Abstract
The knowledge of genetic diversity of tree crop is very important for breeding and improvement program for the purpose of improving the yield and quality of its produce. Genetic diversity study and analysis of genetic relationship among 20 Moringa oleifera were carried out with the aid of twelve primers from, random amplified polymorphic DNA marker. The seeds of twenty M. oleifera genotypes from various origins were collected and germinated and raised in nursery before transplanting to the field at University Agricultural Park (TPU). Genetic diversity parameter, such as Shannon's information index and expected heterozygosity, revealed the presence of high genetic divergence with value of 1.80 and 0.13 for Malaysian population and 0.30 and 0.19 for the international population, respectively. Mean of Nei's gene diversity index for the two populations was estimated to be 0.20. In addition, a dendrogram constructed, using UPGMA cluster analysis based on Nei's genetic distance, grouped the twenty M. oleifera into five distinct clusters. The study revealed a great extent of variation which is essential for successful breeding and improvement program. From this study, M. oleifera genotypes of wide genetic origin, such as T-01, T-06, M-01, and M-02, are recommended to be used as parent in future breeding program.
1. Introduction
Drumstick tree (Moringa oleifera Lam.), a short to medium height tree with luxurious evergreen leaves, was said to have originated from Himalayan tract in northwestern part of India [1–4]. The tree has a true diploid chromosome 2n = 28 with a distinguished tripinnate leaves having yellow or white petiole streaks [5, 6]. Moringa is potentially one of the planet's most valuable plants, at least in humanitarian terms [7] and has been regarded as a wonder tree due to its great economic importance and uses [3, 7]. Its pods were reported to have a protein content ranging from 20 to 30%, with a high vitamin C content. The moringa seeds were found to exhibit the property of natural coagulants/flocculants, which allows for growing of the tree for the purpose of usage by water and sewage treatment plant to clear turbidity in drinking water and sludge in sewage [8]. Similarly, the nutritive value of this plant for animals has been documented by Mendieta-Araica et al. [1], who reported that moringa contains large amount of crude protein, iron, zinc, and high concentration of vitamins A, B, and C in its foliage sample which makes it a very good feed and fodder for animals to browse and graze upon [9].
With respect to oil quality, M. oleifera seed concentrate contains about 35–45% seed oil, having odourless and colourless physical properties [10]. The edible oil is highly nutritious and is extracted by boiling the seeds with water and collecting the oil from the surface of the water [9, 11]. The seed oil has high concentration of oleic acid (>73%) coupled with low polyunsaturated fatty acid, which gives the oil an outstanding and remarkable oxidative stability properties. The suitability of M. oleifera seed oil as biodiesel feed source has been tested and recommended by Da Silva et al. [12], who reported that the oil could be used as pure biodiesel or petrodiesel mixture on engine after converting it to fatty acid methyl esters (FAME) through the process of transesterification in the presence of sodium hydroxide (NaOH) as catalyst.
Moreover, despite the great economic importance of this plant in terms of nutritional, social, and environmental benefits, the genetic diversity pattern, genetic makeup, and agronomical requirement needed for successful breeding and improvement, domestication, and large scale cultivation are yet to be established. This obstacle is an impediment to a successful production and commercialization of moringa and its related products [6]. Also, the knowledge of genetic diversity of tree crop is very important for rational planning of conventional, modern breeding, and improvement program for the purpose of improving the yield and quality of its produce [9, 13].
In other words, the use of molecular markers, such as inter-simple sequence repeat (ISSR), random amplified polymorphic DNA (RAPD), and simple sequence repeat (SSR), has gained popularity as a genetic diversity assessment methods of tree and oil seed crops [14–16]. Molecular methods of genetic diversity study are a fast, efficient, reliable, and simple means of establishing genetic diversity pattern in plant [17]. The RAPD as one of the numerous molecular markers has been reported to be a reliable, reproducible, cost effective, fast, and less tedious marker, which is widely used in the field of plant breeding and molecular genetics due to its outstanding quality [18].
Therefore, this research work will study the genetic diversity of twenty new genotypes of Moringa oleifera from two populations with the aim of studying genetic diversity pattern in relation to their geographical origin, and dissection of germplasms as a means of initiating the breeding programme in the nearest future.
2. Materials and Methods
2.1. Plant Materials
These were made up of seeds of twenty new genotypes of M. oleifera collected from six different countries (Table 1). The moringa genotypes prior to their collection were found in the wild growing in their natural form. The countries of origin are Virgin Island, USA, Thailand, India, Tanzania, Taiwan, and Malaysia. The collection was principally made by the Asia Vegetable Research and Development Center (AVRDC) or World Vegetable Centre, Taiwan (15 accessions classified as international population), and the Institute of Tropical Agriculture, universiti Putra Malaysia (5 accessions classified as Malaysian population). The collected seeds were germinated and raised in nursery, Universiti Putra Malaysia Agricultural Park (TPU) for two months, exposed to the hardening process in the last ten days of nursery, then transplanted out to the University's agricultural experimental farm in Puchong (02°N59.035′, 101°E38.913′), Selangor, Malaysia. Young and disease-free leaves of M. oleifera were collected for each of the genotypes during the early hours of the day; the leaves sample were wrapped in aluminum foil and labeled and kept in the freezer at −10°C.
Table 1.
List of moringa genotypes and their countries of origin.
| Serial number | Genotypes ID | Pedigree/cultivar name | Origin country |
|---|---|---|---|
| 1 | T01 | Virgin Islands Drum Stick | USA |
| 2 | T02 | Ma Rum01 | Thailand |
| 3 | T03 | Ma Rum02 | Thailand |
| 4 | T04 | Ma Rum03 | Thailand |
| 5 | T05 | Ma Rum04 | Thailand |
| 6 | T06 | Ma Rum05 | Thailand |
| 7 | T07 | Ma Rum Khaw Nheaw | Thailand |
| 8 | T08 | Ma Rum06 | Thailand |
| 9 | T09 | Ma Rum07 | Thailand |
| 10 | T10 | Ma Rum K | Thailand |
| 11 | T11 | Tnau-1 | India |
| 12 | T12 | Rca Moringa | Tanzania |
| 13 | T13 | Ma Rum C | Thailand |
| 14 | T14 | Drumstick Tree Pkm-1 | India |
| 15 | T15 | La-Mu W | Taiwan |
| 16 | M01 | ITA-UPM01 | Malaysia |
| 17 | M02 | ITA-UPM02 | Malaysia |
| 18 | M03 | ITA-UPM03 | Malaysia |
| 19 | M04 | ITA-UPM04 | Malaysia |
| 20 | M05 | ITA-UPM05 | Malaysia |
2.2. DNA Extraction Protocol
Six hundred μL of extraction buffer (100 mM of Tris-HCl, 20 mM of EDTA, 1.4 M NaCl, and 5% SDS) was added to 10 mg leaf sample and ground with mortar and pestle without liquid nitrogen according to Ferdous et al. [19]. The finely ground leaf tissue was transferred into 2 mL centrifuge tube. Four hundred μL of 2X CTAB solution, 100 mM of Tris-HCl, 20 mM of ethylenediaminetetraacetic acid di-sodium salt (EDTA), 1.4 M of sodium chloride (NaCl), 2% (w/v) CTAB, and 1% (w/v) of polyvinyl pyrrolidone (PVP) and 400 μL chloroform: isoamyl alcohol : phenol (24 : 1 : 5%) mixture were then added to the leaf tissue containing the extraction buffer. After mixing through vortex and centrifuge, the supernatant was transferred to another tube. Two-third of volume of isopropanol was added and incubated at room temperature for 10 to 15 min. Centrifuged supernatant was then removed and the DNA pellet was washed using 70% ethanol; afterwards the DNA pellet was air-dried and dissolved into 50 μL TE buffer.
2.3. DNA Quantification and Dilution
The quantification of DNA was carried out using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, USA). The DNA was again requantified by running it through 1% agarose gel electrophoresis with 1 × TAE buffer for 30 min and viewed under UV light after staining it with Midori green DNA stain (Nippon Genetics Inc., Germany). The dilution was done with sterile distilled water to ensure that all of the DNA samples have equal concentration of 100 ng/μL.
2.4. RAPD Polymerase Chain Reaction Procedure
According to the company instruction (Promega), 5 μL of 5X Green GoTaq Flexi Buffer, 3 μL MgCl2 solution (25 mM), 0.5 μL PCR nucleotide mix (10 mM each), 0.2 μL primers (0.4 μmol), and 1.0 U of Taq DNA polymerase were used for 25 μL of PCR reaction including 1 μL DNA template directly used after extraction [19]. In RAPD analysis, the following condition was used: initial denaturation at 94°C for 1 min followed by 45 cycles of denaturation done at 94°C for 1 min, annealing was done at 34°C for 1.5 min, and extension was done at 72°C for 2 min and a final extension at 72°C for 5 min [20]. The amplified PCR products were subjected to electrophoresis on 3% (w/v) MetaPhor agarose gel at 75 volt for 70 minutes. The gel was stained with ethidium bromide and visualized under ultraviolet (UV) light.
2.5. Band Scoring
The image of the gel acquired in JPEG format was imported into UVIdoc 99.02 for band scoring. The band sizes were estimated based on DNA ladder (Promega Inc.). The absence and presence of band were scored in a binary model of 0 and 1, respectively. Band scoring was carried out only on those bands that are clear and reproducible and then those that are >50 bp. The data obtained at the end of the scoring was transferred and saved in Microsoft excel sheet.
2.6. Data Analysis
Data of the twelve primers were analyzed to obtain the information on genetic diversity of the 20 moringa accessions (Table 2). Genetic similarity among the genotypes and principal component analysis (PCA) were calculated using NTSYS-pc 2.1. Cluster analysis was also carried out using the unweighted pair-group method with arithmetic average (UPGMA) based on the Nei's genetic distance matrix and dendrogram was drawn to show the clustering pattern of the different genotypes using NTSYS-pc. The percentage polymorphism of the bands (PPB), effective alleles (ne), genetic diversity index (h), Shannon's information index (I), and Nei's gene diversity were calculated using POPGEN 1.31 software. Analysis of molecular variance was conducted using GeneAIEx 6.5 to partition the variation present in the germplasm and at the same time test the variance component for RAPD phenotype.
Table 2.
RAPD polymorphic primers and their sequence ID.
| No. | Synthesis ID | Sequence | μg | nmol |
|---|---|---|---|---|
| 1 | OPA_17 | 5′-GAC CGC TTG T-3′ | 101 | 33.5 |
| 2 | OPA_19 | 5′-CAA ACG TCG G-3′ | 93 | 34.5 |
| 3 | OPB_17 | 5′-AGG GAA CGA G-3′ | 85 | 27.1 |
| 4 | OPBC_10 | 5′-AAC GTC GAG G-3′ | 160 | 52 |
| 5 | OPBD_18 | 5′-ACG CAC ACT C-3′ | 179 | 60.6 |
| 6 | OPBD_19 | 5′-GGT TCC TCT C-3′ | 181 | 61.1 |
| 7 | OPF_20 | 5′-GAG GAT CCC T-3′ | 95 | 31.3 |
| 8 | OPH_19 | 5′-CTG ACC AGC C-3′ | 100 | 33.7 |
| 9 | OPO_3 | 5′-CTG TTG CTA C-3′ | 103 | 34.6 |
| 10 | OPM_6 | 5′-CTG GGC AAC T-3′ | 100 | 32.9 |
| 11 | OPM_8 | 5′-TCT GTT CCC C-3′ | 110 | 37.5 |
| 12 | OPQ_2 | 5′-TCT GTC GGT C-3′ | 104 | 34.5 |
3. Results
3.1. Screening of Primers
A total of 24 RAPD primers were used to study the genetic diversity of twenty genotypes of M. oleifera (Figure 1). Out of these primers, only 12 showed as distinct, reproducible polymorphic bands. A total of 108 polymorphic fragments were generated by these 12 primers with an average of 9.0.
Figure 1.
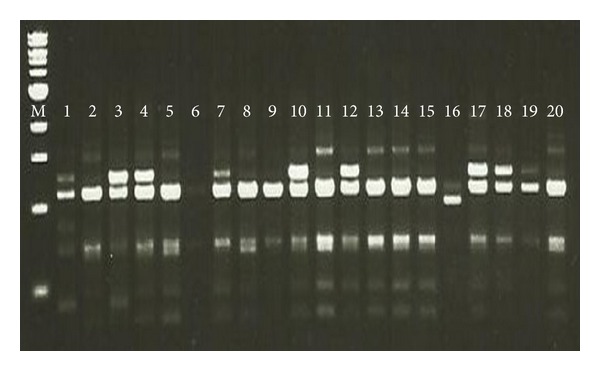
Gel picture of twenty genotypes of Moringa oleifera RAPD profile.
Genetic Diversity within the Two Populations. The mean percentage polymorphic loci in the two populations (international and Malaysian) were calculated to be 75.73 and 32.70, respectively (Table 3). The observed number of alleles in the two populations from Taiwan and Malaysia is 1.50 and 0.71, respectively with 1.26 as the mean value for effective alleles for the two countries. With the Hardy Weinberg equilibrium assumption in place, Shannon's information index and expected heterozygosity for Taiwan population were 0.3 and 0.19 and those of Malaysian population were 0.18 and 0.13, respectively, while mean Nei's gene diversity for the two populations were estimated to be 0.20.
Table 3.
Genetic diversity in M. oleifera germplasm as revealed by RAPD.
| Population | N | Na | Ne | I | He | P (%) | |
|---|---|---|---|---|---|---|---|
| International | 15.000 | 1.500 | 1.308 | 0.295 | 0.188 | 72.73 | |
| Malaysia | 5.000 | 0.709 | 1.220 | 0.184 | 0.125 | 32.73 | |
| Grand total | Mean | 10.000 | 1.105 | 1.264 | 0.239 | 0.156 | 52.73 |
| SE | 0.338 | 0.065 | 0.024 | 0.018 | 0.013 | 20.00 |
Furthermore, in order know the source of genetic variation for these Moringa genotypes, RAPD profile was analyzed using AMOVA. This was aimed to partition all the sources of variation existing in the germplasm into two major groups. The result revealed that 95% of the total genetic variation occurred as a result of variation within the population, while variation among the populations accounted for the remaining 5% of the total genetic variance (Table 4). Also the genetic variance among the population as indicated by the result (F st = 0.16) was significant at 5% probability level when permutation test was conducted.
Table 4.
Genetic divergence differentiation with analysis of molecular variance.
| Source | Degree of freedom | Sum square | Mean square | Estimated variance | variation (%) | F st | P value |
|---|---|---|---|---|---|---|---|
| Among population | 1 | 15.733 | 15.733 | 0.558 | 5 | 0.160 | <0.001 |
| Within population | 18 | 207.867 | 11.548 | 11.548 | 95 | <0.001 | |
|
| |||||||
| Total | 19 | 20 | 233.600 | 12.106 | 100 | ||
3.2. Cluster Analysis
Cluster analysis based on Jaccard's genetic similarity coefficient showed high level of genetic variation among the genotypes from the two countries. The similarity coefficient ranged from 0.38 to 0.89, with T-11 and T-15 genotypes found to have highest genetic similarity (0.89), while T-06 together with T-07 possessed least similarity coefficient (Figure 2).
Figure 2.
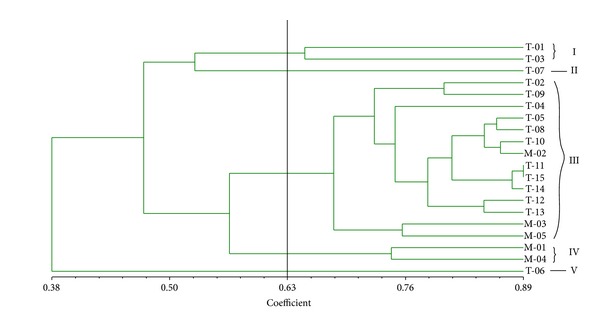
A dendrogram display of twenty accessions of Moringa oleifera.
In addition, a dendrogram was constructed using UPGMA cluster analysis to show the genetic relationship among the twenty genotypes from different geographical backgrounds. From this dendrogram, the twenty genotypes were grouped into five major clusters at a coefficient of 0.63. Cluster III, with highest number of members, had 14 genotypes, followed by cluster I (T-01 and T-03) and cluster IV (M-01 and M-04) with two genotypes each. Clusters II (T-07) and V (T-06) have one genotype each and were ranked the least populated clusters (Table 5).
Table 5.
Name of clusters and their corresponding genotypes.
| Cluster I | T-01, T-03 |
| Cluster II | T-07 |
| Cluster III | T-02, T-04, T-05, T-08, T-09, T-10, T-11, T-12, T-13, T-14, T-15, M-2, M-03, M-04 |
| Cluster IV | M-01, M-04 |
| Cluster V | T-06 |
3.3. Principal Component Analysis
The principal component analysis (PCA) carried out with RAPD M. oleifera profile classified the twenty genotypes of Moringa into five major groups with two genotypes from Malaysia occupying one group. This grouping pattern corresponds with that of clustering analysis as shown by the dendrogram. Dimension one of the PCA ranges from 0.49 to 0.92, while dimension two ranged from −0.65 to 0.26 and dimension three ranged from −0.65 to 0.68 (Figure 3).
Figure 3.

Three-dimensional principal components analysis of 20 accessions of Moringa oleifera.
4. Discussion
Effective and efficient genotyping of any plant species through RAPD requires a careful selection of suitable primer combination in order to get detail and informative result. High level of genetic polymorphism detected by these markers is in agreement with the assumption that outcrossing plant species from natural population will have higher level of genetic diversity when compared to in-breeding plant species. This finding agrees with earlier report on similar out-crossing plant species, such as Jatropha [23] and other oil plant species. High value of Shannon's information index (0.295) for international population as compared to Malaysian population (0.184) suggests that members of this population are more diverse. This is also obvious from the way the accessions cluster together.
Additionally, high level of genetic differentiation in these two populations as reflected by the genetic diversity parameters, such as Shannon's information index, expected heterozygosity, percentage polymorphism, and others, are pointing to the fact that there is wild variability in these populations of Moringa and this is very important for successful crossing and improvement programs in future. This observation follows a similar trend with the result of genetic diversity study on 75 accessions from Sudan and Guinea savanna zones of Nigeria, where six polymorphic primers of RAPD origin gave a total of 42 polymorphic bands [9].
Furthermore, interaction between various ecological and biological factors, such as genetic drift, gene flow, selection, and mating system, affects the genetic structure of any plant populations [24]. The overall genetic variability and differentiation pattern observed in these M. oleifera populations are in agreement with those of other outcrossing plant species [14, 25, 26]. As shown by these results, the two populations of M. oleifera exhibit moderate F st value in order to demonstrate low significant genetic differentiation among the populations. However, higher genetic differentiation and diversity were observed within the populations of M. oleifera and this indicates a relatively restricted variability as expected. This pattern of population structure has been previously reported in other out crossing plant species [27, 28].
Moreover, clustering analysis showing wide range of similarity coefficient showed that, there is high level of genetic variation within the two populations. The M. oleifera genotypes from the two populations were seen clustering together in the same group. This shows that there is no any distinct relationship between the geographical origin and the genetic distance as shown in the dendrogram. This finding implies that the genetic divergence within and between these two populations could not be explained by their geographical distance. This finding also means that isolation as a result of distance cannot be said to have been responsible for the divergence observed in these population [24].
In conclusion, these findings have proven that genetic divergence is very high in these populations and it can therefore be inferred from the data that the Moringa populations will be a very good germplasm material for the future breeding and improvement of this economically important tree crop. Genotypes that are far apart based on their genetic similarity coefficient (like T-01, T-06, M-01, and M-02) should be selected for future breeding.
Acknowledgments
The authors are grateful to Professor M. C. Palada and Dr. Andreas Ebert from the World Vegetable Centre (Asia Vegetable Research and Development Center), Taiwan, for the provision of germplasm. The authors also wish to express their sincere gratitude to the Research Management Unit (RMC) of Universiti Putra Malaysia (UPM) for the funding of this research, and to the management of Bayero University Kano, Nigeria for granting study fellowship to the SR.
References
- 1.Mendieta-Araica B, Spörndly E, Reyes-Sánchez N, Salmerón-Miranda F, Halling M. Biomass production and chemical composition of Moringa oleifera under different planting densities and levels of nitrogen fertilization. Agroforestry Systems. 2012;87(1):81–92. [Google Scholar]
- 2.Pandey A, Pradheep K, Gupta R, Roshini Nayar E, Bhandari DC. ‘Drumstick tree’ (Moringa oleifera Lam.): a multipurpose potential species in India. Genetic Resources and Crop Evolution. 2011;58(3):453–460. [Google Scholar]
- 3.von Maydell HJ. Trees and Shrubs of the Sahel: Their Characteristics and Uses. Verlag Josef Margraf; 1990. [Google Scholar]
- 4.Berger MR, Habs M, Jahn SAA, Schmahl D. Toxicological assessment of seeds from Moringa oleifera and Moringa stenopetala, two highly efficient primary coagulants for domestic water treatment of tropical raw waters. East African Medical Journal. 1984;61(9):712–716. [PubMed] [Google Scholar]
- 5.Ramachandran C, Peter KV, Gopalakrishnan P. (Moringa oleifera): economic. Botany. 1980;34:276–283. [Google Scholar]
- 6.Mgendi MG, Manoko MK, Nyomora AM. Genetic diversity between cultivated and non-cultivated Moringa oleifera Lam. provenances assessed by RAPD markers. Journal of Cell and Molecular Biology. 2010;8(2):95–102. [Google Scholar]
- 7.National Research Council. Lost Crops of Africa. Vol. 1. Washington, DC, USA: National Academy Press; 2006. [Google Scholar]
- 8.Madrona GS, Serpelloni GB, Vieira AMS, Nishi L, Cardoso KC, Bergamasco R. Study of the effect of saline solution on the extraction of the Moringa oleifera seed’s active component for water treatment. Water, Air, and Soil Pollution. 2010;211(1–4):409–415. [Google Scholar]
- 9.Abubakar BY, Wusirika R, MuA’zu S, Khan AU, Adamu AK. Detection of genetic variability using random amplified polymorphic DNA marker in some accession of Moringa oleifera Lam from Northern Nigeria. International Journal of Botany. 2011;7:237–242. [Google Scholar]
- 10.Ayerza (h) R. Seed and oil yields of Moringa oleifera variety Periyakalum-1 introduced for oil production in four ecosystems of South America. Industrial Crops and Products. 2012;36(1):70–73. [Google Scholar]
- 11.Somali MA, Bajneid MA, Al-Fhaimani SS. Chemical composition and characteristics of Moringa peregrina seeds and seeds oil. Journal of the American Oil Chemists’ Society. 1984;61(1):85–86. [Google Scholar]
- 12.Da Silva JPV, Serra TM, Gossmann M, Wolf CR, Meneghetti MR, Meneghetti SMP. Moringa oleifera oil: studies of characterization and biodiesel production. Biomass and Bioenergy. 2010;34(10):1527–1530. [Google Scholar]
- 13.Rafii MY, Arolu IW, Omar MHA, Latif MA. Genetic variation and heritability estimation in Jatropha curcas L. population for seed yield and vegetative traits. Journal of Medicinal Plants Research. 2012;6:2178–2183. [Google Scholar]
- 14.Rafii MY, Shabanimofrad M, Puteri Edaroyati MW, Latif MA. Analysis of the genetic diversity of physic nut, Jatropha curcas L. accessions using RAPD markers. Molecular Biology Reports. 2012;36(6):6505–6511. doi: 10.1007/s11033-012-1478-2. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Qin Y, Du G Yan Z. Genetic diversity and differentiation of the endangered tree Elaeagnus mollis Diels (Elaeagnus L.) as revealed by simple sequence repeat (SSR) markers. Biochemical Systematics and Ecology. 2012;40:25–33. [Google Scholar]
- 16.Tanya P, Taeprayoon P, Hadkam Y, Srinives P. Genetic diversity among Jatropha and Jatropha-related species based on ISSR markers. Plant Molecular Biology Reporter. 2011;29(1):252–264. [Google Scholar]
- 17.Abdelmigid HM. Efficiency of random amplified polymorphic DNA, (RAPD) and inter-simple sequence repeats (ISSR) markers for genotype fingerprinting and genetic diversity studies in canola (Brassica napus) African Journal of Biotechnology. 2012;11:6409–6419. [Google Scholar]
- 18.Bardakci F. Random amplified polymorphic DNA, (RAPD) markers. Turkish Journal of Biology. 2001;25:185–196. [Google Scholar]
- 19.Ferdous J, Hanafi MM, Rafii MY, Muhammad K. A quick DNA extraction protocol: without liquid nitrogen in ambient temperature. African Journal of Biotechnology. 2012;11(27):6956–6964. [Google Scholar]
- 20.Resmi DS, Celine VA, Rajamony L, Sony KB. Detection of genetic variability in Dramstick (Moringa oleifera Lam.) using RAPD markers. In: Keshvachandran R, Nazeem PA, editors. Recent Trends in Horticultural Biotechnology. New Delhi, India: New India Publishing Agency; 2007. pp. 587–592. [Google Scholar]
- 21.Kimura M, Crow JF. The number of alleles that can be maintained in a finite population. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewontine RC. Testing the theory of natural selection. Nature. 1972;236:181–182. [Google Scholar]
- 23.Arolu IW, Rafii MY, Hanafi MM, Mahmud TMM, Latif MA. Molecular characterization of Jatropha curcas germplasm using inter simple sequence repeat (ISSR) markers in Peninsular Malaysia. Australian Journal of Crop Science. 2012;6(12):1666–1673. [Google Scholar]
- 24.Verma S, Rana TS. Genetic diversity within and among the wild populations of Murraya koenigii (L.) Spreng., as revealed by ISSR analysis. Biochemical Systematics and Ecology. 2011;39(2):139–144. [Google Scholar]
- 25.El Hadj Ali IB, Guetat A, Boussaid M. Genetic diversity, population structure and relationships of Tunisian Thymus algeriensis Boiss. et Reut. and Thymus capitatus Hoffm. et link assessed by isozymes. Industrial Crops and Products. 2012;36(1):149–163. [Google Scholar]
- 26.Nejatzadeh-Barandozi F, Naghavi MR, Enferadi ST, Mousavi A, Mostofi Y, Hassani ME. Genetic diversity of accessions of Iranian Aloe vera based on horticultural traits and RAPD markers. Industrial Crops and Products. 2012;37(1):347–351. [Google Scholar]
- 27.Kumar S, Kumaria S, Sharma SK, Rao SR, Tandon P. Genetic diversity assessment of Jatropha curcas L. germplasm from Northeast India. Biomass and Bioenergy. 2011;35(7):3063–3070. [Google Scholar]
- 28.Parvaresh M, Talebi M, Sayed-Tabatabaei BE. Molecular diversity and genetic relationship of pomegranate (Punica granatum L.) genotypes using microsatellite markers. Scientia Horticulturae. 2012;138:244–252. [Google Scholar]


