Abstract
Recent studies of prostate cancer and other tumor types have revealed significant support as well as unexpected complexities for the application of concepts from normal stem cell biology to cancer. In particular, the cell of origin and cancer stem cell models have been proposed to explain the heterogeneity of tumors during the initiation, propagation, and evolution of cancer. Thus, the basis of intertumor heterogeneity has emerged from studies investigating whether stem cells and/or non-stem cells can serve as cells of origin for cancer and give rise to tumor subtypes that vary in disease outcome. Furthermore, analyses of putative cancer stem cells have revealed the genetically diverse nature of cancers and expanded our understanding of intratumor heterogeneity and clonal evolution. Overall, the principles that have emerged from these stem cell studies highlight the challenges to be surmounted to develop effective treatment strategies for cancer.
Keywords: cell of origin, cell of mutation, cancer stem cell, tumor-initiating cell, clonal evolution
Introduction
The conceptual basis of the application of stem cell biology to cancer research arises from the fundamental observation of tumor heterogeneity. Two types of tumor heterogeneity lie at the heart of current challenges in identification of effective cancer biomarkers, prediction of treatment response, and design of targeted therapies. Intertumor heterogeneity can be observed between tumors of the same tissue type arising in different patients, which can vary in their prognosis and/or therapeutic response. In addition, there is the intratumor heterogeneity that can be observed within a single patient tumor, which has long been noted in terms of histopathological features, and more recently at the molecular level. Central concepts of stem cell biology can be invoked to explain the basis of both types of tumor heterogeneity, and thereby yield experimentally testable hypotheses with clinical relevance.
Although stem cell biologists who work on regenerative medicine frequently utilize similar concepts and methodologies to those who study cancer, these two groups of researchers have fundamentally divergent objectives. Thus, the principal objectives of many stem cell biologists include the development of methods for the identification, isolation, and propagation of adult tissue stem cells as an approach for disease treatment, or for the differentiation of pluripotent cells to desired cell types for applications in regenerative medicine. In contrast, cancer biologists pursue stem cell research to establish approaches for improved cancer prognosis and to generate targeted therapies through the eradication of cancer stem cells. However, despite great interest in the field, it remains to be determined whether the application of stem cell concepts can provide clinically useful insights into cancer biology.
Here, we review several recent papers that illustrate basic principles as well as complexities of understanding the relationship of stem cells to cancer biology. We will focus on analyses of prostate cancer, but also refer to studies of other cancers to highlight essential points.
The cell of origin and intertumoral heterogeneity
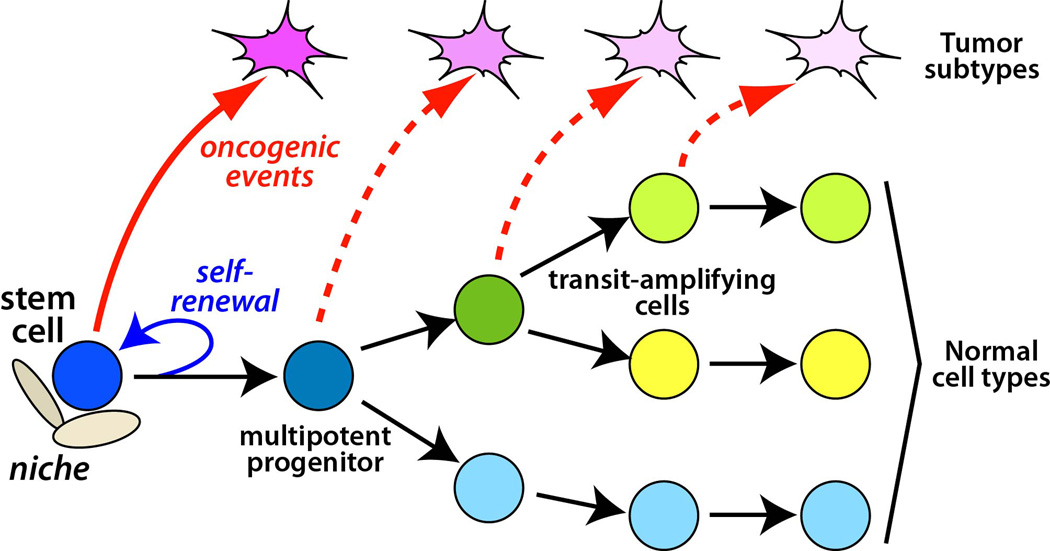
The cell of origin of a cancer is defined as the normal cell type from which a tumor arises following oncogenic transformation [1]. Given their inherent ability to self-renew, adult stem cells are believed to represent excellent targets for oncogenic transformation. However, in many cases cancer can also arise from cell types that are not stem cells, which thereby acquire the property of self-renewal upon transformation. The cell of origin model further proposes that distinct cell types of origin within a given tissue can give rise to corresponding tumor subtypes, which have different treatment responses and/or patient outcomes (Figure 1). Thus, the transformation of different cell types of origin within a given tissue may be an important source of intertumor heterogeneity. Notably, this model may have clinical significance, since it may be possible to design therapies targeted to key signaling pathways or master regulators of the cell type of origin that are retained in the corresponding tumor subtype.
Figure 1.
Cell of origin and relationship to tumor subtypes. The lineage hierarchy of a normal tissue is schematically depicted with a stem cell at the top of the hierarchy giving rise to a multipotent progenitor, downstream transit-amplifying cells, and differentiated cell types. The stem cell is capable of self-renewal, and its properties are regulated by a neighboring niche. Oncogenic transformation of different cell types of origin such as a stem cell (red arrow) or downstream progenitors (dashed arrows) can give rise to distinct tumor subtypes.
Methods to identify the cell of origin
Cells of origin can be identified experimentally in mouse models using two distinct approaches. One methodology involves lineage-tracing in genetically-engineered mice, using a Cre driver that is expressed specifically within the desired cell type of the tissue of interest. (In many cases, this approach utilizes a tamoxifen-inducible Cre recombinase that allows for temporal specificity of activity.) In combination with a suitable Cre reporter allele that typically leads to expression of a fluorescent protein after recombination, the consequences of tumor suppressor deletion and/or oncogene activation on the transforming potential of a desired cell type can be examined. Should a tumor form, histopathological and molecular analyses can be performed to evaluate the similarities or differences of the mouse tumor to tumor subtypes from human patients.
A second approach to analyze cell types of origin examines whether mouse or human cells can act as cells of origin by flow-sorting of enzymatically dissociated cells, followed by genetic manipulation using infection by lentiviral vectors to induce oncogenic transformation. The resulting cells can then be transplanted into immunodeficient mice, either as an orthotopic graft or in the context of ectopic tissue locations and/or in combination with a heterologous supporting cell type. This method requires reliable cell-surface markers for isolating a purified cell population, as well as a suitable in vivo graft model to test for tumor formation. An additional caveat is that viral infection may result in non-physiological levels of oncogene expression, influencing the ability of a targeted cell to become a cell of origin. However, this methodology has the advantage of being amenable for human cells and does not require genetically-engineered mice.
An important complicating factor in experimental analyses of the cell of origin is the distinction between a cell of mutation, where the oncogenic event takes place, versus an actual cell of origin. For example, a genetic manipulation could create an oncogenic alteration in a stem cell (cell of mutation), but phenotypic transformation might not take place until the stem cell differentiated into a specific downstream progeny cell type (cell of origin). This distinction is significant because many studies in the literature have concluded that stem cells represent a cell of origin for a given cancer, but have not excluded the possibility that the stem cells were only the cell of mutation. Furthermore, this distinction may also have clinical implications if the properties of a tumor subtype are dictated at least in part by the actual cell of origin.
A recent study has used an elegant genetic lineage-marking system termed MADM (mosaic analysis with double markers) to resolve the cell of mutation versus the cell of origin for malignant glioma [2]. This system has the advantage of combining Cre-mediated recombination with fluorescent labeling of sister cells in which one sister is homozygous mutant for a tumor suppressor gene and expresses one fluorescent protein (e.g., GFP), while the other sister is wild-type and expresses a different fluorescent protein (e.g., RFP). This system was used with a neural stem cell (NSC) specific Cre driver to simultaneously mutate p53 and Nf1 as well as label NSCs under conditions where relatively few cells were targeted, allowing analyses at single-cell resolution. To determine the actual cell origin, these authors measured the ratio of mutant green and wild-type red cells in neurons, astrocytes, oligodendrocytes, and oligodendrocyte precursor cells (OPCs) in brains at pre-malignant stages. These analyses revealed that mutant green OPCs, but not NSCs, were over-represented, suggesting that OPCs were the cell of origin for glioma. This finding was then confirmed by conditional deletion of p53 and Nf1 using a Cre driver expressed specifically in OPCs and not in NSCs. Thus, these results appear to differ from findings of previous studies that have suggested that NSCs are the cell of origin for glioma [3–5], and highlight the necessity of detailed genetic analyses in evaluating the cell of origin model for specific cancers.
Both luminal cells and basal epithelial cells are cells of origin for prostate cancer
In the case of the prostate, there are two major epithelial cell types, corresponding to a columnar layer of secretory luminal cells and an underlying layer of basal cells. Normal prostate maintenance is dependent on androgens, and as a consequence, androgen deprivation leads to tissue regression due to apoptosis of most of the luminal cells. However, upon androgen restoration, the mouse prostate can regenerate, and can also undergo multiple cycles of regeneration in response to further androgen deprivation/restoration, indicating the existence of a stem cell population(s) in the regressed prostate.
With respect to prostate cancer, there has been considerable interest in identifying whether luminal or basal epithelial cells can serve as cells of origin, and whether these cells of origin correspond to stem cells. A luminal cell of origin would be consistent with the exclusively luminal phenotype of prostate adenocarcinoma, whereas a basal cell of origin would imply that transformed basal cells acquire luminal features during tumor progression. In particular, lineage-tracing studies have identified castration-resistant Nkx3.1-expressing cells (CARNs) as a rare luminal stem cell population that can display bipotentiality and self-renewal, and can also serve as a cell of origin for prostate cancer in vivo [6]. Furthermore, CARNs can also generate prostate ducts in single-cell tissue reconstitution assays [6], which can determine the potential of isolated populations of prostate epithelial cells to form prostate tissue or serve as a cell of origin for cancer. In such transplantation assays, prostate epithelial cells are combined with rodent embryonic urogenital sinus mesenchyme (UGM) cells and implanted under the kidney capsule of immunodeficient male mice, followed by analysis after two to three months of growth [7].
Moreover, basal cells can also serve as a cell of origin for prostate cancer, as has been demonstrated by lentiviral infection and/or tissue reconstitution assays using purified basal cells isolated from mouse and human prostate [8–10]. Similarly, lineage-tracing studies using luminal-specific and basal-specific inducible Cre drivers for deletion of the Pten tumor suppressor have shown that both luminal and basal epithelial compartments contain cells of origin for prostate cancer [11].
At present, however, it is unclear whether these cells of origin correspond to stem cells or more downstream progenitors. Notably, lineage-tracing studies using specific tamoxifen-inducible Cre drivers have suggested that both luminal and basal compartments are largely or wholly sustained by unipotent progenitors [11,12]. These lineage-tracing studies have been conducted using luminal-specific Cre drivers including PSA-CreERT2 and K8-CreERT2, as well as the basal-specific K14-CreER transgenic mice [11–13]. However, it is important to note that such lineage-tracing experiments are dependent on the promoters and mouse lines used, and that certain promoters could conceivably be biased towards labeling specific subsets of cells that may not be representative of the entire basal or luminal population. Although it is conceivable that tamoxifen itself could alter the results, particularly since prolonged administration of the synthetic estrogen diethylstilbestrol leads to squamous metaplasia [14], it appears unlikely that the relatively short duration of tamoxifen treatment used in lineage-tracing studies would exert a substantial effect.
If luminal and/or basal stem cells constitute a rare population in the prostate epithelium, then the cells of origin identified in both lineage-tracing and tissue reconstitution assays are likely to correspond to non-stem cells. While it is often presumed that non-stem cells of origin correspond to downstream progenitors in the lineage hierarchy, there is little experimental evidence to address whether terminally differentiated cells might also act as cells of origin in the appropriate assay. Thus, the relationship between differentiation status and ability to serve as cell of origin is not fully understood. More generally, the identification of cell types of origin may be dependent on the specific oncogene/tumor suppressor gene being activated/inactivated. For example, it may be the case that Pten deletion can transform both luminal and basal epithelial cells, consistent with its central role in human prostate cancer, but other tumor suppressors could conceivably display more limited activities in transforming either luminal or basal cells.
A related question is whether prostate tumors arising from luminal or basal cells of origin can give rise to distinct tumor subtypes. While deletion of Pten in luminal or basal cells give rise to prostate lesions with indistinguishable histological phenotypes, the resulting tumors might nonetheless have distinct molecular properties that could be associated with differing patient outcomes and/or treatment responses. In the case of human prostate cancer, a significant minority of patients diagnosed with intermediate grade tumors will have aggressive tumors that have unfavorable outcomes, yet these tumors appear to be histologically and molecularly indistinguishable from the majority of tumors with an indolent phenotype [15]. Therefore, it is likely that human prostate cancer can be stratified into distinct subtypes, although the specific molecular features of these subtypes have yet to be identified, despite substantial efforts to do so [16]. Moreover, it remains to be determined whether these putative subtypes would arise from oncogenic transformation of distinct cell types of origin, as predicted by the cell of origin model.
Why does the cell of origin matter?
Although the clinical significance of the cell of origin model is currently unknown for prostate cancer, the relevance of this model has been demonstrated by studies of the mammary lineage hierarchy and cell types of origin for breast cancer. There is considerable evidence that major subtypes of breast cancer that differ in the histopathological features, molecular profiles, treatment response, and patient outcome arise through transformation of distinct cell types within the mammary epithelial lineage hierarchy [17]. Interestingly, there is not a simple relationship between the phenotype of the cell of origin and the histopathological features of the resulting tumor. For example, BRCA1 mutant breast cancers display an estrogen receptor (ER) negative “basal-like” phenotype, and therefore have previously been thought to originate from mammary epithelial stem cells, which have a basal phenotype. However, deletion of Brca1 in a p53+/− genetic background using a basal-specific cytokeratin 14-Cre driver resulted in tumors that did not resemble human breast cancers [18]. In contrast, deletion of Brca1 in a p53+/− background using a Blg-Cre driver expressed predominantly in luminal cells gave rise to tumors that phenocopied human BRCA1 mutant breast cancers, suggesting that these tumors arose from luminal progenitors. These in vivo results in mouse models are also consistent with data from colony-forming assays and bioinformatic analyses of epithelial subsets isolated from human BRCA1 mutation carrier and control breast tissue [19]. Overall, these findings demonstrate that the cell of origin can determine the histopathological features of tumors, but not always as expected, and also suggest that mammary stem cells are not a frequent cell of origin for human breast cancers. Notably, however, a recent lineage-tracing study has demonstrated that the multipotency and developmental potential of the mammary stem cell identified in transplantation assays may be context dependent [20]. Thus, a re-evaluation of previous studies on the mammary epithelial lineage hierarchy and cell of origin may be necessary.
Cancer stem cells and tumor-initiating cells
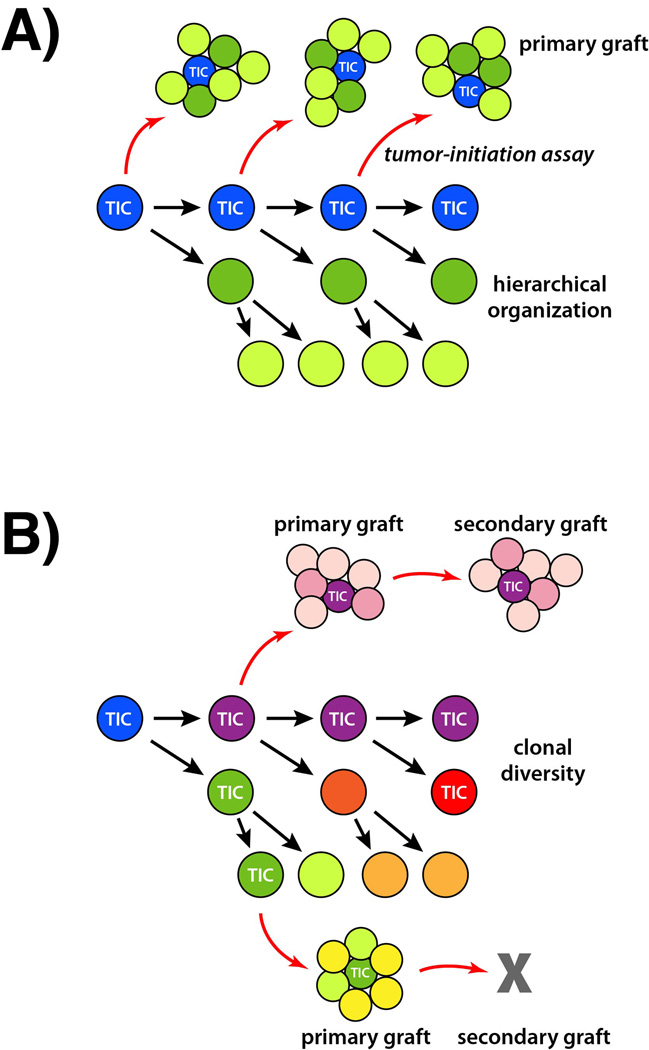
Over the past decade, the relationship between stem cell concepts and cancer biology has been dominated by the cancer stem cell model (Figure 2A). This model proposes that cells within a tumor are generated through a lineage hierarchy, similar to a normal tissue, and are sustained by a population of self-renewing stem cells [21,22]. Furthermore, such cancer stem cells may also differentiate to produce phenotypically distinct cell types with more limited proliferative potential. The cancer stem cell model is attractive because of its potential therapeutic implications, since cancer stem cells might not be affected by conventional therapies, and thus could contribute to cancer recurrence and long-term propagation. Nonetheless, the cancer stem cell model has remained highly controversial because of difficulties in identifying robust cancer stem cell markers as well as development of reliable assays [23].
Figure 2.
The cancer stem cell model and clonal evolution. A: The traditional cancer stem cell model postulates that a tumor-initiating cell (TIC) gives rise to distinct tumor cell types through a lineage hierarchy. Within the tumor, only the TIC and not its progeny can generate successful tumor grafts in a tumor-initiating assay (red arrows). The resulting grafts recapitulate the hierarchical organization and phenotype of the parental tumor. B: In a clonal evolution model, a tumor may contain multiple distinct tumor-initiating cells that in turn generate clonal diversity and resulting intratumor heterogeneity. These different tumor-initiating cells may differ in their tumor-propagating capabilities in serial grafting assays.
Experimental approaches to identify putative cancer stem cells typically utilize flow cytometry to isolate candidate cell populations from either human or mouse tumors. The resulting cell populations can be grafted into immunodeficient mice to assess whether they can form tumors that recapitulate the phenotype of the parental tumor. If such candidate tumor-initiating cells can be identified, limiting dilution assays can then evaluate their frequency of cancer stem cells, while serial transplantation can be performed to assess long-term proliferative potential. Using this general approach, populations of tumorigenic cells have been identified from a wide range of human cancers [23]. However, the most convincing evidence for cancer stem cells and hierarchical organization of tumors comes from studies of hematological malignancies, whereas the published evidence for their existence in solid tumors has been more controversial [24–26]. Recently, new evidence from lineage-tracing studies in mouse models has provided direct evidence for a lineage hierarchy in intact solid tumors [27–29]. These in vivo studies have shown that stem cell-like tumor cells that can give rise to more differentiated cell types with varying proliferative potentials exist in intestinal adenomas, skin papillomas, and squamous cell carcinomas, as well as in gliomas [27–29]. It remains to be determined whether similar cancer stem cells can be identified in other tumor types.
It is worth noting that basic terminology is often poorly defined in the cancer stem cell literature. Unfortunately, the use of the nomenclature “tumor-initiating cell” has led to some confusion in the field, as this term has also been applied to the cell of origin, resulting in a lack of distinction between a cancer cell and a normal cell. To avoid such linguistic confusion, some investigators favor alternative terms such as cancer-repopulating cell, which may also represent a more accurate description of the grafting assay. In addition, although putative cancer stem cells are sometimes assumed to resemble their cell type of origin, there is little basis for a phenotypic resemblance if the tumor is generated from a non-stem cell origin. Similarly, hierarchical organization of a tumor does not necessarily imply that it arose from a normal stem cell.
It is now evident that there are numerous methodological and interpretative pitfalls associated with conventional tumor-initiation assays. For example, rigorous controls are necessary to establish the properties of the putative non-cancer stem cell population isolated by flow cytometry. It is crucial to demonstrate that the inability of this population to form tumors is not simply due to the presence of dead/dying cells, or growth disadvantages resulting from specific assay conditions. It is also essential to demonstrate that the cell surface markers used to isolate the putative cancer stem cell population are robust, and can be used in a highly reproducible fashion.
In several notable instances, the expression patterns of cell surface markers that have been considered to be “universal” stem cell and/or tumor-initiating cell markers have been misleading. For example, studies of the CD133 marker have shown its widespread expression in a wide range of differentiated epithelial cell types in adult mice as well as in primary colon cancers: most CD133− cells correspond to stromal or inflammatory cells, whereas metastatic colon cancers consist of both CD133+ and CD133− epithelial cell populations that do not differ in tumor-initiating capability [30]. Furthermore, markers such as CD133 and CD271 do not appear to identify stable melanoma cell populations that differ in tumor-initiation frequency, supporting the interpretation that malignant melanomas lack hierarchical organization and do not follow a cancer stem cell model [31,32].
Moreover, systematic methodological improvements can also greatly improve the efficiency of the tumor-initiation assay. Thus, technical modifications to the grafting methodology as well as the use of severely immunodeficient mice as graft recipients can increase the detection frequency of tumor-initiating cells in malignant melanoma to approximately 30% [31,32]. On the other hand, it is also evident that the same methods do not lead to the identification of a high frequency of tumor-initiating cells for many other tumor types [33]. Thus, the overall frequency of tumor-initiating cells appears to be determined by the tumor type, but is also influenced by assay conditions that need to be optimized for each tumor type.
Identification of cancer stem cells and tumor-initiating cells in the prostate
Efforts have been made to identify tumor-initiating cells and markers of tumor-initiating cells in human and mouse prostate cancers using prostate cancer cell lines, xenograft lines, and primary prostate cancer tissues. Several types of transplantation assays have been used, including orthotopic grafting of cells into the prostate, as well as grafting of cells to ectopic locations. For example, a higher frequency of tumor formation has been achieved by grafting of the LAPC9 xenograft line subcutaneously rather than orthotopically to the dorsal prostate [34], while in contrast, the CWR22 xenograft line forms tumors more readily after grafting to orthotopic rather than subcutaneous sites [35]. One advantage of ectopic grafting is that endogenous prostate epithelial cells will not compete with the prostate cancer cells being grafted. Moreover, subcutaneous grafts also allow for easy visualization of the resulting tumors. However, the major caveat of ectopic grafting is that cancer cells are removed from their endogenous microenvironment, which may have positive or negative effects on their tumorigenic properties. For instance, non-transformed BPH-1 prostate cells can be stimulated by cancer associated fibroblasts to induce prostate tumor formation in grafts [10,36].
Several studies have suggested that established tumor xenografts display tumor-initiating frequencies that are significantly higher than for the primary tumors from the same tissue, perhaps consistent with the fact that most xenograft lines have been selected for growth under cell culture conditions. Indeed, in the case of prostate cancer, tumor-initiating cells enriched by flow-sorting for CD44+ α2β1+ cells can be identified from xenografts much more readily than from primary tumors [34]. However, recent work has developed a new method that greatly improves the survival and growth of tumor-initiating populations from primary prostate tumors, by co-grafting of neonatal mouse urogenital mesenchyme along with human primary prostate cancer tissues [37]. Interestingly, this methodology failed to demonstrate a difference in tumor-initiating frequency between α2β1-integrin high and low populations from primary prostate tumors [37], which is notable since α2β1-integrin high cells were previously reported as being enriched for prostate epithelial stem cells in primary human prostate tissue [38].
To date, it has been a challenge to identify cell surface markers for isolating tumor-initiating populations from primary human prostate cancers. However, recent analysis of a mouse model has identified tumor-initiating properties of a Lin−Sca-1+CD49fhi cell population from Pb-Cre4; Ptenflox/flox mice [39]. In this study, sorted prostate epithelial cells were recombined with urogenital sinus mesenchyme, followed by subcutaneous grafting [39]. However, one caveat is that the long-term proliferative potential of Lin−Sca-1+CD49fhi cells in vivo is unknown, as serial transplantation assays were not performed.
Clonal evolution and intratumor heterogeneity
A major factor contributing to tumor heterogeneity is clonal evolution, in which cancers are believed to evolve by acquiring genetic mutations that provide a selective advantage [40]. As tumors undergo competition for space and nutrient resources, either during cancer growth or in response to treatment, different populations of cancer cells can dramatically expand or contract in their proportion within a given tumor. In principle, clonal evolution could be driven by competition between genetically and/or epigenetically distinct cancer stem cell types that each generate a hierarchically organized population (clone) of cancer cells, thereby generating intratumor heterogeneity (Figure 2B).
The clonal evolution model suggests that cancers are composed of a complex mixture of cell populations that generate phenotypic diversity and can respond to treatment with outgrowth of resistant clones. For example, acute lymphoblastic leukemia (ALL) typically consists of genetically diverse clones with leukemia-initiating function that evolve in a complex branching fashion [41]. When samples from Philadelphia chromosome ALL patients were grafted into immunodeficient mice, some grafts caused disease in recipient mice before 15 weeks, whereas others did not. To study the evolution of tumor diversity, the resulting grafts were regrafted, followed by DNA copy number analyses to trace the evolution of subclones in secondary grafts. Although the clone identified at patient diagnosis could be detected in many aggressive secondary grafts, new copy number alterations as well specific genetic differences explaining functional heterogeneity were identified in other grafts [41]. In a similar approach, the evolution of distinct clones in ETV6-RUNX1 fusion positive ALL patients could be traced using fluorescence in situ hybridization to detect common genomic copy number alterations [42]. In both studies, the presence of multiple tumor subclones with different gene deletions suggested that tumors with leukemia-initiating function were continuing to evolve in a multi-clonal branching manner. These findings highlight the difficulty in achieving an effective therapeutic response by targeting of specific cancer cell populations, since tumors do not correspond to a static entity. Instead, intratumor heterogeneity results in the ability of tumors to respond to selective pressures following treatment with the emergence of clones that display therapeutic resistance.
In support of this view, other studies have identified distinct tumor-initiating cell populations in prostate cancers that differ in their treatment response. For prostate cancer, androgen-deprivation therapy remains the mainstay of treatment strategies, yet many prostate cancers ultimately develop castration-resistance and disease relapse. Notably, a recent study using prostate cancer cell lines and xenografts has identified castration-resistant tumor-initiating cells that express low levels of prostate-specific antigen (PSA), which is a marker of differentiated luminal cells, as well as a castration-sensitive tumor-initiating population that expresses high PSA [43]. In particular, following infection with a lentiviral vector containing a PSA promoter driving GFP expression, human LAPC xenograft lines could be sorted into distinct GFP-bright PSA+ and GFP-dim PSA−/lo populations. Following orthotopic grafting into immunodeficient male hosts, both PSA−/lo and PSA+ cell populations displayed tumor-initiating capability, although the PSA+ population generated more and larger tumors. In contrast, following serial grafting, PSA−/lo derived xenografts grew faster after the third passage, and could be passaged for more serial generations, whereas PSA+ derived grafts displayed diminishing potential. These findings suggest an important experimental distinction between tumor-initiation and long-term tumor-propagation that may not be readily observed in the absence of extensive serial passaging. Moreover, these findings also imply that tumor-initiation and tumor-propagation capabilities are not absolutely correlated.
Additional complexity in tumor-initiating populations has been identified by lineage-tracing strategies to follow the self-renewal of individual human primary colon cancer cells in serial transplantation assays in mice [44]. Following growth of tumor spheres derived from primary colon cancers, dissociated cells were infected with lentiviral vectors that integrated at unique sites, followed by renal grafting in immunodeficient mice. Serial passaging of the resulting tumors identified distinct clones that could not be readily propagated in secondary grafts, perhaps derived from “tumor transient-amplifying cells”, as well as a smaller number of clones containing tumor-propagating cells (termed “long-term tumor-initiating cells”) that could form tumors in serial grafts. These tumor-propagating cells were also more likely to generate metastases relative to the tumor transient-amplifying cells. Most interestingly, this marking system could identify “delayed contributing tumor-initiating cells” that quantitatively contributed to secondary or tertiary grafts, but were undetectable in the primary graft, consistent with activation of a previously dormant tumor-initiating population [44].
Although there is increasing evidence of genetic differences that contribute to intratumor heterogeneity, the extent to which epigenetic mechanisms are involved remains unclear. However, indirect evidence for epigenetic regulation has been provided by work suggesting plasticity of putative cancer stem cells [45]. In cultured SUM human breast cancer cell lines derived from primary tumors, basal, luminal, and “stem-like” populations were defined by flow-sorting using the cell-surface markers CD24, CD44 and EpCAM. Following isolation of each subpopulation and their expansion in culture, the relative proportions of the basal, luminal, and stem-like populations was restored to equilibrium over time. Similarly, after orthotopic implantation of sorted populations into mammary glands of immunodeficient mice, all three populations were able to seed tumors. Mathematical modeling provided quantitative assessments of the ability of non-stem cells to become cancer stem-like cells, indicating that interconversion readily occurs between cell states. Whether these findings apply to prostate cancer or other solid tumors remains unknown. However, these findings provide additional evidence for complex population dynamics underlying intratumor heterogeneity, and further suggest that simultaneous combinatorial therapies targeting multiple cell types will be required to prevent cancer relapse.
Conclusions
The application of concepts from stem cell research to cancer biology has provided important insights into the basis for tumor heterogeneity. In particular, the cell of origin of a cancer may affect treatment response and patient outcome, as recent work has shown that the cell of origin can influence the resulting tumor subtype and thereby give rise to intertumor heterogeneity. Therefore, careful analysis of the cell type of origin (versus the cell of mutation) may improve cancer prognosis and assist the development of targeted therapies. For example, in prostate cancer, identification of cell types of origin, which may be inherently castration-resistant, may lead to therapies targeted to such cells and thereby improve the treatment of castration-resistant lethal disease.
With respect to intratumor heterogeneity, there is now significant evidence that at least some cancers are hierarchically organized and follow a cancer stem cell model. However, although much effort has been devoted to identifying putative cancer stem cell populations, the cancer stem cell model remains controversial and requires further validation. Finally, recent studies have demonstrated the extraordinary genetic complexity that underlies intratumor heterogeneity, indicating that evolutionary selection pressures act upon multiple co-existing populations of tumor cells, perhaps generated by distinct tumor-initiating populations. This process of clonal evolution suggests that combinatorial approaches targeting distinct tumor cell populations will be necessary for successful cancer treatment.
Acknowledgments
This work was supported by an NIH training grant T32DK007328 (M.S.) and by grants from the NIH and the DOD Prostate Cancer Research Program (M.M.S.).
References
- 1.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 2.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, et al. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, et al. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, et al. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldstein AS, Drake JM, Burnes DL, Finley DS, Zhang H, et al. Purification and direct transformation of epithelial progenitor cells from primary human prostate. Nat Protoc. 2011;6:656–667. doi: 10.1038/nprot.2011.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a Cell of Origin for Human Prostate Cancer. Science. 2010;329:568–571. doi: 10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, et al. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci USA. 2010;107:2610–2615. doi: 10.1073/pnas.0913873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor RA, Toivanen R, Frydenberg M, Pedersen J, Harewood L, et al. Human Epithelial Basal Cells are Cells of Origin of Prostate Cancer, Independent of CD133 Status. Stem Cells. 2012 doi: 10.1002/stem.1094. N/A-N/A. [DOI] [PubMed] [Google Scholar]
- 11.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21:253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Pascal LE, Isharwal S, Metzger D, Ramos Garcia R, et al. Regenerated luminal epithelial cells are derived from preexisting luminal epithelial cells in adult mouse prostate. Mol Endocrinol. 2011;25:1849–1857. doi: 10.1210/me.2011-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Zhang B, Han SJ, Shore AN, Rosen JM, et al. Targeting CreERT2) expression to keratin 8-expressing murine simple epithelia using bacterial artificial chromosome transgenesis. Transgenic Res. 2012 doi: 10.1007/s11248-012-9598-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Risbridger GP, Wang H, Frydenberg M, Cunha G. The metaplastic effects of estrogen on mouse prostate epithelium: proliferation of cells with basal cell phenotype. Endocrinology. 2001;142:2443–2450. doi: 10.1210/endo.142.6.8171. [DOI] [PubMed] [Google Scholar]
- 15.Martin NE, Mucci LA, Loda M, Depinho RA. Prognostic determinants in prostate cancer. Cancer J. 2011;17:429–437. doi: 10.1097/PPO.0b013e31823b042c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur SR, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC medical genomics. 2010;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7:403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 19.Lim E, Vaillant F, Wu D, Forrest NC, Pal B, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15:907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 20.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 21.Clarke MF, Fuller M. Stem cells and cancer: two faces of Eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 23.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 25.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138:822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 28.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012 doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Li Y, Yu TS, McKay RM, Burns DK, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012 doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, et al. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7:279–282. doi: 10.1016/j.stem.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: the CD44+alpha2beta1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796–6805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 35.Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappaB signalling. Nature communications. 2011;2:162. doi: 10.1038/ncomms1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, et al. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toivanen R, Berman DM, Wang H, Pedersen J, Frydenberg M, et al. Brief report: a bioassay to identify primary human prostate cancer repopulating cells. Stem Cells. 2011;29:1310–1314. doi: 10.1002/stem.668. [DOI] [PubMed] [Google Scholar]
- 38.Collins AT, Habib FK, Maitland NJ, Neal DE. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114:3865–3872. doi: 10.1242/jcs.114.21.3865. [DOI] [PubMed] [Google Scholar]
- 39.Mulholland DJ, Xin L, Morim A, Lawson D, Witte O, et al. Lin-Sca-1+CD49fhigh stem/progenitors are tumor-initiating cells in the Pten-null prostate cancer model. Cancer Res. 2009;69:8555–8562. doi: 10.1158/0008-5472.CAN-08-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Notta F, Mullighan CG, Wang JC, Poeppl A, Doulatov S, et al. Evolution of human BCR-ABL1 lymphoblastic leukaemia-initiating cells. Nature. 2011;469:362–367. doi: 10.1038/nature09733. [DOI] [PubMed] [Google Scholar]
- 42.Anderson K, Lutz C, van Delft FW, Bateman CM, Guo Y, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 43.Qin J, Liu X, Laffin B, Chen X, Choy G, et al. The PSA(−/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dieter SM, Ball CR, Hoffmann CM, Nowrouzi A, Herbst F, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]