (MyHC type IIb) in GA muscles were quantified by qPCR, revealing that there were no differences between anti miR-128- or scrambled miRNA-transduced mice. The expression values were normalized with 18s rRNA expression, and are represented as mean±s.e. (n = 3).
TNF-α is a regulator of miR-128a and upregulates IRS1/Akt signaling pathway by suppressing miR-128a
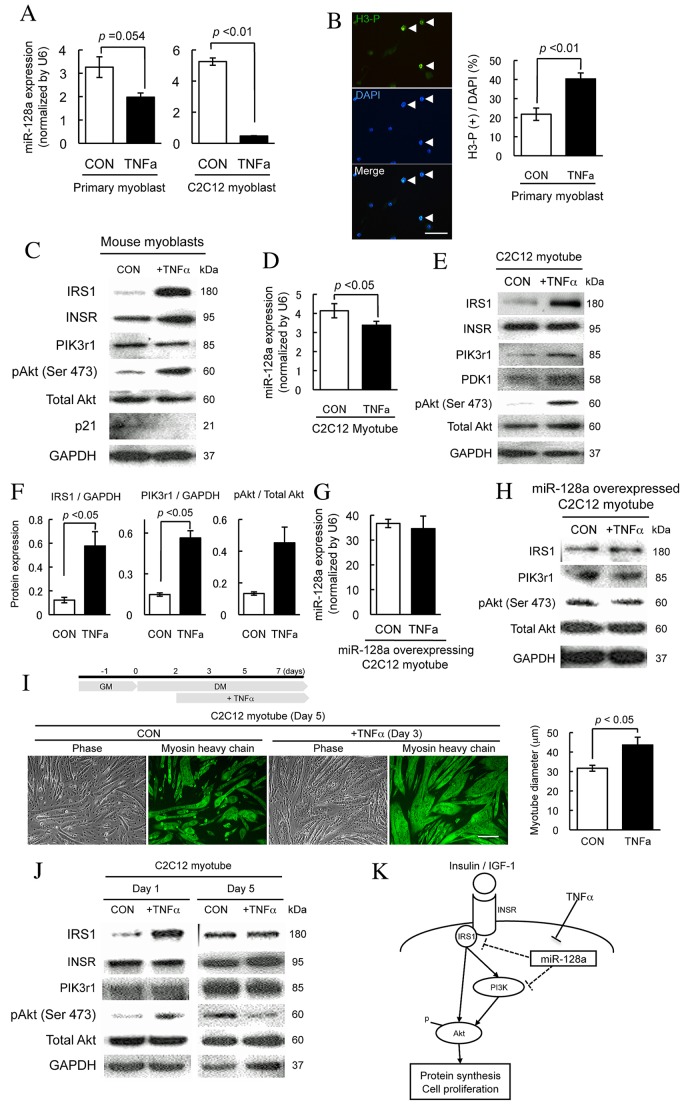
MiR-128a expression is decreased in early phase of skeletal muscle regeneration as shown in Fig. 1C and previously published results (Chen et al., 2010), suggesting that the factors or cytokines secreted by inflammatory cells such as neutrophils and macrophages may regulate miR-128a expression. TNF-α is a major pro-inflammatory cytokine that plays an important role in muscle regeneration by promoting myoblast proliferation and differentiation (Alter et al., 2008; Chen et al., 2007; Li, 2003; Li and Schwartz, 2001; Warren et al., 2002). TNF-α expression is upregulated immediately following CTX injection in mouse muscles (Chen et al., 2007). Furthermore, TNF-α is also known to regulate miR-128a by targeting the adenosine 2B receptor (A2BAR) in epithelial cells (Kolachala et al., 2010), which in turn regulates insulin signaling and myoblast proliferation (Alter et al., 2008; Li, 2003; Yamasaki et al., 1996). We first examined the effects of TNF-α on miR-128a expression in myoblasts. Primary myoblasts and C2C12 myoblasts cultured with growth medium were treated with mouse TNF-α recombinant protein (20 ng/ml), and then analyzed miR-128a expression by qPCR. TNF-α-treated samples showed a decrease in miR128a expression levels during the proliferation phase of primary myoblasts (P = 0.054) and C2C12 myoblast (P<0.01) (Fig. 6A). To further investigate the link between TNF-α and the regulation of miR-128a expression, we assessed whether the decline of miR-128a expression levels observed after TNF-α treatment might stimulate cell proliferation by promoting IRS1 and Akt activation. TNF-α treatment significantly increased myoblast proliferation, as indicated by the increased number of phospho-histone-H3-positive cells in TNF-α-treated myoblasts when compared with non-treated myoblasts (Fig. 6B). Protein expression of IRS1 and phosphorylated Akt was elevated after TNF-α treatment (Fig. 6C), consistent with previous reports that TNF-α promotes glucose uptake and cell proliferation (Li, 2003; Yamasaki et al., 1996). These observations imply that miR-128a and its target genes might be regulated under the control of TNF-α during myoblast proliferation. Next we examined the effects of TNF-α treatment in C2C12 myotubes to determine if TNF-α decreases miR-128a expression and promotes increase of IRS1/Akt signaling. According to previous reports, TNF-α inhibits myoblast fusion but at the same time also increases protein amounts in myotubes mediated by the PI3K/Akt signaling pathway (Alter et al., 2008; Plaisance et al., 2008). Treatment with TNF-α decreased miR-128a expression in C2C12 myotubes, and increased protein levels of IRS1, PIK3r1 and phosphorylated Akt (Fig. 6D,F). Although these observations support the fact that TNF-α regulates miR-128a expression and IRS1/Akt signaling, it did not address whether TNF-α directly or indirectly regulates IRS1/Akt signaling during myogenesis. To clarify this concept, C2C12 myoblasts overexpressing miR-128a from a lentiviral vector were induced in differentiation medium for 5 days, followed by 24 hours of treatment with/without TNF-α in serum-free medium, after which expression of miR-128a target genes were evaluated. As shown in Fig. 6G,H, miR-128a expression levels as well as the protein levels of IRS1, PIK3r1 and phosphorylated Akt were not altered in TNF-α-treated myotubes in which miR-128a is overexpressed. This finding suggested that increase of IRS1, PIK3r1 and phosphorylated Akt levels required the subsequent downregulation of miR-128a expression levels by TNF-α. Lastly, C2C12 myotubes were cultured with/without TNF-α to determine if the reduction of miR-128a levels caused by TNF-α treatment would induce the changes of IRS1 and pAkt levels and subsequent myotube hypertrophy, as observed in miR-128 inhibited myotubes (Fig. 4D–G). C2C12 myoblasts were induced into myotube with differentiation medium for 2 days, and then differentiated myotubes were cultured under the differentiation conditions with/without TNF-α (Fig. 6I), because TNF-α is known to hinder the early phase of myoblast differentiation (Alter et al., 2008; Coletti et al., 2002). The inhibition of myoblast fusion and the reduction of miR-128a was observed with the presence of TNF-α (Fig. 6A; supplementary material Fig. S5A). When miR-128a overexpressing myoblasts were treated with TNF-α, the inhibition of myoblast fusion was observed while miR-128a expression levels were not altered (Fig. 6G; supplementary material Fig. S5B). These data suggest that TNF-α inhibits myoblast fusion regardless of miR-128a expression levels. TNF-α treatment resulted in significantly thicker myotubes than control myotubes 3 days following initial exposure to TNF-α or mock controls (Fig. 6I). Furthermore, to assess the protein levels of miR-128a targets and Akt signaling after treatment of TNF-α, myotubes underwent western blot analysis. One day after TNF-α treatment, the levels of IRS1 and phosphorylated Akt were higher in TNF-α-treated myotubes than in mock control myotubes (Fig. 6J). Five days after the initiation of TNF-α treatment, IRS1 levels in TNF-α-treated myotubes had declined to the same level of that observed in control myotubes. Futhermore, phospho-Akt levels in TNF-α-treated myotubes were lower than mock controls even though INSR and PIK3r1 expression levels remained the same in both control and TNF-α-treated myotubes (Fig. 6J). When TNF-α treatment was performed in miR-128a overexpressing myotubes, no significant changes in myotube size was observed (supplementary material Fig. S5C). These observations indicated a strong regulation of genes related to the insulin-signaling pathway by miR-128a and TNF-α during myogenesis. Additionally, to evaluate the effect of insulin addition and to determine if IRS1 stimulation by TNF-α is sensitive to insulin, C2C12 myoblasts were differentiated into myotubes for 5 days, then treated with TNF-α and/or insulin. Our results indicated that increased IRS1 levels by TNF-α treatment are subsequently decreased by the addition of exogenous insulin. This finding suggested that the sensitivity of IRS1 to insulin in myotubes was increased after treatment of TNF-α (supplementary material Fig. S6). Collectively, these observations suggest that TNF-α might promote myotube hypertrophy caused by reduction of miR-128a levels. Furthermore, the successive elevation of IRS1, pAkt and TNF-α levels are upstream of IRS1/Akt insulin signaling that is mediated through miR-128a expression during myogenesis (Fig. 6K).
Fig. 6.
TNF-α decreases miR-128a expression, and promotes myoblast proliferation through the elevation of IRS1. (A) Primary myoblasts and C2C12 myoblasts cultured in growth medium were treated with TNF-α (TNFa) (20 ng/ml) for 24 hours. Total RNAs containing miRNAs were extracted from TNF-α-treated C2C12 myoblasts, and then subjected to analysis of the miR-128a levels by qPCR. This analysis revealed that miR-128a expression was suppressed by TNF-α. The expression values were normalized with U6 expression, and are represented as mean±s.e. (n = 3). (B) Primary myoblasts in growth medium were treated with TNF-α (20 ng/ml) for 3 days, stained with phospho-histone H3 (H3-P) antibody and DAPI, and the number of H3-P-positive cells calculated. The ratio of H3-P-positive cells to total nuclei was significantly increased in TNF-α-treated myoblasts than in control (CON) myoblasts, indicating that TNF-α promoted myoblast proliferation. Values are represented as mean±s.e. (n = 3). Scale bar: 50 µm. (C) The protein levels of miR-128a targets (IRS1, INSR and PIK3r1), Akt and p21 in myoblasts, which were treated with/without TNF-α for 3 days, were evaluated by western blot analysis, revealing that the expression levels of IRS1 and the level of Akt phosphorylation were increased in myoblasts treated with TNF-α in comparison with non-treated myoblasts. (D) C2C12 myotubes, which were induced to differentiate for 5 days and then cultured with serum-free medium supplemented with or without TNF-α (20 ng/ml) for 24 hours. Total RNAs were extracted from TNF-α-treated C2C12 myotubes, and then subjected to analysis of the miR-128a levels by qPCR. This analysis indicated that miR-128a expression was suppressed by TNF-α in C2C12 myotubes. The expression values were normalized with U6 expression, and are represented as mean±s.e. (n = 3). (E,F) The protein levels of miR-128a targets (IRS1, INSR, PDK1 and PIK3r1) and Akt in C2C12 myotubes, which were treated with or without TNF-α, were explored by western blot analysis. The results of western blot analysis were quantified, suggesting that the expression levels of IRS1, PIK3r1 and Akt phosphorylation were increased in C2C12 myotubes treated with TNF-α relative to non-treated myotubes. Quantitative data were normalized by GAPDH, and are shown as mean±s.e. MiR-128a was overexpressed by using a lentivirus vector, and myoblasts were induced to differentiate in differentiation medium for 5 days. MiR-128a-overexpressing myotubes were cultured with serum-free medium supplemented with or without TNF-α (20 ng/ml) for 24 hours, and then were evaluated for miR-128a expression by qPCR. The results show that miR-128a levels were not affected by TNF-α treatment in miR-128a-overexpressing myotubes. The expression values were normalized with U6 expression, and are represented as mean±s.e. (n = 3). (H) Western blot analysis to verify the protein levels of IRS1, PIK3r1 and phosphorylated Akt was performed in miR-128a-overexpressing myotubes that were cultured with serum-free medium supplemented with or without TNF-α for 24 hours. The results indicated that there was no effect on miR-128a-overexpressing myotubes of TNF-α treatment, revealing that the elevation of IRS1, PIK3r1 and phosphorylated Akt levels by TNF-α treatment was mediated by the decline of miR-128a expression. (I) Schematic protocol of the experiment. C2C12 myoblasts were induced into myotubes with differentiation medium for 2 days, and then myotubes were cultured under the differentiation condition with or without TNF-α (20 ng/ml). Three days after TNF-α treatment, myotubes were stained with myosin heavy chain (MyHC) antibody (green), and myofiber diameter was measured and quantified. Represented data show that TNF-α-treated myotubes were thicker than control myotubes. Values are shown as mean±s.e. Scale bar: 50 µm. (J) The protein levels of IRS1, INSR, PIK3r1 and Akt in myotubes, which were treated with or without TNF-α for 1 and 5 days, were evaluated by western blot analysis, revealing that protein levels of IRS1 and Akt phosphorylation were higher in myotubes treated with TNF-α compared with control myotubes 1 day after differentiation. Five days after treatment, IRS1 level in TNF-α-treated myotubes had declined to the same level as that in control myotubes, and phospho-Akt levels were reduced whereas INSR and PIK3r1 levels were not altered in TNF-α-treated myotubes. All quantitative results were normalized by GAPDH, and are shown as mean±s.e. from four independent experiments. (K) A model for the regulatory network involving miR-128a and IRS/Akt signaling in myogenesis. The model proposes that TNF-α regulates IRS1, PIK3r1 and phosphorylation of Akt levels through miR-128a.
Discussion
MiR-128a regulates myoblast proliferation and differentiation through IRS1/Akt signaling
Our study has identified miR-128a as a novel regulator of insulin signaling and characterized its function in skeletal muscle. Recent studies have shown that miR-128a expression is reduced in several tumor cell lines compared with normal cells through suppression of cell proliferation by targeting E2F3, Bmi-1 or p70S6K1 (Godlewski et al., 2008; Shi et al., 2012; Zhang et al., 2009). Our findings have highlighted miR-128a function in myogenesis, and identified novel muscle-enriched target gene, IRS1. Small RNAi-based gene silencing experiments revealed that insulin signaling pathways are dependent on Irs1 and are required for myotube formation and glucose uptake through the phosphorylation of Akt (Bouzakri et al., 2006). In addition, phosphorylated Akt promotes myoblast proliferation in cooperation with mammalian target of rapamycin (mTOR) or S6K1 (Naguibneva et al., 2006), suggesting that IRS1 is a key factor to induce myoblast proliferation and myotube hypertrophy by increasing phospho-Akt levels, and thus miR-128a inhibited myoblasts might be resulting from the increase of IRS1/Akt phosphorylation.
Additionally, we demonstrated that miR-128a expression levels increased during the myogenic differentiation of primary myoblasts (Fig. 1C). These observations suggest that miR-128a might also act as a negative regulator of insulin signaling in myotubes. MiR-128a has additional, non-insulin signaling pathway predicted target genes which include: Foxo1, Gsk3b and Pten (Table 1). These three factors are also known to negatively regulate the phosphorylation of Akt. Foxo1 (forkhead transcription factor O1) and GSK3 (glycogen synthase kinase 3β) are downstream targets of PI3K/Akt signaling in muscle (Rommel et al., 2001; Sandri et al., 2004). Activation of Foxo1 caused by PI3K/Akt phosphorylation induces the ubiquitin ligase, Atrogin-1, resulting in muscle atrophy (Sandri et al., 2004), and activation of GSK3β inhibits protein synthesis by inhibiting the translation initiation factor eIF2B (Rommel et al., 2001). In addition, miR-128a was recently reported to suppress Akt activity by targeting PTEN in pituitary tumors (Palumbo et al., 2012). Moreover, another miRNA, let-7, was also recently shown to regulate the insulin signaling pathway by targeting multiple genes including Igfr1, Insr and Irs2 in skeletal muscle (Zhu et al., 2011). Thus, it is likely that insulin signaling is regulated by several miRNAs in addition to miR-128a, which may also be IRS1-dependent and induce the phosphorylation of Akt during myotube hypertrophy.
TNF-α as a positive regulator of insulin signaling through repression of miR-128a
There is increasing evidence that TNF-α, a major pro-inflammatory cytokine produced by activated macrophages, has an important role in muscle regeneration by promoting myoblast proliferation and differentiation (Alter et al., 2008; Chen et al., 2007; Li, 2003; Li and Schwartz, 2001; Warren et al., 2002). TNF-α has also been shown to induce muscle wasting through the inhibition of protein synthesis, selective proteolysis, and the activation of nuclear factor-kappaB (NF-κB) and caspase-dependent pathways (Coletti et al., 2002; Li and Reid, 2000). Prior studies have demonstrated that TNF-α-deficient mdx dystrophic mice showed decreased muscle mass and increased dystrophic disease progression, further implying that TNF-α expression may be essential for normal skeletal muscle regeneration (Spencer et al., 2000). Our findings suggest that TNF-α promotes IRS1/Akt signaling through the suppression of miR-128a expression in myoblasts and myotubes. It has been reported that TNF-α-treated myoblasts increased total DNA content by accelerating cell cycle progression in a dose-dependent manner, whereas the high concentrations of TNF-α induced cell death (Li, 2003; Li and Reid, 2000). Additionally, TNF-α is thought to be secreted from myoblasts and myotubes, and to have a role as an autocrine factor in the activation of p38 (Chen et al., 2007). These observations and our results indicate that TNF-α might regulate insulin signaling through miR-128a expression.
MiRNAs are important contributors to mechanisms of disease and offer the potential to generate novel target therapies (Eisenberg et al., 2009; Williams et al., 2009). Several miRNAs have been implicated in disease progression and pathology, including muscular dystrophy (Cacchiarelli et al., 2010; Eisenberg et al., 2007). Our results suggest that inhibition of miR-128a might promote muscle regeneration and hypertrophy by inhibiting the expression of insulin signaling factors. Our findings demonstrate that miR-128a is likely to have an important role in cell growth and proliferation of skeletal muscle, and understanding its function can offer new insight into mechanisms of muscle regeneration, further opening up the potential for designing novel target therapies for muscle diseases.
Materials and Methods
Animals and cardiotoxin (CTX) injection
Animals were housed in the Animal Resources Children's Hospital (ARCH) and all procedures were approved by the Children's Hospital Boston Animal Facilities/IACUC protocols. C57BL/6 mice were originally purchased from Jackson Laboratory (Bar Harbor, ME), housed and bred according to standard procedures.
To induce muscle regeneration, cardiotoxin (CTX, 10 µM; Sigma, St. Louis, MO) was injected into the TA muscle. CTX-injected TA muscles were harvested 1, 3, 7 and 14 days after injection, while the contra-lateral leg was used as day 0 uninjured control.
Isolation of muscle satellite cells
Mononuclear cells were prepared from fore and hind limb muscles of 8- to 12-week-old female C57BL/6 mice. In brief, muscles were digested with 1.2 U/ml neutral protease/dispase and 5 mg/ml collagenase IV (Worthington Biochemical, Lakewood, NJ) for 60 minutes at 37°C. After elimination of RBCs by RBC lysis solution (Qiagen, Hilden, Germany), cells were resuspended with 2% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) in PBS, and incubated on ice for 30 minutes in the presence of PE-conjugated anti-CD31 antibody (1∶200, clone 390; BD Pharmingen, Franklin Lakes, NJ), FITC-conjugated anti-CD45 (1∶200, clone 30-F11; BD Pharmingen), and APC-conjugated Vcam1 (1∶200, clone 429; BioLegend, San Diego, CA). Propidium iodide was added to samples at a concentration of 2 µg/ml to exclude dead cells. Cells were detected in a FACS Aria flow cytometer (BD Bioscience, Franklin Lakes, NJ).
Cell cultures
Satellite cells from mouse muscle were cultured in growth medium consisting of high-glucose (4.5 g/L) Dulbecco's modified Eagle's medium (DMEM; cellgro, Manassas, VA) with 20% FBS (Atlanta Biological), 2.5 ng/ml basic fibroblast growth factor (Invitrogen), and penicillin (100 U/ml)-streptomycin (100 mg/ml) (Gibco-BRL, Gaithersburg, MD) on culture dishes coated with Matrigel (BD Biosciences). For myotube formation, the medium was replaced to DMEM supplemented with 5% horse serum and penicillin-streptomycin to promote myoblast differentiation (differentiation medium). TNF-α was purchased from R&D Systems (Minneapolis, MO), dissolved with 1% BSA in PBS, and then supplemented to the culture medium (20 ng/ml).
RNA extraction, TaqMan miRNA expression assay and SYBR Green-based quantitative real-time PCR
Total RNAs containing miRNA were extracted from satellite cells, myoblasts and myotubes using a miRNeasy RNA isolation kit (Qiagen). TaqMan microRNA expression assay was performed with 50 ng of total RNA by using TaqMan microRNA reverse transcription kit and TaqMan microRNA kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. For gene expression analysis, first strand cDNA was produced using a QuantiTect Reverse Transcription Kit (Qiagen), and synthesized cDNA was mixed with SYBR Green PCR Master Mix. Specific primers used for PCR are listed in supplementary material Table S2. The expression levels of miRNA and mRNA were quantified on an ABI 7900HT real-time PCR machine (Applied Biosystems) following the manufacturer's instructions. MiRNA expression was normalized against the expression of U6, and individual mRNA expression was normalized against the expression of 18 s rRNA. mRNA levels were quantified by comparative Ct (ΔΔCt) method (Schmittgen and Livak, 2008).
Functional and pathway analysis of microRNA predicted target genes
To identify the predicted miR-128a target genes, TargetScan (http://www.targetscan.org/) was used. To investigate the molecular function of the predicted target genes, biological pathway analysis of a large gene list was carried out to obtain gene lists on the basis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases in the Database for Annotation, Visualization and Integrated Discovery tool (DAVID: http://david.abcc.ncifcrf.gov) (Dennis et al., 2003; Huang et al., 2009). P-values for all of the different genes in GO terms and KEGG pathways were calculated by DAVID, and the false discovery rate (FDR) was calculated to correct the enrichment P-value based on the Benjamini–Hochberg multiple testing correction method; the significance was defined as P<0.01 and FDR<0.05.
Luciferase reporter assay
The 3′ UTR of mouse Irs1, Insr and Pik3r1 genes were amplified by PCR and cloned into a modified version of the pGL2Basic vector (containing a novel multi-cloning site between the luciferase open-reading frame and the SV40 polyA signal; gift from Dr J.A. Kreidberg) (Alexander et al., 2011) using the sequence-specific primers listed in supplementary material Table S2. The predicted miR-128a binding sites located in the 3′ UTR were mutated using the QuikChange II Site-Directed Mutagenesis Kit (Strategene, La Jolla, CA), and the primers used in this experiment were listed in supplementary material Table S2. For luciferase reporter assay, 3×104 HEK293T cells were initially plated in a 48-well plate 24 hours prior to transfection. The cells were transfected using Lipofectamine 2000 (Invitrogen) with 30 ng of reporter construct and 100 ng of miRNA overexpression plasmids or scrambled miRNA controls (Origene, Rockville, MD). Luciferase activity was measured 48 hours after transfection with the Dual Reporter Assay System (Promega, Madison, WI) on a single-tube luminometer (Berthold Technologies, Bad Wildbad, Germany). Transfections were performed in triplicate and the individual experiments were repeated three times.
Lentiviral miRNA overexpression and inhibition
Lentivirus-based expression plasmids containing green fluorescent protein (GFP) that overexpress either pre-miR-128a or anti miR-128 or miRNA control, were purchased from System Biosciences (Mountain View, CA). Lentiviral vectors, along with packaging plasmids (MDL/RRE, Rev, and VSV-G) were transfected into HEK 293T cells using Lipofectamine 2000 (Invitrogen). Three days after transfection, viral supernatants were collected and filtered through 45 micron filters (VWR, West Chester, PA), mixed with Lenti-X lentivirus concentrator (Clontech, Palo Alto, CA), and incubated overnight at 4°C. The following day, the virus co-precipitate was concentrated by centrifugation at 1,500×g for 60 minutes at 4°C. Viral pellets were re-suspended in PBS. To overexpress or inhibit miRNA expression, the viruses were added to myoblasts in culture. Seventy-two hours after induction, miR-128a overexpressed or inhibited myoblasts were collected based on GFP expression via flow cytometry (BD Biosciences).
Western blotting
Proteins from cultured cells were extracted with RIPA buffer (Sigma) supplemented with Complete Mini protease inhibitor cocktail (Roche, Meylan, France) and PhosStop phosphatase inhibitor (Roche). Proteins (30 µg per lane) were separated by 4–20% gradient Tris-glycine gels (Invitrogen), and transferred to polyvinylidene difluoride membrane (Invitrogen) using 15 V for 1 hour. Following transfer, the membranes were blocked with 5% nonfat milk in Tris-buffered saline containing Tween 20 (TBS-T) for 1 hour at room temperature, before being incubated overnight at 4°C with primary antibodies diluted with 5% BSA (BSA; Sigma) in TBS-T. The following antibodies were used for immunoblotting; anti-IRS1, anti-INSR, anti-PDK1, anti-phospho Akt (Ser 473), anti-Akt (Cell Signaling Technology, Danvers, MA), anti-PIK3r1 (Abcam, Cambridge, MA), p21, GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA). After serial washes with TBS-T, the membranes were incubated with anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology) in TBS-T containing 5% milk for 1 hour at room temperature. The signals obtained from the western blot analysis were quantified with the ImageJ program (NIH).
Immunocytochemical analysis
Myoblasts and myotubes were cultured on 8-well Lab-Tek Chamber Slides (Nunc, Rochester, NY, USA). Cultured cells were fixed in 4% paraformaldehyde (PFA) for 5 minutes, stained with anti-phospho-histone H3 antibody (Upstate Biotechnology, Lake Placid, NY) or mouse anti-myosin heavy chain (clone: MF20; Developmental Studies Hybridoma Bank, Iowa City, IA) at 4°C overnight and then incubated with the secondary antibody conjugated with Alexa 488 or Alexa 568 (Molecular Probes, Eugene, OR). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). The fusion indices were calculated as the percentage of nuclei in myosin heavy chain-positive myocyte or myotubes per total nuclei in the dish.
Preparation of minicircle vectors and intravenous injection
Minicircle producer plasmid vector was purchased from System Biosciences. It contained an EF1a promoter and an H1-promoter with scrambled miRNAs (CON) or antisense miR-128 (anti miR-128) located between attB and attP sites of the parental plasmid. Minicircle DNA vectors were generated by site-specific intramolecular recombination between attB and attP mediated by PhilC31 integrase, which allowed for the plasmid backbone to be degraded in bacteria and purified following manufacturer's instructions. Briefly, parental plasmids were transformed with E. coli strain ZYCY10P3S2T (System biosciences) and single colonies were cultured at 37°C. Bacterial cultures were spun down at 1,500 g for 15 minutes, resuspended with LB broth containing 1% L-(+)-arabinose (System biosciences) and then cultured at 30°C for 16 hours. Minicircle vectors were extracted using PureLink™ HiPure Plasmid Purification Kit (Invitrogen). Prepared 50 µg of minicircle vectors carrying anti miR-128 or scrambled miRNA (CON) were injected into C57B6 wild-type mice via tail vein using 29-gauge insulin syringe (Terumo Medical Products, Somerset, NJ).
Histological analyses
GA, QC and TA muscles were isolated from mice and frozen in liquid nitrogen-cooled isopentane (Sigma-Aldrich). Muscle tissues were sectioned on a cryostat into 6-µm cross sections and stained with Hematoxylin and Eosin (H&E). The maximum myotube diameter from the largest branch point was measured from at least 80 myotubes with ImageJ program (NIH).
Statistics
All quantitative data are represented as means±SE. Analysis was performed between different groups using two-tailed Student's t-test and nonparametric Mann–Whitney U-test. A probability of less than 5% (P<0.05) was considered statistically significant. A chi-square test was performed to assess significant differences in frequency distribution of myofiber diameter following anti miR-128 treatment.
Supplementary Material
Acknowledgments
The authors declare no competing financial interests. We would like to thank members of the Kunkel laboratory, Dr Emanuela Gussoni for her helpful advice, and Noreen Francis and Richard Bennett, who are members of the Stem Cell Core Facility at Children's Hospital Boston, for technical support.
Footnotes
Author contributions
N.M. was responsible for conception and design of experiments, collection and assembly of data, data analysis and interpretation, manuscript writing. M.S.A., Y.S.-M., J.A.M., G.K. were responsible for provision of experimental materials, data analysis and discussion of results. L.M.K. was responsible for conception and design, data analysis, financial support, manuscript writing and final approval of manuscript.
Funding
This work was funded by the Bernard F. and Alva B. Gimbel Foundation (to L.M.K.); and the National Institutes of Health [grant number 5P50NS040828]. All sequencing was accomplished in the IDDRC Molecular Genetics Core Facility funded by the National Institute of Child Health and Human Development [grant number 2P30HD018655-26]. The Stem Cell Core Facility at Children's Hospital Boston is funded by the National Institutes of Health [grant number NIH-P30-HD18655]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.119966/-/DC1
References
- Alexander M. S., Casar J. C., Motohashi N., Myers J. A., Eisenberg I., Gonzalez R. T., Estrella E. A., Kang P. B., Kawahara G., Kunkel L. M. (2011). Regulation of DMD pathology by an ankyrin-encoded miRNA. Skelet Muscle 1, 27 10.1186/2044-5040-1-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alter J., Rozentzweig D., Bengal E. (2008). Inhibition of myoblast differentiation by tumor necrosis factor alpha is mediated by c-Jun N-terminal kinase 1 and leukemia inhibitory factor. J. Biol. Chem. 283, 23224–23234 10.1074/jbc.M801379200 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bouzakri K., Zachrisson A., Al-Khalili L., Zhang B. B., Koistinen H. A., Krook A., Zierath J. R. (2006). siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 4, 89–96 10.1016/j.cmet.2006.04.008 [DOI] [PubMed] [Google Scholar]
- Cacchiarelli D., Martone J., Girardi E., Cesana M., Incitti T., Morlando M., Nicoletti C., Santini T., Sthandier O., Barberi L. et al. (2010). MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab. 12, 341–351 10.1016/j.cmet.2010.07.008 [DOI] [PubMed] [Google Scholar]
- Chen Z. Y., He C. Y., Ehrhardt A., Kay M. A. (2003). Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol. Ther. 8, 495–500 10.1016/S1525-0016(03)00168-0 [DOI] [PubMed] [Google Scholar]
- Chen J. F., Mandel E. M., Thomson J. M., Wu Q., Callis T. E., Hammond S. M., Conlon F. L., Wang D. Z. (2006). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. E., Jin B., Li Y. P. (2007). TNF-alpha regulates myogenesis and muscle regeneration by activating p38 MAPK. Am. J. Physiol. Cell Physiol. 292, C1660–C1671 10.1152/ajpcell.00486.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. F., Tao Y., Li J., Deng Z., Yan Z., Xiao X., Wang D. Z. (2010). microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 190, 867–879 10.1083/jcb.200911036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung T. H., Quach N. L., Charville G. W., Liu L., Park L., Edalati A., Yoo B., Hoang P., Rando T. A. (2012). Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524–528 10.1038/nature10834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletti D., Yang E., Marazzi G., Sassoon D. (2002). TNFalpha inhibits skeletal myogenesis through a PW1-dependent pathway by recruitment of caspase pathways. EMBO J. 21, 631–642 10.1093/emboj/21.4.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins C. A., Olsen I., Zammit P. S., Heslop L., Petrie A., Partridge T. A., Morgan J. E. (2005). Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 122, 289–301 10.1016/j.cell.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Crist C. G., Montarras D., Pallafacchina G., Rocancourt D., Cumano A., Conway S. J., Buckingham M. (2009). Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. USA 106, 13383–13387 10.1073/pnas.0900210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003). DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 10.1186/gb-2003-4-5-p3 [DOI] [PubMed] [Google Scholar]
- Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A. A., Lidov H. G., Kang P. B., North K. N., Mitrani-Rosenbaum S. et al. (2007). Distinctive patterns of microRNA expression in primary muscular disorders. Proc. Natl. Acad. Sci. USA 104, 17016–17021 10.1073/pnas.0708115104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg I., Alexander M. S., Kunkel L. M. (2009). miRNAS in normal and diseased skeletal muscle. J. Cell. Mol. Med. 13, 2–11 10.1111/j.1582-4934.2008.00524.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert J. C., Berglund E. B., Rosenthal N. (1996). Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J. Cell Biol. 135, 431–440 10.1083/jcb.135.2.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryburg D. A. (1994). Insulin-like growth factor I exerts growth hormone- and insulin-like actions on human muscle protein metabolism. Am. J. Physiol. 267, E331–E336 [DOI] [PubMed] [Google Scholar]
- Fukada S., Uezumi A., Ikemoto M., Masuda S., Segawa M., Tanimura N., Yamamoto H., Miyagoe-Suzuki Y., Takeda S. (2007). Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells 25, 2448–2459 10.1634/stemcells.2007-0019 [DOI] [PubMed] [Google Scholar]
- Ge Y., Sun Y., Chen J. (2011). IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 192, 69–81 10.1083/jcb.201007165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgantas R. W., 3rd, Hildreth R., Morisot S., Alder J., Liu C. G., Heimfeld S., Calin G. A., Croce C. M., Civin C. I. (2007). CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. USA 104, 2750–2755 10.1073/pnas.0610983104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J., Nowicki M. O., Bronisz A., Williams S., Otsuki A., Nuovo G., Raychaudhury A., Newton H. B., Chiocca E. A., Lawler S. (2008). Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 68, 9125–9130 10.1158/0008-5472.CAN-08-2629 [DOI] [PubMed] [Google Scholar]
- Greco S., De Simone M., Colussi C., Zaccagnini G., Fasanaro P., Pescatori M., Cardani R., Perbellini R., Isaia E., Sale P. et al. (2009). Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 23, 3335–3346 10.1096/fj.08-128579 [DOI] [PubMed] [Google Scholar]
- Hoffman E. P., Brown R. H., Jr, Kunkel L. M. (1987). Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 10.1016/0092-8674(87)90579-4 [DOI] [PubMed] [Google Scholar]
- Hoffman E. P., Fischbeck K. H., Brown R. H., Johnson M., Medori R., Loike J. D., Harris J. B., Waterston R., Brooke M., Specht L. et al. (1988). Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N. Engl. J. Med. 318, 1363–1368 10.1056/NEJM198805263182104 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jackman R. W., Kandarian S. C. (2004). The molecular basis of skeletal muscle atrophy. Am. J. Physiol. Cell Physiol. 287, C834–C843 10.1152/ajpcell.00579.2003 [DOI] [PubMed] [Google Scholar]
- Kim H. K., Lee Y. S., Sivaprasad U., Malhotra A., Dutta A. (2006). Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 174, 677–687 10.1083/jcb.200603008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolachala V. L., Wang L., Obertone T. S., Prasad M., Yan Y., Dalmasso G., Gewirtz A. T., Merlin D., Sitaraman S. V. (2010). Adenosine 2B receptor expression is post-transcriptionally regulated by microRNA. J. Biol. Chem. 285, 18184–18190 10.1074/jbc.M109.066555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J., Loedige I., Filipowicz W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610 [DOI] [PubMed] [Google Scholar]
- Lawlor M. A., Rotwein P. (2000). Coordinate control of muscle cell survival by distinct insulin-like growth factor activated signaling pathways. J. Cell Biol. 151, 1131–1140 10.1083/jcb.151.6.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. J., Baek M., Gusev Y., Brackett D. J., Nuovo G. J., Schmittgen T. D. (2008). Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 14, 35–42 10.1261/rna.804508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. P. (2003). TNF-alpha is a mitogen in skeletal muscle. Am. J. Physiol. Cell Physiol. 285, C370–C376 [DOI] [PubMed] [Google Scholar]
- Li Y. P., Reid M. B. (2000). NF-kappaB mediates the protein loss induced by TNF-alpha in differentiated skeletal muscle myotubes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 279, R1165–R1170 [DOI] [PubMed] [Google Scholar]
- Li Y. P., Schwartz R. J. (2001). TNF-alpha regulates early differentiation of C2C12 myoblasts in an autocrine fashion. FASEB J. 15, 1413–1415 [DOI] [PubMed] [Google Scholar]
- Liu J., Luo X. J., Xiong A. W., Zhang Z. D., Yue S., Zhu M. S., Cheng S. Y. (2010). MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J. Biol. Chem. 285, 26599–26607 10.1074/jbc.M110.115824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luz M. A., Marques M. J., Santo Neto H. (2002). Impaired regeneration of dystrophin-deficient muscle fibers is caused by exhaustion of myogenic cells. Braz. J. Med. Biol. Res. 35, 691–695 10.1590/S0100-879X2002000600009 [DOI] [PubMed] [Google Scholar]
- Mauro A. (1961). Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9, 493–495 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A. P., Neve R. L., Colletti-Feener C., Bertelson C. J., Kurnit D. M., Kunkel L. M. (1986). Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature 323, 646–650 10.1038/323646a0 [DOI] [PubMed] [Google Scholar]
- Motohashi N., Alexander M. S., Casar J. C., Kunkel L. M. (2012). Identification of a novel microRNA that regulates the proliferation and differentiation in muscle side population cells. Stem Cells Dev. 21, 3031–3043 10.1089/scd.2011.0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly V., Aamiri A., Périé S., Mamchaoui K., Barani A., Bigot A., Bouazza B., François V., Furling D., Jacquemin V. et al. (2005). Myoblast transfer therapy: is there any light at the end of the tunnel? Acta Myol. 24, 128–133 [PubMed] [Google Scholar]
- Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A. (2006). The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 8, 278–284 10.1038/ncb1373 [DOI] [PubMed] [Google Scholar]
- Palumbo T., Faucz F. R., Azevedo M., Xekouki P., Iliopoulos D., Stratakis C. A. (2013). Functional screen analysis reveals miR-26b and miR-128 as central regulators of pituitary somatomammotrophic tumor growth through activation of the PTEN-AKT pathway. Oncogene. 32, 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance I., Morandi C., Murigande C., Brink M. (2008). TNF-alpha increases protein content in C2C12 and primary myotubes by enhancing protein translation via the TNF-R1, PI3K, and MEK. Am. J. Physiol. Endocrinol. Metab. 294, E241–E250 10.1152/ajpendo.00129.2007 [DOI] [PubMed] [Google Scholar]
- Rao P. K., Kumar R. M., Farkhondeh M., Baskerville S., Lodish H. F. (2006). Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA 103, 8721–8726 10.1073/pnas.0602831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C., Bodine S. C., Clarke B. A., Rossman R., Nunez L., Stitt T. N., Yancopoulos G. D., Glass D. J. (2001). Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3, 1009–1013 10.1038/ncb1101-1009 [DOI] [PubMed] [Google Scholar]
- Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004). Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 10.1016/S0092-8674(04)00400-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Shi Z. M., Wang J., Yan Z., You Y. P., Li C. Y., Qian X., Yin Y., Zhao P., Wang Y. Y., Wang X. F. et al. (2012). MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS ONE 7, e32709 10.1371/journal.pone.0032709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L., Gräfe A., Seiler A., Schumacher S., Nitsch R., Wulczyn F. G. (2005). Regulation of miRNA expression during neural cell specification. Eur. J. Neurosci. 21, 1469–1477 10.1111/j.1460-9568.2005.03978.x [DOI] [PubMed] [Google Scholar]
- Spencer M. J., Marino M. W., Winckler W. M. (2000). Altered pathological progression of diaphragm and quadriceps muscle in TNF-deficient, dystrophin-deficient mice. Neuromuscul. Disord. 10, 612–619 10.1016/S0960-8966(00)00160-7 [DOI] [PubMed] [Google Scholar]
- Stefani G., Slack F. J. (2008). Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 10.1038/nrm2347 [DOI] [PubMed] [Google Scholar]
- Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M. et al. (2008). Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 36, 2690–2699 10.1093/nar/gkn032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Ge Y., Drnevich J., Zhao Y., Band M., Chen J. (2010). Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J. Cell Biol. 189, 1157–1169 10.1083/jcb.200912093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L. B., Liu N., Williams A. H., McAnally J., Gerard R. D., Richardson J. A., Olson E. N. (2006). A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 103, 18255–18260 10.1073/pnas.0608791103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. (2007). Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 10.1126/science.1139089 [DOI] [PubMed] [Google Scholar]
- Venkataraman S., Alimova I., Fan R., Harris P., Foreman N., Vibhakar R. (2010). MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS ONE 5, e10748 10.1371/journal.pone.0010748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S., Kato Y., Okutsu M., Miyaki S., Suzuki K., Yan Z., Schiaffino S., Asahara H., Ushida T., Akimoto T. (2011). Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J. Biol. Chem. 286, 38456–38465 10.1074/jbc.M111.271270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. P., Diaugustine R. P., Zeringue E., Bunger M. K., Schmitt M., Archer T. K., Richards R. G. (2010). An IGF1/insulin receptor substrate-1 pathway stimulates a mitotic kinase (cdk1) in the uterine epithelium during the proliferative response to estradiol. J. Endocrinol. 207, 225–235 10.1677/JOE-10-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. L., Hulderman T., Jensen N., McKinstry M., Mishra M., Luster M. I., Simeonova P. P. (2002). Physiological role of tumor necrosis factor alpha in traumatic muscle injury. FASEB J. 16, 1630–1632 [DOI] [PubMed] [Google Scholar]
- Williams A. H., Liu N., van Rooij E., Olson E. N. (2009). MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 21, 461–469 10.1016/j.ceb.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. F., Tellam R. L. (2008). MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 283, 9836–9843 10.1074/jbc.M709614200 [DOI] [PubMed] [Google Scholar]
- Wright W. E. (1985). Myoblast senescence in muscular dystrophy. Exp. Cell Res. 157, 343–354 10.1016/0014-4827(85)90119-3 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Yamaguchi Y., Takino H., Matsuo H., Matsumoto K., Uotani S., Akazawa S., Yamashita S., Nagataki S. (1996). TNF-alpha stimulates glucose uptake in L6 myoblasts. Diabetes Res. Clin. Pract. 32, 11–18 10.1016/0168-8227(96)01221-1 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chao T., Li R., Liu W., Chen Y., Yan X., Gong Y., Yin B., Liu W., Qiang B. et al. (2009). MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J. Mol. Med. (Berl) 87, 43–51 10.1007/s00109-008-0403-6 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ying Z. Z., Tang Z. L., Long L. Q., Li K. (2012). MicroRNA-148a promotes myogenic differentiation by targeting the ROCK1 gene. J. Biol. Chem. 287, 21093–21101 10.1074/jbc.M111.330381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shyh-Chang N., Segrè A. V., Shinoda G., Shah S. P., Einhorn W. S., Takeuchi A., Engreitz J. M., Hagan J. P., Kharas M. G. et al. DIAGRAM Consortium; MAGIC Investigators(2011). The Lin28/let-7 axis regulates glucose metabolism. Cell 147, 81–94 10.1016/j.cell.2011.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.