Abstract
The RNA-dependent RNA polymerase of hepatitis C virus (HCV) is necessary for the replication of viral RNA and thus represents an attractive target for drug development. Several structural classes of nonnucleoside inhibitors (NNIs) of HCV RNA polymerase have been described, including a promising series of benzothiadiazine compounds that efficiently block replication of HCV subgenomic replicons in tissue culture. In this work we report the selection of replicons resistant to inhibition by the benzothiadiazine class of NNIs. Four different single mutations were identified in separate clones, and all four map to the RNA polymerase gene, validating the polymerase as the antiviral target of inhibition. The mutations (M414T, C451R, G558R, and H95R) render the HCV replicons resistant to inhibition by benzothiadiazines, though the mutant replicons remain sensitive to inhibition by other nucleoside and NNIs of the HCV RNA polymerase. Additionally, cross-resistance studies and synergistic inhibition of the enzyme by combinations of a benzimidazole and a benzothiadiazine indicate the existence of nonoverlapping binding sites for these two structural classes of inhibitors.
Hepatitis C virus (HCV) chronically infects about 3% of the human population, causing a slowly evolving liver disease that leads to cirrhosis, liver failure, and occasionally hepatocellular carcinoma (39). Given the size of the HCV epidemic and the limited efficacy of the present therapy based on alpha interferon (16), the development of new, safer, and more effective drugs is of paramount importance and is presently an area of intensive research. The strategy most widely applied for developing novel anti-HCV therapeutics aims at identifying small molecule inhibitors of viral enzymes. The nonstructural protein 5B RNA-dependent RNA polymerase (NS5B RdRp) is an important target of drug discovery activities largely because it is essential for viral replication and also due to the clinical successes of inhibitors of other viral polymerases. In addition, the extensive structural and biochemical characterization of this enzyme provides the basis for drug design efforts as well as for elucidating the mechanism of action of inhibitors and for rapidly optimizing their potency.
The NS5B protein was initially identified as an RdRp based on the presence of the signature GDD (Gly-Asp-Asp) motif characteristic for this class of enzymes (11). Its function was confirmed when an active form of the full-length protein was purified from baculovirus-infected insect cells (5). Subsequently, attempts to improve solubility, stability, and activity lead to the expression of C-terminal-truncated forms lacking the hydrophobic membrane anchor contained within the last 21 amino acids (1, 17, 25, 33, 35). In vitro, the enzyme shows little, if any, specificity for the HCV genome and can catalyze the synthesis of RNA by using a variety of homo- or heteropolymeric RNA templates both with and without a primer. In the absence of primer, NS5B can initiate RNA synthesis either by using the 3′-terminal OH group of the template as primer (5, 25) or by means of an authentic de novo initiation mechanism using a nucleotide complementary to the base at the 3′ end of the template as primer (29, 32, 41). It is generally believed that in infected cells HCV replication is initiated with a de novo mechanism that guarantees the maintenance of the genome integrity.
A significant advance in the understanding of the NS5B polymerase was provided by crystallographic studies of several truncated forms of the apoenzyme and of complexes with nucleotides or RNA template (1, 2, 6, 7, 24, 31). The NS5B polymerase has the canonical right-hand shape, with the characteristic fingers, palm, and thumb subdomains. In the crystal, the apoenzyme and the complexes with nucleoside triphosphates (NTPs) or RNA have a closed conformation due to the presence of two extended loops—the fingertips—that connect the fingers and thumb domains and completely encircle the active site cavity. Two structural elements peculiar to the NS5B polymerase structure are a β-hairpin, which protrudes from the thumb into the active site, and a C-terminal region, located immediately before the transmembrane domain, that folds from the surface of the thumb towards the active site and establishes a series of hydrophobic interactions with a shallow pocket located between palm and thumb subdomains (1). These elements are probably involved in proper positioning of the 3′ terminus of template RNA and are considered crucial for template selection. Indeed, although the overall conformation of the enzyme may be maintained during catalysis, local rearrangements of the β-hairpin and of the C-terminal region seem to be necessary to allow template access and, subsequently, to allow the extrusion of the nascent double-stranded RNA. Biochemical characterizations of deleted enzymes lacking either one of these domains have confirmed that they play an important role in modulating the initial steps of polymerization (1, 23). Deletion of these elements increased primer-dependent elongation activity by more than one order of magnitude and had only a minor effect on de novo initiation of RNA synthesis. These results suggested that the function of these elements is to discriminate against double-stranded RNA, thus favoring de novo initiation of RNA synthesis from the 3′ end of single-stranded template. Consistently, superposition of NS5B structures with and without the C-terminal extension shows that removal of the C-terminal extension results in a wider RNA binding cleft and an outward rotation of the thumb domain (1).
Several inhibitors of the NS5B RdRp have been reported in the scientific and patent literature that can be classified into three categories according to their chemical structure and putative mechanism of action: nucleoside analogues, pyrophosphate mimics, and NNIs (12, 13, 37). Among the substrate analogues, 2′-substituted nucleosides act as chain terminators and effectively inhibit replication of HCV subgenomic replicons (9). The pyrophosphate mimics include two classes of compounds, dihydroxypyrimidine carboxylic acids and diketoacid derivatives, which are believed to inhibit the RdRp activity through an interaction with the catalytic metal ions in the enzyme active site (S. Altamura, L. Tomei, J. O. Koch, P. J. S. Neuner, and V. Summa, 10 February 2000, Published International Patent Application, patent WO 00006529, and C. Gardelli, C. Giuliano, S. Harper, U. Koch, F. Narjes, J. M. Ontoria, M. Poma, S. Ponzi, I. Stansfield, and V. Summa, 24 January 2002, Pubished International Patent Application, patent WO 0206246).
Among the numerous nonnucleoside compounds documented to have anti-NS5B activity, two series of compounds have been shown to inhibit the NS5B polymerase allosterically by binding on the protein surface in a narrow cleft in the thumb domain (27, 38). In addition, two series of compounds based on a benzimidazole scaffold (3, 4, 20-22) and a series of benzothiadiazine derivatives (15) have been reported to inhibit NS5B, with a 50% inhibitory concentration (IC50) below 1 μM. Compounds of both the benzimidazole and the benzothiadiazine series were found to effectively inhibit replication of subgenomic replicons with no apparent cytotoxicity (14, 18, 34). Extensive biochemical characterization suggested that benzimidazole and benzothiadiazine derivatives inhibited the enzyme with an allosteric mechanism. Both classes have been shown to bind directly the NS5B polymerase and to inhibit RNA synthesis noncompetitively with respect to NTPs. Single-turnover experiments indicated that these two classes of compounds blocked the RdRp activity prior to the formation of an elongation complex (18, 34). Despite these similarities, it is not clear whether the mechanism of inhibition and/or the site(s) where these two classes of compounds bind on the enzyme are similar.
In this work we report an extensive investigation on the mode of action of benzothiadiazine inhibitors based on a combination of biochemical analysis and characterization of escape mutants. Several lines of experimental evidence, including binding studies, kinetic analysis, and cross-resistance studies, indicate that benzothiadiazine compounds inhibit the enzyme by interacting with a site distinct from that of benzimidazole analogues.
MATERIALS AND METHODS
Inhibitors.
Compound 1 [1-butyl-3-(1,1-dioxido-2H-1,2,4-benzothiadiazin-3-yl)-4-hydroxyquinolin-2(1H)-one] was purchased from Interbioscreen (Moscow, Russia), and compound 2 [3-(1,1-dioxido-2H-1,2,4-benzothiadiazin-3-yl)-4-hydroxy-1-isobutylquinolin-2(1H)-one] was purchased from Asinex (Moscow, Russia). Compound A (2-[4-({4′-chloro-4-[(4-hydroxypiperidin-1-yl) carbonyl]-1,1′-biphenyl-2-yl}methoxy)-2-fluorophenyl]-1-cyclohexyl-1H-benzimidazole-5-carboxylic acid) and compound B (N-{[1-cyclohexyl-2-(3-furyl)-1H-benzimidazol-5-yl]carbonyl}-5-hydroxy-l-tryptophan) were synthesized as described previously (3, 21).
Plasmids.
Plasmid pHCVNeo17.B (36) encodes an HCV replicon identical to I377neo/NS3-3′/wt (26) (EMBL-GenBank No. AJ242652) but containing two tissue culture-adaptive mutations, namely replacement of triplet GAA (nucleotides [nt] 2329 to 2331), coding for glutamic acid 176 in NS3, with GGA, coding for glycine, and insertion of an extra AAA triplet (coding for lysine) after the triplet GTG (nt 4840 to 4842), coding for valine 67 in NS5A. All other replicon plasmids are derived from pHCVNeo17.B and contain the following mutations: pHCVNeo17.BR3, replacement of triplet CAT (nt 7882 to 7884), coding for histidine 95 in NS5B, with triplet CGT (coding for arginine); plasmid pHCVNeo17.BR4, replacement of the triplet ATG (nt 8839 to 8841), coding for methionine 414 in NS5B, with ACG (coding for threonine); plasmid pHCVNeo17.BR5, replacement of triplet TCT (nt 8951 to 8953), coding for cysteine 451 in NS5B, with triplet CGT (coding for arginine); plasmid pHCVNeo17.BR6, replacement of triplet GGA (nt 9271 to 9273), coding for glycine 558 in NS5B, with triplet AGA (coding for arginine); plasmid pHCVNeo18.B, replacement of triplet GAG (nt 2096 of the neomycin phosphotransferase gene), coding for glutamate 182, with triplet TAG (coding for aspartate) (40). Plasmid pT7-NS5BΔC21 contains the HCV-BK sequence coding for NS5B protein lacking the 21 C-terminal residues (retaining residues 1 to 570) in the pT7-7 expression vector (8). pT7-NS5BΔC55 codes for NS5B protein lacking 55 C-terminal residues (retaining residues 1 to 536) (35). pT7-NS5BΔC21-ΔLoop codes for a NS5BΔC21 carrying a deletion of residues C445 to Y452 and substitutions D444G and S453G. Plasmids pT7-NS5BΔC21-H95R, pT7-NS5BΔC21-M414T, pT7-NS5BΔC21-C451R, and pT7-NS5BΔC21-G558R contain the same point mutations indicated in the corresponding replicon plasmids.
NS5B expression and purification.
Expression in Escherichia coli BL21(DE3) and purification of the HCV NS5B proteins were carried out as described previously (9).
Polymerase assays.
Primer elongation reactions were carried out in 50 μl of a solution of 20 mM Tris-HCl [pH 7.5], 0.01% Triton X-100, 10 mM NaCl, 1 mM dithiothreitol, 0.1 μg of bovine serum albumin/μl, 2 mM MgCl2, 10 μM UTP, 2 μCi of 3H-UTP (47 Ci/mmol; Amersham), 20 nM purified NS5B, and 0.5 μM poly(A) · oligo(U)18. Compounds were dissolved in 100% dimethyl sulfoxide (DMSO), and serial dilutions were made in DMSO. Unless otherwise specified, compounds, polymerase, and template RNA were incubated at room temperature for 60 min before the addition of NTPs. Elongation reactions were allowed to proceed for 30 min at RT, and the activity was measured as the radioactivity present in the acid-insoluble material.
De novo assays were performed in 50 μl of a solution of 15 mM Tris [pH 7], 20 mM MgCl2, 0.05% Triton X-100, 10 μg of BSA/ml, 1 mM DTT, 6 μM 3H-GTP (2 × 106 cpm; 7.8 Ci/mmol; Amersham), 2 mM GMP, 50 nM poly(C) (50-mer), and 50 nM HCV NS5BΔC21 enzyme. The enzyme was preincubated in reaction buffer at room temperature for 15 min with the compound and template RNA. The reaction was started by the addition of the 3H-GTP and GMP mixture and was incubated at room temperature for 60 min. The RNA products were precipitated by addition of 50 μl of 20% trichloroacetic acid (TCA), filtered on GF/B filter plates, and read by a Top Count liquid scintillation counter (Packard).
IC50 values were calculated by using a three-parameter logistic equation, and inhibition data were fitted by using Kaleidagraph software (Synergy Software, Reading, Pa.).
Kinetic parameters were calculated from a least-square fit of initial rates as a function of substrate concentration assuming Michaelis-Menten kinetics.
Polymerase-inhibitor interaction.
The polymerase-inhibitor complex was monitored essentially as already described (34). Polymerase (10 μM) and compound (10 μM) were mixed in 60 μl of incubation buffer containing 20 mM Tris-HCl (pH 7.5), 3 mM DTT, 100 mM NaCl, 0.03% n-octyl glucoside, 10% glycerol, 0.01% DMSO, 15 μM poly(A) · oligo(U)18. Following 30 min of incubation at room temperature the mixture was applied to a gel filtration G-25 spin column (Pharmacia) prewashed with the incubation buffer lacking the RNA. The eluate, containing the protein-inhibitor complex and the free protein, was recovered by centrifugation for 2 min at 1,450 × g. The eluting protein was quantified by Bradford assay (Bio-Rad), while the presence of the inhibitor was assessed by mass spectrometry, as described previously (34).
Tissue culture, replication analysis, selection, and sequencing of resistant replicons.
Huh-7, HBI10A, and NM15 cells were cultured and transfected by electroporation with in vitro-transcribed RNAs as previously described (36). NM15 cells were obtained by G418 selection of Huh-7 cells transfected with replicon pHCVNeo18.B, a modified replicon with a point mutation in the neomycin phosphotransferase protein that renders the enzyme less active, resulting in a tighter correlation between G418 resistance and replication level (L. Bartholomew, A. Cellucci, and G. Migliaccio, unpublished data). Transient transfection assays were performed by using cells highly competent for HCV replication, obtained by curing HBI10A cells of the endogenous replicons with human interferon-α2b as described previously (36). The effect of compounds on viral replication was monitored by Cell-enzyme-linked immunosorbent assay (ELISA) (36). Clones resistant to compounds 1 and 2 were selected as described previously (36). NM15 cells were plated in 15-cm-diameter tissue culture dishes at a density of 3 × 103/cm2 and were cultured in the presence of 0.8 mg of G418/ml and increasing concentrations of compounds. Approximately 15 days after beginning selection, small colonies of cells resistant to the inhibitor and the antibiotic became visible and were isolated. Replicon RNAs extracted from resistant clones were retrotranscribed and amplified by PCR, and their sequence was determined by direct automated sequencing of the PCR products (36).
RESULTS
Biochemical characterization of benzothiadiazine inhibitors.
NNIs of the NS5B polymerase based on a benzothiadiazine scaffold have been described in the scientific and patent literature (14, 15, 18). Based on a benzimidazole structure, benzothiadiazines inhibit the NS5B polymerase activity prior to the RNA elongation phase of the reaction in a manner similar to that of other NNI. To gain a better understanding of the mechanism of inhibition of this series, we selected two representative compounds and studied details of the inhibition of several enzyme variants by using different assay methods.
Effect of template RNA.
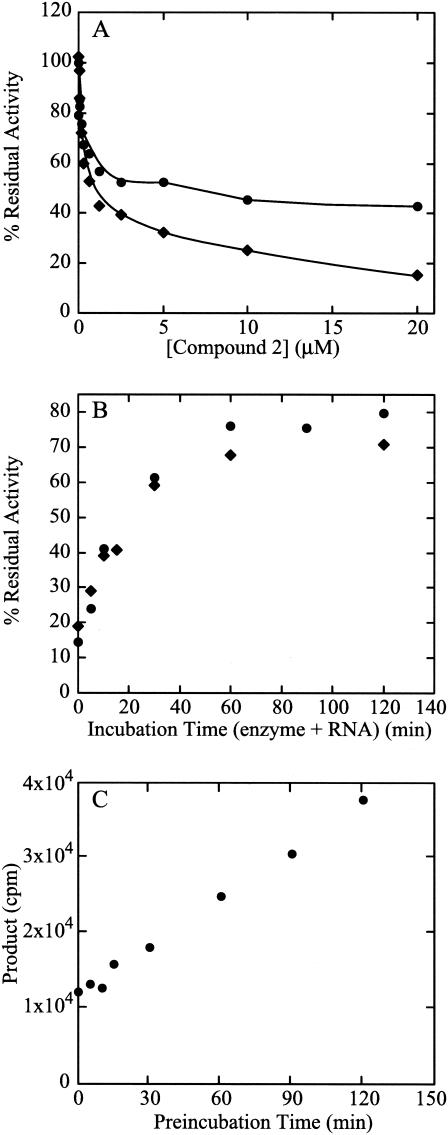
Benzothiadiazines have been reported to inhibit the NS5B polymerase with a mechanism that is noncompetitive with NTPs or RNA (14, 18). Changing the order of assay reagents did not affect inhibitor potency (18). However, by changing the order of reagent addition in the polymerase reaction, we observed that template RNA influenced the ability of compounds 1 (data not shown) and 2 (Fig. 1A) to cause complete inhibition of the ΔC21 enzyme activity. When inhibitors were added to the reaction prior to or together with the RNA template, complete inhibition was gradually reached at concentrations above 20 μM. Conversely, when the enzyme was preincubated with the poly(A) · oligo(U)18 template prior to inhibitor addition, inhibition curves were altered and a significant fraction of the polymerase activity was not inhibited even at very high compound concentrations (Fig. 1A). Similar results were obtained with the heteromeric RNA template t500 (8) (data not shown). In agreement with previous results (18), the IC50 for the inhibited fraction of the enzyme activity was not significantly changed by the preincubation with template. These observations were confirmed by varying the time of the preincubation of the RNA template and enzyme (Fig. 1B). The polymerase activity resistant to inhibition by 15 μM compound 1 or compound 2 increased as a function of enzyme-template preincubation time, suggesting that preincubation with template partially protected the enzyme from inhibition by both inhibitors. Enzyme activity in the absence of inhibitor also increased as preincubation time increased (Fig. 1C). In these experiments the inhibited fraction of the activity that remains after longer preincubations of enzyme and template likely results from those polymerase molecules that dissociated from the template during the reaction and were thus susceptible to inhibition. Indeed, in agreement with previous results (18), we could confirm that in the presence of heparin as a trapping reagent for free enzyme, preincubation with template completely protected the enzyme from inhibition (data not shown).
FIG. 1.
Effect of template RNA. (A) Order of addition. Increasing amounts of compound 2 (20 nM to 20 μM) were added to polymerase and to poly(A) · oligo(U)18 RNA that were (•) or were not (⧫) preincubated 20 min at room temperature. (B) Preincubation time course. The NS5BΔC21 and the poly(A) · oligo(U)18 RNA were preincubated from 0 to 120 min before the addition of 15 μM compound 1 (⧫) or 2 (•). (C) Effect of preincubation of template RNA and enzyme on enzyme activity. Polymerase activity is reported as the radioactivity incorporated in the absence of inhibitor compounds with increasing time of preincubation of enzyme and RNA.
Activity on different enzyme forms.
To further investigate the mechanism of inhibition of this series, we tested each inhibitor on different forms of the polymerase. Remarkably, compared to inhibition of the ΔC21 enzyme form, the inhibitors displayed at least one order of magnitude-reduced inhibitory potency with truncated enzymes lacking either the C-terminal 34 residues of the ΔC21 enzyme form (ΔC55) or the β-hairpin (ΔLoop), indicating that these elements were crucial for the inhibitory activity of benzothiadiazines (Table 1).
TABLE 1.
Structure and activity of benzothiadiazine compounds on different forms of the NS5B polymerase
Selection and characterization of resistant mutants.
The effect of compounds 1 and 2 on the replication of HCV subgenomic replicons was confirmed by using replicon clones HBI10A and NM15 and by monitoring expression of the NS3 protein by Cell-ELISA (36). Incubation of either clone with compounds 1 and 2 resulted in a dose-dependent reduction of the level of NS3 protein with IC50 values comparable to those reported previously (Table 2) (14). Similar results were obtained when viral replication was measured by monitoring expression of viral RNA by in situ RNase protection (data not shown) (9). Cytotoxicity assays and [14C]thymidine incorporation experiments showed that compounds 1 and 2 were not toxic and had no effect on cellular growth rate at concentrations up to 50 μM (data not shown), thus indicating that the decreased expression of viral RNA and proteins reflected a direct effect on viral replication.
TABLE 2.
Effect of replication inhibitors on parental and benzothiadiazine-resistant replicon clones
| Clonea | NS5B mutation | IC50 (μM) as determined by Cell-ELISAb
|
|||
|---|---|---|---|---|---|
| Compound 1 | Compound 2 | Compound A | 2′-C-methyl-adenosine | ||
| HB110A | 3.2 ± 1.1 | 9.8 ± 3 | 0.35 ± 0.05 | 0.32 ± 0.1 | |
| NM15 | 4.7 ± 1.6 | 11.4 ± 3.6 | 0.63 ± 0.22 | 0.39 ± 0.15 | |
| P2 | H95R | >20 | >50 | 0.28 ± 0.04 | 0.38 ± 0.08 |
| P6 | M414T | >20 | >50 | 0.25 ± 0.16 | 0.51 ± 0.11 |
| P13 | H95R | >20 | >50 | Not tested | Not tested |
| P17 | C451R | >20 | >50 | 0.13 ± 0.02 | 0.29 ± 0.12 |
| P20 | G558R | >20 | >50 | 0.2 ± 0.12 | 0.28 ± 0.12 |
| Q13 | H95R | >20 | >50 | 0.23 ± 0.11 | 0.38 ± 0.15 |
| Q14 | H95R | >20 | >50 | 0.24 ± 0.1 | 0.75 ± 0.3 |
P clones were selected with compound 1, and Q clones were selected with compound 2.
Data are average of at least duplicate determinations.
To obtain definitive evidence of the link between enzyme inhibition and antiviral effect, we selected and characterized replicons resistant to compounds 1 and 2 by adopting a strategy already used for selecting mutants resistant to other replication inhibitors (30, 34, 36). Cell clones resistant to both compounds were obtained by culturing NM15 cells in the presence of G418 and either compound at concentrations increasing from 4- to 10-fold (compound 2) or 4- to 20-fold (compound 1) higher than their respective IC50. Resistance selection experiments yielded several clones resistant to both compounds 1 and 2 that maintained the same rate of cell doubling as the parental cells and expressed HCV RNA and proteins at comparable levels (data not shown). Independently of the compound used for selection, replication of the cognate replicons was resistant to both inhibitors: selected clones displayed IC50 values more than fivefold higher than those of parental cells (Table 2). Unexpectedly, incubation of cells with compounds 1 and 2 increased in a dose-dependent fashion the expression of the NS3 protein in clones P17 and P20 but not in the other clones, suggesting that the compounds activated rather than inhibited replication of the cognate replicons (data not shown). These clones were still sensitive to inhibition by alpha interferon (19), the nucleoside inhibitor 2′-C-methyl-adenosine (9), and the benzimidazole-based nonnucleoside inhibitor compound A (34), demonstrating that resistance was specific for benzothiadiazine-based compounds (Table 2 and data not shown). Correspondingly, replicon clones selected with 2′-C-methyl-adenosine and with compound A remained sensitive to inhibition by the two benzothiadiazine inhibitors (9, 34 and data not shown).
To ascertain whether resistance was due to replicon mutations, we determined the sequences of the NS5B coding region of replicons extracted from resistant clones and compared them to that of replicons extracted from parental NM15 cells (Table 2). Remarkably, all replicons derived from resistant clones contained a single amino acid mutation in the NS5B polymerase. Four clones (P2, P13, Q13, and Q14) contained replacement of histidine 95 with arginine (H95R), while mutations in the other clones were different and included the following replacements: methionine 414 with threonine (M414T), cysteine 451 with arginine (C451R), and glycine 558 with arginine (G558R).
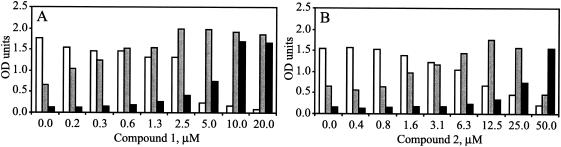
The role of these mutations was investigated by segregating them in a replicon vector containing the adaptive mutations present in the parental replicon (36) and testing the resulting replicons in transient transfection assays. As expected, the parental replicon (B) was inhibited by both compounds with IC50 values comparable to those observed for HBI10A and NM15 cells (Table 3). In contrast, replicons containing NS5B substitutions H95R and M414T replicated at levels comparable to the control replicon and were clearly resistant to both compounds. The IC50 values measured for these replicons were similar to those observed in the resistant cells, indicating that each of the tested substitutions was by itself sufficient to recapitulate the resistance phenotype displayed by the corresponding clones. Consistent with the results obtained with clones P17 and P20, the replicons containing the C451R or the G558R mutation replicated to low or undetectable levels in the absence of compounds, respectively, but their replication was stimulated in a dose-dependent fashion by both compounds 1 and 2. At high compound concentrations replication was comparable to that of the control replicon (Fig. 2). A much less pronounced activation was also observed with the H95R mutant replicon (data not shown). As expected, the replicons containing NS5B substitutions H95R, M414T, and C451R remained sensitive to inhibition by the benzimidazole-based inhibitor compound A, confirming that these mutations were specifically responsible for resistance to benzothiadiazine inhibitors (Table 3). The effect of compound 1 on the G558R replicon could not be estimated because of the inefficient replication of this mutant in the absence of benzothiadiazine compounds.
TABLE 3.
Replication efficiency and sensitivity to inhibitors of mutant pHCVNEO17 replicons
| Clone | NS5B mutation | Replication efficiencya | IC50 (μM) for:
|
||
|---|---|---|---|---|---|
| Compound 1 | Compound 2 | Compound A | |||
| B | 19 | 1.9 ± 0.4 | 11.5 ± 1 | 0.37 | |
| R3 | H95R | 17.9 | >20 | >50 | 0.2 |
| R4 | M414T | 12 | >20 | >50 | 0.32 |
| R5 | C451R | 5-18b | >20b | >50b | 0.1 |
| R6 | G558R | 2-15b | >20b | >50b | NAc |
Replication efficiency and replicon IC50 were determined by Cell-ELISA. Replication efficiency is expressed in arbitrary units.
Replication efficiency increased in the presence of compounds in a dose-dependent fashion (see Fig. 3).
NA, not applicable.
FIG. 2.
Benzothiadiazine compounds stimulate replication of C451R and G558R mutant replicons. 10AIFN cells were transfected with control (white), C451R (gray), and G558R (black) replicons and were cultured in the absence or in the presence of the indicated concentrations of either compound 1 (A) or compound 2 (B). HCV replication was monitored by cell-ELISA as indicated in Materials and Methods.
Characterization of mutant polymerases.
To confirm that the above mutations were sufficient for resistance, we expressed and purified recombinant ΔC21 polymerases containing the H95R, M414T, C451R, and G558R mutations and compared them to the wild-type ΔC21 enzyme. All the mutant polymerases displayed catalytic efficiencies greater than that of the wild-type enzyme both in the primer elongation (Table 4) and in the de novo reaction (data not shown). The increased catalytic efficiency in each case was primarily the result of an increase in kcat, because Km values for NTP and for the RNA template were unchanged (Table 4). Unexpectedly, the purified mutant polymerases exhibited different responses to inhibition depending on the individual mutation. While the M414T mutation conferred a reduced susceptibility to inhibition by both compound 1 and 2 (and was therefore confirmed as sufficient for resistance to both inhibitors), the H95R substitution did not protect the purified enzyme against inhibition. Moreover, the protective effect of both C451R and G558R mutations was marginal and was mainly evident in de novo assays. Interestingly, in the de novo reaction both compounds 1 and 2 inhibited the wild-type ΔC21 polymerase with IC50 values 5- to 10-fold lower than those measured in the primer elongation assay.
TABLE 4.
Catalytic efficiency and sensitivity to benzothiadiazine compounds of mutant enzymes
| Protein | Km UTP (μM) | Km RNAa (nM) | kcat (min−1) | IC50 (μM) of compound in:
|
|||
|---|---|---|---|---|---|---|---|
| Primer elongation
|
de novo reaction
|
||||||
| Compound 1 | Compound 2 | Compound 1 | Compound 2 | ||||
| ΔC21 | 10.8 ± 2 | 32 ± 6 | 0.43 ± 0.03 | 0.2 ± 0.1 | 0.3 ± 0.1 | 0.017 ± 0.02 | 0.06 ± 0.03 |
| ΔC21-H95R | 10 ± 1.5 | 25 ± 5 | 2.8 ± 0.60 | 0.04 ± 0.01 | 0.035 ± 0.02 | 0.018 ± 0.02 | 0.085 ± 0.04 |
| ΔC21-M414T | 8.3 ± 0.7 | 35 ± 3 | 1.85 ± 0.25 | 2.2 ± 0.3 | 5.9 ± 0.8 | 1 ± 0.02 | 3 ± 0.05 |
| ΔC21-C451R | 11.3 ± 1.3 | 44 ± 8 | 6.2 ± 0.10 | 0.05 ± 0.02 | 1 ± 0.05 | 0.1 ± 0.02 | 0.2 ± 0.02 |
| ΔC21-G558R | 11.6 ± 1.3 | 45 ± 3 | 5.15 ± 0.15 | 0.27 ± 0.03 | 0.95 ± 0.09 | 0.1 ± 0.01 | 0.5 ± 0.03 |
RNA is poly(A)·oligo(U)18.
To further investigate the mechanism of resistance we utilized a direct binding assay based on separation of the polymerase-inhibitor complex from the unbound inhibitor by gel filtration chromatography. In agreement with previous results (14), this assay confirmed that compound 1 bound the ΔC21 enzyme both in the absence and in the presence of template RNA (Table 5). In line with the reduced inhibitory potency measured in activity assays, binding of compound 1 to the ΔC55 form of the polymerase was not observed. As expected on the basis of the significant decrease of inhibition efficacy, mutant M414T was unable to interact with the compound both in the absence and in the presence of RNA (Table 5). In contrast, interaction of compound 1 with the H95R, C451R, and G558R mutants was still observed, partially explaining the modest effect of these substitutions on inhibitor potency and the enhancement of replication of mutant replicons, which suggests that these mutations do not abolish inhibitor binding capability. Due to the low solubility of compound 1 in aqueous buffers, the compound eluted with the each mutant enzyme could not be estimated with accuracy sufficient to evaluate binding stoichiometry. Nevertheless, the differences measured by liquid chromatography-mass spectrometry analysis were of amplitudes sufficient to permit an unequivocal evaluation of the binding ability of the different mutant enzymes (data not shown).
TABLE 5.
Binding capability of compound 1 to wild-type and mutant polymerases
| Protein | Compound 1 binding |
|---|---|
| ΔC55 | − |
| ΔC55a | − |
| ΔC21 | + |
| ΔC21a | + |
| ΔC21-M414T | − |
| ΔC21-M414Ta | − |
| ΔC21-C451R | + |
| ΔC21-C451Ra | + |
| ΔC21-G558R | + |
| ΔC21-G558Ra | + |
| ΔC21-H95R | + |
| ΔC21-H95Ra | + |
| ΔC21 + compound Ba | + |
Protein plus poly(A)·oligo(U)18.
Synergistic inhibition.
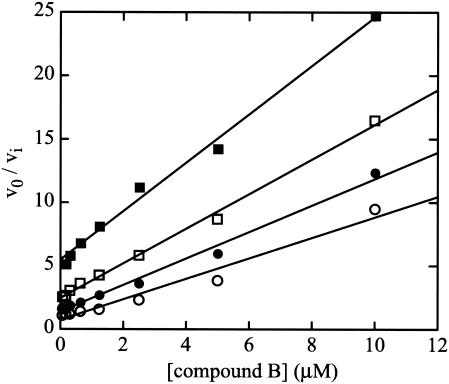
The potential for synergistic inhibition of enzyme activity by combinations of nonnucleoside inhibitors was assessed in enzyme-catalyzed kinetic reactions. As shown in Fig. 3, synergistic inhibition of the wild-type enzyme is evident in reaction mixtures containing combinations of a benzimidazole (compound B) and a benzothiadiazine (compound 1), as demonstrated by the nonparallel lines in the Yonetani-Theorell plots. Synergistic inhibition of enzyme activity can result from the simultaneous binding of two inhibitors and occurs in reactions measuring either primer-dependent (data not shown) or de novo RNA synthesis (Fig. 3). The possibility of simultaneous binding of the two compounds was further verified by binding competition experiments using compound 1 and a saturating concentration of compound B (100 μM). The experiment was performed only in the presence of RNA because of the inability of compound B to interact with the enzyme in the absence of RNA (34). Binding of compound 1 to the ΔC21 enzyme was still observed even in the presence of high molar excess of compound B, further strengthening the conclusion that the two compounds bind the enzyme at different sites (Table 5).
FIG. 3.
Synergistic inhibition of polymerase activity by combinations of a benzothiadiazine and a benzimidazole. Inhibition by the benzimidazole, compound B, of the catalytic activity of ΔC21 in assays following de novo initiation of synthesis was determined in the absence (○) or the presence of 0.015 (•), 0.03 (□), or 0.06 (▪) μM compound 1. Data are plotted as the ratio of reaction rates in the absence (v0) and in the presence (vi) of inhibitor(s). The nonparallel best-fit lines indicate synergistic inhibition by the combination of inhibitors. A similar pattern of lines was evident for inhibition of primer-dependent assays of polymerase activity in the presence of combinations of compound 1 and compound B.
DISCUSSION
The identification of several structural classes of NNIs of HCV NS5B polymerase that are active in the cell-based replicon assay has previously been disclosed (3, 14, 21, 27, 38). Representative compounds from these structural classes have been shown to be noncompetitive with the NTP substrate, suggesting that their binding site(s) on the polymerase does not overlap the enzyme active site. Indeed, crystallographic data establish that the binding site of two classes of NNIs, phenylalanine derivatives and a dihydropyranone, lies at the base of the thumb region of the polymerase distal to the active site (28, 38). Additionally, substitution of proline 495 of NS5B polymerase with leucine or alanine (P495L/A) renders both the enzyme and the corresponding replicon resistant to inhibition by a third class of NNI based on a benzimidazole scaffold, supporting the existence of another binding site for this class of compounds (34). In this paper the mode of action of benzothiadizine inhibitors and their binding site on the HCV polymerase were explored by a combination of biochemical and genetic analyses, with particular emphasis on the comparison with benzimidazole-based inhibitors.
Basic similarities exist in the mechanism of inhibition by benzothiadiazine and benzimidazole compounds. Experiments to determine the mechanism of inhibition have indicated that benzimidazoles (34) and benzothiadiazines (18) inhibit the enzyme prior to the elongation phase of the reaction. In addition, results presented in this and in previous work (34) indicated that the activity of both classes of inhibitors is affected by the presence of the template RNA. Indeed, as in the case of benzimidazoles, enzyme activity resistant to benzothiadiazines increased as the time of preincubation of enzyme and template increased, suggesting that the binding of the template protected the enzyme from inhibition in the absence of catalysis and that this protection occurred relatively slowly. Enzyme activity also increased as a result of the preincubation with template, suggesting that the protection against inhibition and increase in activity may have resulted from the same or a related change. However, binding of the template does not prevent benzimidazoles (34) or benzothiadiazines from binding to the enzyme, as shown in the direct compound binding experiments. Thus, the model that emerges from these data are one where template or inhibitor binding influences the distribution of enzyme between an active and an inactive enzyme conformation, that this conformational change occurs slowly and prior to NTP binding and catalysis.
However, several differences in the inhibition by benzimidazoles and benzothiadiazines are apparent. Benzimidazoles are equally effective inhibitors of the ΔC55 and ΔC21 forms of the enzyme, whereas benzothiadiazines are much more potent inhibitors of the ΔC21 polymerase. In addition, synergistic inhibition by their combination, highlighted by the nonparallel lines in the Yonetani-Theorell plot, and the lack of competition in the direct binding experiment indicate the ability of both inhibitors to bind simultaneously to the enzyme, supporting the existence of separate binding sites. Moreover, increasing concentrations of GTP abrogate inhibition by benzimidazoles (34) but do not have the same effect on the inhibition by benzothiadiazines. Lastly, the resistance substitutions for the two classes of compounds are different, providing additional evidence that they do not interact with the enzyme at the same site.
Replicons resistant to inhibition by benzothiadiazines were selected in the presence of compound and the mutations were mapped to the polymerase, genetically validating the target of benzothiadiazines. Four different single mutations were identified that lie in different regions of the enzyme (Fig. 4). M414T is in an alpha-helical region of the thumb domain, with its side chain directed into the active site. C451R is in the β-hairpin; H95R is in the fingers and may interact with the bound template; G558R is close to the C-terminal end of the ΔC21 enzyme. The fact that individual mutations located in different regions of the enzyme give rise to resistance suggests there may be multiple mechanisms for resistance. This interpretation is reinforced by the observation that the replicons containing each of these mutations display different replication and resistance phenotypes and that the corresponding recombinant polymerases show different degrees of sensitivity to the inhibitors.
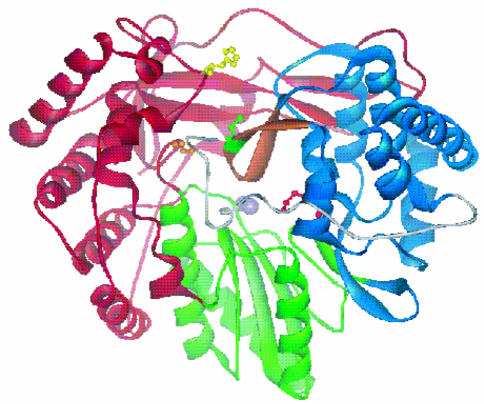
FIG. 4.
Position of the sites of resistance mutations within the structure of ΔC21 polymerase. The fingers region of the enzyme is shown in red, the palm in green, and the thumb in blue. The β-hairpin is shown in orange. The metal ions that are required for activity are represented by the gray spheres. The 34-residue C-terminal region of the ΔC21 enzyme form is colored in gray. The side chains of amino acids H95 (yellow), M414 (red), C451 (green), and G558 (orange) are also shown. The structural coordinates are from Lesburg et al. (24).
Replicons containing M414T and H95R showed a similar efficiency of replication compared to that of the wild type and were completely resistant to inhibition by benzothiadiazines. Surprisingly, replicons containing C451R and G558R showed significant levels of replication only in the presence of benzothiadiazines, suggesting that within the context of the replication complex the active conformation of the polymerase was achieved only in the presence of the compound.
Biochemical data from studies of the purified M414T mutant enzyme demonstrate that it is 50-fold more resistant to inhibition by either benzothiadiazine in the assay measuring RNA synthesis following de novo initiation. These results biochemically confirm the target of the inhibition and suggest that the primary mechanism of resistance of M414T is a reduction in the binding affinity of the benzothiadiazines. This interpretation was confirmed by assays assessing the direct binding of the compound to the M414T mutant enzyme that indirectly suggest that methionine 414 is part of the binding site for benzothiadiazine compounds. Additionally, the M414T polymerase displays an increased catalytic efficiency compared to that of the wild-type enzyme, as do the other resistant mutants in this study. Since neither Km for NTPs nor for the RNA template is significantly different for the mutants than it is for the wild-type enzyme, the increased catalytic efficiency of these mutants could be due to a higher amount of active protein that can productively interact with RNA and/or to a faster rate of the conformational change leading to a productive polymerase-RNA complex.
At variance with the M414T polymerase, the enzymes containing the other three resistance mutations show a complex pattern of sensitivity to the benzothiadiazine inhibitors. The enzymes containing either the C451R or the G558R mutations were not clearly resistant to inhibition in the primer elongation assay but showed a moderate level of resistance in the de novo assay. The location of these mutations in the β-hairpin (C451R) and C-terminal region near the β-hairpin (G558R) is consistent with the results of deletion mutants that demonstrate that these regions are important determinants for inhibition by benzothiadiazines. Both mutations might have a role in modulating the movement of the β-hairpin and/or C-terminal region in a way that leads to decreased inhibition by benzothiadiazines as well as increased catalytic activity. This altered movement or position of the β-hairpin/C-terminal region might explain why replicons containing C451R or G558R do not replicate efficiently in the absence of compound. Replicons containing NS5B with the β-hairpin deleted also replicate poorly, though the corresponding enzyme is more active than the wild-type enzyme in both primer-dependent and de novo initiation assays (10). The higher inhibitory potency, together with the more obvious effect of resistance mutations, in the de novo rather than in primer elongation assays, might indicate the de novo initiation of RNA synthesis as the primary target of inhibition by benzothiadiazine compounds.
Surprisingly, the H95R mutant enzyme was as sensitive or more sensitive than the wild type to inhibition by benzothiadiazines, making it difficult to rationalize the resistant phenotype observed with the corresponding mutant replicon. It is conceivable that some of the functions of the HCV polymerase in the replication complex, such as specific recognition of the viral genome and interaction with other factors which might modulate its activities, are not entirely reproduced in the in vitro assays, thus limiting the biochemical dissection of the mutations’ effect.
Interestingly, alignment of the sequences of full-length HCV genomes available in the public databases revealed a different degree of conservation of the NS5B residues involved in resistance to benzothiadiazine inhibitors. In fact, while histidine 95 and glycine 558 are highly conserved (>99% and 96%, respectively), the other two residues are much more variable. The amino acid at position 414 is methionine in 86% of the natural isolates and is replaced by glutamine in genotype 2 isolates. The amino acid at position 451 is even less conserved and is cysteine only in 52% of the natural viruses and is replaced by valine in genotype 2 isolates and threonine, isoleucine, tyrosine, or histidine in viruses belonging to other genotypes. Although the effects of these natural variations on the activity of benzothiadiazine compounds have not been investigated, their occurrence suggests that some HCV isolates might be resistant to this class of inhibitors.
The discovery of NNIs of HCV RNA polymerase that are active in the cell-based replicon assay has created the possibility of developing direct inhibitors of polymerase activity as therapies for HCV infection. A greater understanding of the mechanism of inhibition would assist in the design of more potent compounds with the required pharmacokinetic and safety parameters to justify clinical investigation of their efficacy.
Acknowledgments
We are grateful to Michael Rowley and Frank Narjes for continuous helpful discussions and suggestions.
REFERENCES
- 1.Adachi, T., H. Ago, N. Habuka, K. Okuda, M. Komatsu, S. Ikeda, and K. Yatsunami. 2002. The essential role of C-terminal residues in regulating the activity of hepatitis C virus RNA-dependent RNA polymerase. Biochim. Biophys. Acta 1601:38-48. [DOI] [PubMed] [Google Scholar]
- 2.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 3.Beaulieu, P. L., G. Fazal, J. Gillard, G. Kukolj, and V. Austel. January2002. Preparation of benzimidazolecarboxylates and related compounds as viral polymerase inhibitors. WO patent 0204425.
- 4.Beaulieu, P. L., G. Fazal, S. Goulet, G. Kukolj, M. Poirier, and Y. S. Tsantrizos. January2003. Preparation of benzimidazoles as viral polymerase inhibitors. WO patent 0307945.
- 5.Behrens, S. E., L. Tomei, and R. De Francesco. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15:12-22. [PMC free article] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroll, S. S., V. Sardana, Z. Yang, A. R. Jacobs, C. Mizenko, D. Hall, L. Hill, J. Zugay-Murphy, and L. C. Kuo. 2000. Only a small fraction of purified hepatitis C RNA-dependent RNA polymerase is catalytically competent: implications for viral replication and in vitro assays. Biochemistry 39:8243-8249. [DOI] [PubMed] [Google Scholar]
- 9.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 10.Cheney, I. W., S. Naim, V. C. Lai, S. Dempsey, D. Bellows, M. P. Walker, J. H. Shim, N. Horscroft, Z. Hong, and W. Zhong. 2002. Mutations in NS5B polymerase of hepatitis C virus: impacts on in vitro enzymatic activity and viral RNA replication in the subgenomic replicon cell culture. Virology 297:298-306. [DOI] [PubMed] [Google Scholar]
- 11.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 12.De Francesco, R., and C. M. Rice. 2003. New therapies on the horizon for hepatitis C: are we close? Clin. Liver Dis. 7:211-242. [DOI] [PubMed] [Google Scholar]
- 13.De Francesco, R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antivir. Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 14.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 15.Dhanak, D., A. C. Kaura, and A. Shaw. November2001. Preparation of 3-(1,1-dioxo-2H-benzo-1,2,4-thiadiazin-3-yl)-2-quinolones as novel anti-infectives. WO patent 0185172.
- 16.Di Bisceglie, A. M., and J. H. Hoofnagle. 2002. Optimal therapy of hepatitis C. Hepatology 36:S121-S127. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu, B., V. K. Johnston, L. L. Gutshall, T. T. Nguyen, R. R. Gontarek, M. G. Darcy, R. Tedesco, D. Dhanak, K. J. Duffy, C. C. Kao, and R. T. Sarisky. 2003. Arresting initiation of hepatitis C virus RNA synthesis using heterocyclic derivatives. J. Biol. Chem. 278:16602-16607. [DOI] [PubMed] [Google Scholar]
- 19.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hashimoto, H., K. Mizutani, and A. Yoshida. January2003. Fused cyclic compounds and medicinal use thereof. WO patent 0300254.
- 21.Hashimoto, H., K. Mizutani, and A. Yoshida. July2001. Preparation of heterocyclic compounds as remedies for hepatitis C. WO patent 0147883.
- 22.Hashimoto, H., K. Mizutani, and A. Yoshida. March2003. Preparation of substituted 1-cyclohexyl-2-phenylbenzimidazole-5-carboxylic acids as remedies for hepatitis C. U.S. patent 2,003,050,320.
- 23.Hong, Z., C. E. Cameron, M. P. Walker, C. Castro, N. Yao, J. Y. Lau, and W. Zhong. 2001. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285:6-11. [DOI] [PubMed] [Google Scholar]
- 24.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Love, R. A., H. E. Parge, X. Yu, M. J. Hickey, W. Diehl, J. Gao, H. Wriggers, A. Ekker, L. Wang, J. A. Thomson, P. S. Dragovich, and S. A. Fuhrman. 2003. Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme. J. Virol. 77:7575-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love, R. A., X. Yu, W. Diehl, M. J. Hickey, H. E. Parge, J. Gao, and S. Fuhrman. November2002. Crystallization properties and structure of hepatitis C virus (HCV) NS5B RNA polymerase and its mutants, and uses in drug screening. EP patent 1256628.
- 29.Luo, G., R. K. Hamatake, D. M. Mathis, J. Racela, K. L. Rigat, J. Lemm, and R. J. Colonno. 2000. De novo initiation of RNA synthesis by the RNA-dependent RNA polymerase (NS5B) of hepatitis C virus. J. Virol. 74:851-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. Characterization of resistance to non-obligate chain terminating ribonucleoside analogs which inhibit HCV replication in vitro. J. Biol. Chem., in press. [DOI] [PubMed]
- 31.O'Farrell, D., R. Trowbridge, D. Rowlands, and J. Jager. 2003. Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation. J. Mol. Biol. 326:1025-1035. [DOI] [PubMed] [Google Scholar]
- 32.Oh, J. W., T. Ito, and M. M. Lai. 1999. A recombinant hepatitis C virus RNA-dependent RNA polymerase capable of copying the full-length viral RNA. J. Virol. 73:7694-7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt-Mende, J., E. Bieck, T. Hugle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 34.Tomei, L., S. Altamura, L. Bartholomew, A. Biroccio, A. Ceccacci, L. Pacini, F. Narjes, N. Gennari, M. Bisbocci, I. Incitti, L. Orsatti, S. Harper, I. Stansfield, M. Rowley, R. De Francesco, and G. Migliaccio. 2003. Mechanism of action and antiviral activity of benzimidazole-based allosteric inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 77:13225-13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomei, L., R. L. Vitale, I. Incitti, S. Serafini, S. Altamura, A. Vitelli, and R. De Francesco. 2000. Biochemical characterization of a hepatitis C virus RNA-dependent RNA polymerase mutant lacking the C-terminal hydrophobic sequence. J. Gen. Virol. 81:759-767. [DOI] [PubMed] [Google Scholar]
- 36.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, R. De Francesco, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, M. P., and Z. Hong. 2002. HCV RNA-dependent RNA polymerase as a target for antiviral development. Curr. Opin. Pharmacol. 2:534-540. [DOI] [PubMed] [Google Scholar]
- 38.Wang, M., K. K. Ng, M. M. Cherney, L. Chan, C. G. Yannopoulos, J. Bedard, N. Morin, N. Nguyen-Ba, M. H. Alaoui-Ismaili, R. C. Bethell, and M. N. James. 2003. Non-nucleoside analogue inhibitors bind to an allosteric site on HCV NS5B polymerase. Crystal structures and mechanism of inhibition. J. Biol. Chem. 278:9489-9495. [DOI] [PubMed] [Google Scholar]
- 39.Wasley, A., and M. J. Alter. 2000. Epidemiology of hepatitis C: geographic differences and temporal trends. Semin. Liver Dis. 20:1-16. [DOI] [PubMed] [Google Scholar]
- 40.Yenofsky, R. L., M. Fine, and J. W. Pellow. 1990. A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 87:3435-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhong, W., A. S. Uss, E. Ferrari, J. Y. Lau, and Z. Hong. 2000. De novo initiation of RNA synthesis by hepatitis C virus nonstructural protein 5B polymerase. J. Virol. 74:2017-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]