Abstract
Human PiT2 (PiT2) is a multiple-membrane-spanning protein that functions as a type III sodium phosphate cotransporter and as the receptor for amphotropic murine leukemia virus (A-MuLV). Human PiT1 (PiT1), another type III sodium phosphate cotransporter, is a highly related protein that functions as a receptor for gibbon ape leukemia virus but not for A-MuLV. The ability of PiT1 and PiT2 to function as discrete viral receptors with unique properties presumably is reflected in critical residue differences between these two proteins. Early efforts to map the region(s) within PiT2 that is important for virus binding and/or entry relied on infection results obtained with PiT1-PiT2 chimeric cDNAs expressed in Chinese hamster ovary (CHOK1) cells. These attempts to localize the PiT2 virus-binding site were hampered because they were based on infectivity, not binding, assays, and therefore, receptors that bound but failed to facilitate virus entry could not be distinguished from receptors that did not bind virus. Using a more accurate topological model for PiT2 as well as an A-MuLV receptor-binding assay, we have identified extracellular domain one (ECD1) of the human PiT2 receptor as being important for A-MuLV binding and infection.
PiT1 and PiT2 are type III sodium-dependent phosphate transporters that also function as receptors for the mammalian gammaretroviruses gibbon ape leukemia virus (GALV) and amphotropic murine leukemia virus (A-MuLV), respectively (10, 18, 19, 34, 36). While these receptors have similar cellular functions and structures, they do not overlap in their virus receptor functions; this has been attributed to critical amino acid differences between PiT1 and PiT2.
Early structural predictions for the arrangement of the PiT receptors in the plasma membrane were based on Kyte-Doolittle hydropathy analyses (8). Both proteins were initially predicted to be nearly identical in structure, each comprising 10 transmembrane (TM) domains. Additionally, the observed absence of a signal peptide for both proteins was used to assign cytoplasmic locations for the N and C termini; both were initially predicted to contain five extracellular domains (ECDs) and four intracellular domains, with all potential N-linked glycosylation sites being positioned within intracellular domains (8).
In order to understand how differences in amino acid composition between PiT1 and PiT2 affect receptor function, researchers have used chimeric PiT1-PiT2 proteins to map regions that are critical for GALV (2, 5, 9, 21, 22, 27, 32) and A-MuLV (12, 13, 14, 21, 28, 30) entry. Previous studies based on Kyte-Doolittle hydropathy models of PiT1 and PiT2 have demonstrated that replacement of the second ECD (ECD2) of PiT1 with the corresponding region of PiT2 results in a chimeric protein which functions as an A-MuLV receptor (12). This result was supported by studies by Lundorf et al. that showed that substitution of PiT2 residues from ECD2 and flanking regions for the corresponding residues of Pho-4, a sodium-dependent phosphate transporter from the filamentous fungus Neurospora crassa, confers A-MuLV receptor function to Pho-4 (13).
It has recently been experimentally determined that both the N and C termini of PiT2 are extracellular, thus reorienting the former first, second, and third ECDs to the cytosol and the former first and second intracellular domains to extracellular positions (26). The reorientation of the N-terminal third of the PiT2 protein was further validated by experiments demonstrating that PiT2 is a glycoprotein carrying an N-linked oligosaccharide in the more recently predicted ECD1 (26). It should be noted that chimeric PiT1-PiT2 receptor studies implicating the former ECD2 as critical to A-MuLV infectivity utilized the earlier Kyte-Doolittle-based PiT receptor topology. The current topological model positions the region formerly designated ECD2 in the cytosol. Interestingly, the region currently designated ECD1 was still present in these chimeras, although it was unclear at the time that this domain was extracellular. Thus, previous results may have been misinterpreted to implicate the former ECD2 as being important for PiT2-mediated A-MuLV infection when, in fact, the receptor function was being mediated by what is now called ECD1.
For A-MuLV receptor function studies, CHOK1 cells have been the preferred cell line, based on the observation that CHOK1 cells are resistant to infection by both GALV and A-MuLV. It was suggested that the reason these cells are refractory to A-MuLV infection is because they express nonfunctional receptors or receptors masked by a tunicamycin-sensitive inhibitor (3, 16, 17, 35). However, it was shown more recently that simply overexpressing the endogenous PiT2 receptor in CHOK1 cells (PiT2CHO) resulted in susceptibility to both GALV and A-MuLV, while expression of the endogenous PiT1 receptor (PiT1CHO) resulted in susceptibility to GALV (30). These findings suggest that the block to A-MuLV infection of CHOK1 cells is not due to the absence of functional receptors, and therefore, CHOK1 cells may not be the best system for investigating A-MuLV receptor function.
As stated above, studies attempting to identify regions of PiT2 that are important for A-MuLV entry used virus infectivity assays to measure receptor function. None of these studies explored the binding capability of A-MuLV to various chimeric receptors, raising the possibility that certain nonfunctional A-MuLV receptors retained the ability to bind A-MuLV while not facilitating entry into target cells. Therefore, while previous studies were useful for mapping receptor regions important for A-MuLV entry, regions of the receptor that are critical for A-MuLV virus binding were not directly assessed.
The purpose of this study was to map the region of PiT2 required for A-MuLV binding and/or entry into host cells, using chimeric PiT1-PiT2 receptors based on the experimentally validated topological models of PiT2, proposed by Salaün and coworkers (25, 26), and PiT1, proposed by Farrell et al. (6). Herein we report that the first ECD of PiT2 is critical for A-MuLV binding and entry.
MATERIALS AND METHODS
Cell lines.
The following cell lines were used in experiments for this study: BHK SN-10 Syrian hamster kidney cells (provided by Noel Bouck, Northwestern University, Chicago, Ill.), CHOK1 Chinese hamster ovary cells (ATCC CCL 61), and 293T human embryonic kidney cells (provided by Cell Genesys, Foster City, Calif.). All cells, with the exception of CHOK1, were maintained in Dulbecco's modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 4 mM glutamine. CHOK1 cells were maintained in alpha minimal essential medium and supplemented as described above.
Production of retrovirus vectors and stable cell lines.
A-MuLV enveloped retrovirus vectors were harvested in supernatants from 293T cells 48 to 72 h after transfection by the calcium phosphate precipitation method (Promega, Madison, Wis.), as previously described (33). The CHOK1 and BHK SN-10 cell lines stably expressing PiT2 or chimeric PiT1-PiT2 receptors were made by transducing cells with vesicular stomatitis virus G pseudotyped retrovirus vectors, using a pLNSX-derived genome expressing the appropriate receptor cDNA, as previously described (6). The assessment of receptor function for the various cell lines was carried out by exposing cells to retrovirus vector-containing supernatant that had been passed through a 0.45-μm-pore-size filter (Millipore, Bedford, Mass.) and then adjusted to contain 10 μg of Polybrene/ml. Twenty-four hours later, the medium was changed and cells were cultured for an additional 24 to 48 h before analysis for expression of β-galactosidase by histochemical staining with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), as previously described (35). Titers were determined after serial dilution of each vector and averaging of the number of blue CFU (BFU) obtained for each cell line tested in three or more independent experiments. All chimeric receptors described for this study were constructed as both hemagglutinin (HA)-tagged and untagged versions and tested for functionality. The presence of the HA tag within each of the receptor constructs was determined not to have altered receptor function (data not shown).
Chimeric receptor cDNAs.
To generate the chimeric PiT1-PiT2 receptor cDNAs C1A and C1E, specific regions of PiT1 were replaced with the corresponding regions of PiT2 between restriction enzyme sites NheI (nucleotide [nt] 196) and AccI (nt 642). For construction of C1A, PiT1 residues 121 to 214 (the region between the HindIII and AccI sites) were replaced with PiT2 residues 106 to 199. A PiT2 cDNA fragment was amplified from a PiT2 plasmid by use of a sense primer incorporating a HindIII site at position 315 and an antisense primer incorporating an AccI site at position 597. A PiT1 fragment was amplified from a PiT1 plasmid by use of a sense primer incorporating the existing NheI site at position 151 and an antisense primer introducing a HindIII site at position 360. The resulting fragments were each cloned into the TOPO-TA cloning vector pCR2.1 (Invitrogen, San Diego, Calif.), excised with HindIII and AccI (for PiT2) or NheI and HindIII (for PiT1), and then cloned into the pSP72 plasmid (Promega), containing the full-length PiT1 gene, between the NheI and AccI sites of PiT1 by a three-way ligation. The resulting C1A cDNA was subcloned into the retroviral vector plasmid pLNS-PiT1 (19) between the HindIII and PflMI sites. C1E contains PiT2 residues 56 to 140 in place of PiT1 residues 71 to 155 between NheI (nt 196) and SacI (nt 460). Primers were designed to introduce SacI sites at position 460 in PiT1 and position 415 in PiT2 by PCR mutagenesis as described above.
The chimeric receptor cDNAs C1F and C1G were made by two rounds of PCR mutagenesis. In the first round, complementary primers were designed which incorporated the individual nucleotide changes desired to mutate specific amino acids. Two fragments were generated by use of these primers with an upstream primer incorporating the NheI site and a downstream primer incorporating the AccI site. These products were annealed, after which a second round of PCR was performed, using the outer primers only. C1F was generated from C1E subcloned into the pSP72 plasmid (pSP72-C1E) by changing the glutamine at position 106 to serine and the valine at position 111 to phenylalanine. C1G was similarly generated from the pSP72 subclone of C1F (pSP72-C1F) by changing the PiT2 threonine residue at position 72 (nt 214 and 215) in C1F back to the original valine residue present in PiT1. The resulting fragments incorporating these amino acid changes were subcloned into the TOPO-TA cloning vector pCR2.1, then into pSP72-C1E or pSP72-C1F, and finally into pLNS-PiT1, as described above, to create pLNS-C1F and pLNS-C1G. C1F contains PiT2 residues 56 to 91 in place of PiT1 residues 71 to 106, and C1G contains PiT2 residues 66 to 91 in place of PiT1 residues 81 to 106. The chimeric receptor K7 was made by replacing PiT1 sequences between the PstI site (nt 1552) and the 3′ end of the cDNA with PiT2 sequences. C1 contains PiT2 sequence between the NheI and BglII sites; BglII was introduced into PiT1 at nt 1167 by PCR mutagenesis, as described above.
PCR products containing mutations were ligated into pCR2.1 and sequenced by use of the Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Biosciences, Piscataway, N.J.) and Cy-5-labeled primers (Integrated DNA Technologies, Inc., Coralville, Iowa) on an AlfExpress automated sequencer (Amersham Biosciences).
Binding assays.
Fluorescence-activated cell sorting-based binding assays using soluble A-MuLV or GALV envelope SU proteins encoding the receptor-binding domain (RBD) fused to a double HA epitope tag (YPYDVPDYA) derived from influenza virus HA were performed as described previously (6), with the following exception. In order to decrease nonspecific binding, HA-tagged RBDs were incubated with target cells at 4°C rather than 37°C. Detection of HA-tagged receptors on the cell surface was accomplished in a similar assay using HA.11 monoclonal antibody (Covance/Babco, Richmond, Calif.), as described previously (11).
RESULTS
Stable expression of the human ortholog of PiT2 renders BHK SN-10 cells susceptible to binding and infection by A-MuLV.
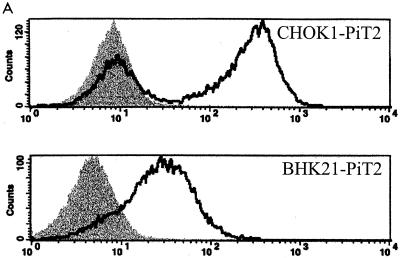
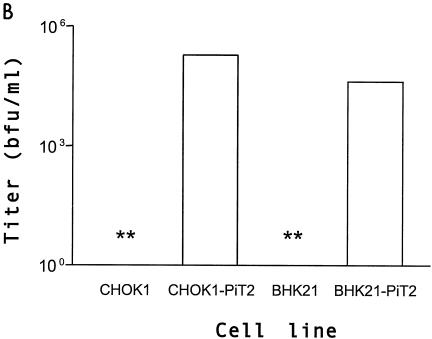
Previous studies designed to assess PiT2 A-MuLV receptor function have shown that CHOK1 cells are resistant to A-MuLV infection but can be rendered transiently susceptible to A-MuLV by calcium phosphate-mediated transfection with a PiT2 expression plasmid. One caveat to assessing A-MuLV receptor function in CHOK1 cells is that the resistance to infection of CHOK1 cells has been attributed to low endogenous receptor expression levels and/or receptor masking (30) that can be overcome by expression of endogenous receptor or by treating the cells with tunicamycin (16, 17, 35). Therefore, we sought an alternative cell line to CHOK1 for the assessment of A-MuLV binding and infectivity. Syrian hamster-derived BHK SN10 cells are resistant to A-MuLV infection, fail to bind A-MuLV envelope proteins (Fig. 1A), and are not rendered susceptible to A-MuLV following treatment with tunicamycin (35). Expression of PiT2 in CHOK1 cells resulted in A-MuLV binding. However, as seen in Fig. 1A, a portion of the CHOK1 cells that were transduced with PiT2 failed to express receptor or were modified in such a way as to be masked. Expression of PiT2 in these cells (BHK SN-10-PiT2) allows efficient binding by A-MuLV SU and renders these cells susceptible to infection by A-MuLV enveloped vectors (Fig. 1B), but not by GALV (data not shown). The data obtained from these experiments indicate that BHK SN-10 cells are a reasonable candidate cell line for studying the effects of various chimeric PiT1-PiT2 receptors on A-MuLV binding and entry.
FIG. 1.
Comparison of CHOK1 and BHK SN-10 cells stably expressing the human ortholog of PiT2. (A) Flow cytometric histograms of HA-tagged A-MuLV RBD binding. Binding was carried out at 4°C as described in Materials and Methods, followed by incubation with 5 μg of HA.11 monoclonal antibody, recognizing HA-tagged soluble A-MuLV SU, per ml. Bound tag was detected with goat anti-mouse antibody-fluorescein isothiocyanate (1:50). Shaded areas correspond to negative control cells exposed to HA-tagged A-MuLV SU; areas beneath bold lines correspond to CHOK1 or BHK SN-10 cells stably expressing PiT2 receptors exposed to HA-tagged A-MuLV SU. (B) A-MuLV titers expressed as BFU per milliliter ± the standard errors of the means from at least three independent experiments. **, titer results were zero.
Assessment of PiT1-PiT2 chimeric receptors expressed in CHOK1 and BHK SN-10 cells.
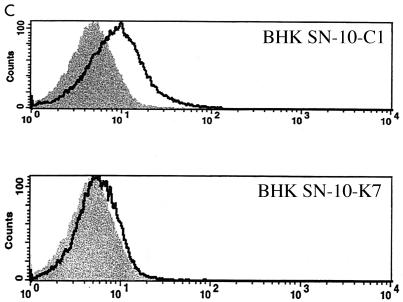
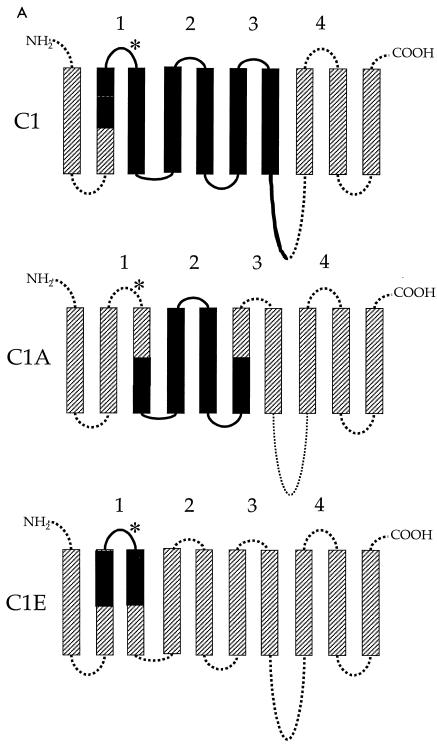
Given that expression of PiT2 in both CHOK1 and BHK SN-10 cells renders them susceptible to A-MuLV infection, expression of the same chimeric PiT1-PiT2 receptors in either cell line would be anticipated to result in similar infectivity patterns. Therefore, we used vesicular stomatitis virus G pseudotyped retroviral vectors to independently transduce CHOK1 and BHK SN-10 cells with vectors encoding two reciprocal PiT1-PiT2 chimeric proteins. Interestingly, we found that a chimeric receptor, C1 (Fig. 2A), rendered both CHOK1 (CHOK1-C1) and BHK SN-10 (BHK SN-10-C1) cells susceptible to A-MuLV infection (Fig. 2B). The C1 chimera contains residues 66 to 495 of PiT2 inserted into a PiT1 protein backbone. The nearly reciprocal chimera, K7, in which PiT2 residues 495 to 653 were substituted for the corresponding residues of PiT1, functions as a GALV receptor (5) and was functional as an A-MuLV receptor in CHOK1 cells (CHOK1-K7), yielding titers that were similar (twofold decrease) to those achieved with CHOK1-C1 cells (Fig. 2B). Surprisingly, when K7 was expressed in BHK SN-10 cells (BHK SN-10-K7), A-MuLV could not infect them (Fig. 2B) and A-MuLV binding was inefficient (Fig. 2C), suggesting that PiT2 residues at the amino terminus of PiT2 are required for A-MuLV binding and infectivity when expressed in BHK SN-10 cells.
FIG. 2.
Functional differences between PiT1-PiT2 receptor chimeras stably expressed in CHOK1 and BHK SN-10 cells. (A) Predicted topologies of chimeric receptor proteins C1 and K7 (see Materials and Methods). TMs are represented by bars (hatched bars, PiT1; solid bars, PiT2) and intra- or extracellular domains are represented by lines (dotted lines, PiT1; solid lines, PiT2). The putative ECDs are numbered 1 through 4. *, N-linked glycosylation site. (B) A-MuLV titers expressed as BFU per milliliter ± the standard errors of the means from at least three independent experiments. Solid bars, CHOK1 cells; open bars, BHK SN-10 cells). **, titer results were zero. (C) Histograms from flow cytometric analysis of BHK SN-10 cells incubated with 5 μg of HA.11 monoclonal antibody, recognizing HA-tagged soluble A-MuLV SU, per ml, followed by goat anti-mouse antibody-fluorescein isothiocyanate (1:50). The x axis represents fluorescence intensity (log scale), and the y axis represents cell number. Shaded areas correspond to negative control BHK SN-10 cells exposed to HA-tagged A-MuLV SU; areas beneath bold lines correspond to BHK SN-10 cells stably expressing chimeric PiT1-PiT2 receptors exposed to HA-tagged A-MuLV SU.
Defining regions of PiT2 that mediate A-MuLV binding and infectivity in BHK SN-10 cells.
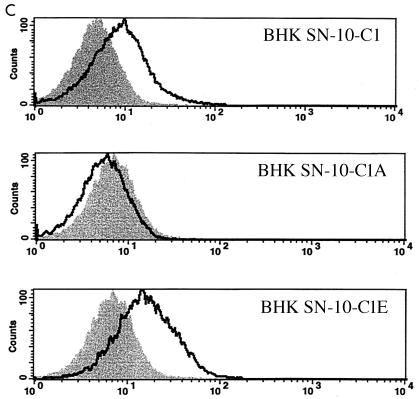
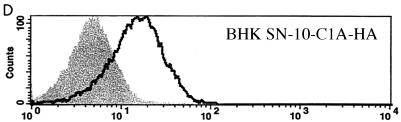
In order to further identify the region(s) of PiT2 that mediates A-MuLV binding and/or entry, additional chimeric PiT1-PiT2 receptors were constructed and expressed in BHK SN-10 cells. The first two chimeric receptors, designated C1A and C1E, divide PiT2 residues 66 to 495 of the chimeric C1 receptor into two parts (Fig. 3A). In C1A, PiT1 residues 121 to 214 were replaced with the corresponding PiT2 residues, 106 to 199. This receptor, when expressed in BHK SN-10 cells (BHK SN-10-C1A), did not function as an A-MuLV receptor and did not bind HA-tagged soluble A-MuLV envelope protein (Fig. 3B and C). In cases for which receptor binding and infectivity were both negative, verification of receptor expression at the cell surface was assessed by flow cytometric analysis. The HA-tagged C1A receptor expressed on the surfaces of BHK SN-10 cells (BHK SN-10-C1A-HA) was detected, thereby indicating that the inability of C1A to function as an A-MuLV receptor is not attributable to the lack of receptor protein expression on the cell surface (Fig. 3D). BHK SN-10 cells expressing the chimeric receptor C1E (BHK SN-10-C1E), in which residues 71 to 155 of PiT1 were replaced with residues 56 to 140 of PiT2 (these residues span regions of TMs 2 and 3 and include the first ECD), conferred infectivity and binding to A-MuLV (Fig. 3B and C). Interestingly, the A-MuLV titer with C1E (3.03 × 103 BFU/ml) was approximately 1/2-log higher than that with C1 (7.09 × 102 BFU/ml) when expressed in BHK SN-10 cells, indicating that regions outside of residues 56 to 140 of PiT2 may play a role in regulating A-MuLV infectivity. HA-tagged C1E, like other HA-tagged receptors, is expressed on the surfaces of BHK cells (data not shown). PiT2 residues 56 to 140 are sufficient to render PiT1 functional as an A-MuLV receptor.
FIG. 3.
Identification of regions within the PiT1-PiT2 C1 chimera that mediate A-MuLV binding and infectivity of BHK SN-10 cells. (A) Chimeras C1A and C1E were constructed to divide C1 into two parts (see Materials and Methods). TMs and ECDs are identified as described for Fig. 2. (B) Infections mediated by BHK SN-10 cells stably expressing C1, C1A, and C1E. A-MuLV titers were expressed as BFU per milliliter ± the standard errors of the means from at least three independent experiments. *, a titer of zero was obtained for C1A. (C) HA-tagged soluble A-MuLV SU binding to BHK SN-10 cells expressing the chimeric receptors was carried out as described for Fig. 1. (D) Direct detection of HA-tagged receptor on the cell surface was done in those cases for which no soluble SU binding or infectivity was detected (chimera C1A). BHK SN-10 cells stably expressing the C1A chimera containing a C-terminal HA tag were subjected to flow cytometric analysis in a manner similar to detection of soluble SU. The shaded area corresponds to BHK SN-10 cells lacking an HA-tagged receptor; areas beneath bold lines correspond to BHK SN-10 cells stably expressing the HA-tagged chimeric C1A receptor.
The first ECD of PiT2 is an important determinant that is required for A-MuLV binding and entry.
The chimeric receptor C1E expressed in BHK SN-10 cells confers A-MuLV binding and infectivity. In order to determine whether the PiT2 region comprising residues 71 to 155 is the minimal domain required for A-MuLV binding and entry, we constructed the following chimeric receptors: (i) C1F, in which PiT1 residues 71 to 106 are replaced with residues 56 to 91 of PiT2, spanning a portion of TM2 and the first ECD; and (ii) C1G, in which the first ECD of PiT1 (residues 81 to 106) is replaced with PiT2 ECD1 (residues 66 to 91) (Fig. 4A). HA-tagged C1F and C1G are expressed in BHK SN-10 cells at levels comparable to HA-tagged C1A (data not shown). BHK SN-10 cells expressing C1F and C1G (BHK SN-10-C1F and BHK SN-10-C1G, respectively) efficiently bind and are infected by A-MuLV (Fig. 4B and C), suggesting that the first ECD of PiT2, when placed in the context of a PiT1 backbone, is sufficient to render BHK SN-10 cells susceptible to A-MuLV binding and infectivity.
FIG. 4.
Identification of the first ECD of PiT2 as an important determinant required for A-MuLV binding and infectivity. (A) Schematic representation of PiT1-PiT2 receptor chimeras C1F and C1G, respectively. TMs and ECDs are identified as described for Fig. 2. (B) A-MuLV titers expressed as BFU per milliliter ± standard errors of the means from at least three independent experiments. (C) Binding of HA-tagged soluble A-MuLV to live BHK SN-10 cells stably expressing receptor chimera C1F or C1G was determined as described for Fig. 1.
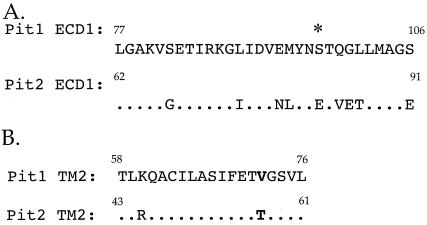
PiT1 does not function as a receptor for A-MuLV in BHK SN-10 cells (data not shown); however, chimeric receptor C1G, in which 30 residues, comprising PiT1 ECD1, are replaced with the corresponding residues from PiT2 ECD1, confers A-MuLV receptor function to PiT1. It should be noted that there is an approximately 100-fold decrease in titer between BHK SN-10-PiT2 and BHK SN-10-C1G cells (Fig. 1 and 4B). This difference is likely due to domains outside of the first ECD of PiT2 that influence the efficiency of A-MuLV binding and subsequent infection. Residues in ECD1 of PiT1 and PiT2 share 70% amino acid identity (Fig. 5A). Thus, the ability to confer A-MuLV receptor function to PiT1 appears to be mediated by, at most, nine amino acids that differ between the two ECDs. While these residues are necessary for conferring A-MuLV binding and infectivity to PiT1, it is not clear at this time if PiT2 ECD1 is the only receptor domain that plays a role in A-MuLV binding and infectivity. Further studies are required to determine how different regions of the receptor interact to confer optimal binding and infectivity.
FIG. 5.
Amino acid sequence comparison of putative ECD1 and TM2 of PiT1 and PiT2. (A) Replacement of PiT1 ECD1 with that of PiT2 renders PiT1 functional as an A-MuLV receptor. The amino acid sequence identity between ECD1 of PiT1 and PiT2 is approximately 70% and includes conservation of the N-linked glycosylation site (*). (B) Alignment of PiT1 and PiT2 TM2 reveals approximately 89% residue identity between the two domains. The bold text represents the difference between PiT1 (valine) and PiT2 (threonine) at PiT1 position 72, which is thought to play a role in determining receptor topology. Numbers indicate where the respective domains begin and end.
DISCUSSION
PiT2, the receptor for A-MuLV, is a symporter that transports inorganic phosphate and sodium ions in a tightly coupled process. We have now identified a region within PiT2 that is critical for both virus binding and entry. The first ECD and a residue present in TM2 of PiT2, when substituted for the corresponding regions of PiT1 (the C1F chimeric receptor), are sufficient to confer A-MuLV binding and infectivity characteristics to BHK SN-10 cells. Thus, 10 residue differences between PiT1 and PiT2 account for the ability of C1F but not PiT1 to function as an A-MuLV receptor. Nine of the residue differences that confer A-MuLV receptor function to PiT1 are part of the first ECD. The 10th residue difference between C1F and PiT1 is a threonine instead of a valine at position 72 of PiT1 TM2 (Fig. 5B). The C1G chimeric receptor, like C1F, contains PiT2 residues throughout ECD1, but it differs from C1F at position 72 such that TM2 is composed entirely of PiT1 residues. This single residue difference between C1F and C1G results in an approximately 1-log decrease in A-MuLV infectivity and a diminished capacity for C1G to function as an A-MuLV receptor. Interestingly, there is no apparent difference in the binding capacities of C1F and C1G, suggesting that binding results may not directly correlate with titer values.
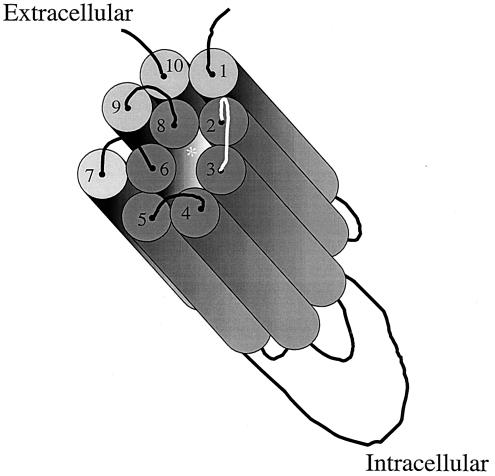
TM2 has been postulated to be one of six amphipathic alpha helices that make up a pore structure within PiT2 (19). These TMs are hypothetically arrayed such that their hydrophobic sides face toward the lipid bilayer of the plasma membrane while their hydrophilic sides form a pore through which inorganic phosphate is transported into the cell. The substitution of a hydrophilic threonine for a hydrophobic valine could alter the positioning of TM2 such that it compromises its contribution to the architecture of the pore, resulting in the reduced A-MuLV binding and titer observed with BHK SN-10 cells expressing C1G (Fig. 5 and 6). In addition, ECD1 is proximal to the region of TM8 that is proposed to harbor a Na+ binding domain (4); therefore, it is conceivable that alterations in ECD1 may also affect receptor binding function by blocking Na+ binding (Fig. 6). The substitution of PiT2 residues at positions 72, 82, 89, 93, 94, 97, 99 to 101, and 106 in ECD1 of PiT1 is requisite for A-MuLV receptor function. It remains unclear whether residues 66 to 91 constitute the sole PiT2 A-MuLV binding domain or if a second domain conserved between PiT1 and PiT2 is required for A-MuLV binding.
FIG. 6.
Hypothetical aqueous pore formed within the PiT receptor by the clustering of six TM amphipathic helices. Each cylinder represents a TM region, with the extracellular and intracellular domains represented as curved lines or loops. When the amphipathic TMs (2, 3, 4, 5, 6, and 8, shown as dark gray cylinders) are oriented so that their hydrophobic sides face the membrane lipid environment and their hydrophilic sides face inward, they form a hexagonal pore consisting of these six alpha helical segments of the PiT1 transporter/receptor. This model is adapted from that proposed by Olah et al. (19) and updated based on the revised topology proposed by Farrell et al. (6). The asterisk represents the position of a proposed Na+ binding domain (4).
All of the gammaretrovirus receptors identified to date are electrochemical potential-driven transport systems that utilize a carrier-mediated process to catalyze uniport (transport of a single species), antiport (transport of two species in opposite directions), or symport (two species transported together in the same direction) of various solutes (7, 24). It is still unclear why gammaretroviral entry requirements converge on this category of transmembrane solute transporters, given the wide variety of other transport systems (e.g., channels, pores, group translocators, and primary active transporters) as well as the multitude of other types of cell surface proteins available on cells.
A second intriguing feature of all gammaretrovirus receptor proteins is the presence of N-linked oligosaccharides in their extracellular domains. PiT1, a receptor for GALV, woolly monkey virus, 10A1 MuLV, and feline leukemia virus type B (FeLV-B), contains an N-linked glycosylation site in ECD1 (6). The receptor for FeLV-C (23, 29) is a glycoprotein, as is XPR, the receptor for polytropic and xenotropic MuLVs (1, 31, 37). In many cases, the glycosylated ECD has been implicated as crucial for virus entry. For example, the third ECD of mCAT, the receptor for E-MuLV, has been demonstrated to be critical for E-MuLV binding and entry. In addition, it has been shown that N-linked glycosylation within ECD3 blocks access to the mCAT binding site (reviewed in reference 20). More recently, ASCT, the receptor for feline endogenous virus (RD114), baboon endogenous virus, human endogenous retrovirus type W, simian retroviruses, avian reticuloendotheliosis virus, and avian spleen necrosis virus, has been shown to contain critical N-linked oligosaccharides present in the second ECD that control retroviral receptor utilization of ASCT (15). Herein we reported that the first ECD of PiT2 plays a critical role in A-MuLV binding and entry and that this domain also contains an N-linked glycosylation site (26). Taken together, these observations suggest that all known gammaretrovirus receptors function as electrochemical potential-driven porters and contain an N-glycosylated ECD. Thus, it remains to be determined if glycosylation of the first ECD of the PiT transporters blocks virus binding and entry and if Na+ binding exerts any regulatory effects on A-MLV receptor function.
Acknowledgments
We are grateful to Howard Mostowski (CBER/FDA) for flow cytometric binding analysis and Julie Overbaugh (Fred Hutchinson Cancer Center) for the HA-tagged viral envelope plasmids. We also thank Nidia Oliveira for critical comments on the manuscript.
REFERENCES
- 1.Battini, J. L., J. F. Rasko, and A. D. Miller. 1999. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 96:1385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaudry, G. J., and M. V. Eiden. 1997. Mutational analysis of the proposed gibbon ape leukemia virus binding site in PiT1 suggests that other regions are important for infection. J. Virol. 71:8078-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaudry, G. J., C. Schmitz, Y.-T. Ting, K. B. Farrell, C. J. Petropoulos, Y. S. Lie, and M. V. Eiden. 1999. Molecular characterization of the gibbon ape leukemia virus (GALV) receptor homologs (HaPIT1 and HaPIT2), present in CHOK1 cells, reveals the basis for their failure to function as GALV receptors. J. Virol. 73:2916-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deguchi, Y., I. Yamato, and Y. Anraku. 1990. Nucleotide sequence of gltS, the Na+/glutamate symport carrier gene of Escherichia coli B. J. Biol. Chem. 265:21704-21708. [PubMed] [Google Scholar]
- 5.Eiden, M. V., K. B. Farrell, and C. A. Wilson. 1996. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J. Virol. 70:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farrell, K. B., J. L. Russ, R. K. Murthy, and M. V. Eiden. 2002. Reassessing the role of region A in PiT1-mediated viral entry. J. Virol. 76:7683-7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hein, S., V. Prassolov, Y. Zhang, D. Ivanov, J. Lohler, S. R. Ross, and C. Stocking. 2003. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cercivolor M813 murine leukemia virus. J. Virol. 77:5926-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johann, S. V., J. J. Gibbons, and B. O'Hara. 1992. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J. Virol. 66:1635-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johann, S. V., M. V. Zeijl, J. Cekleniak, and B. O'Hara. 1993. Definition of a domain of GLVR1 which is necessary for infection by gibbon ape leukemia virus and which is highly polymorphic between species. J. Virol. 67:6733-6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kavanaugh, M. P., D. G. Miller, W. Zhang, W. Law, S. L. Kozak, D. Kabat, and A. D. Miller. 1994. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc. Natl. Acad. Sci. USA 91:7071-7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauring, A. S., M. M. Anderson, and J. Overbaugh. 2001. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for the in vivo tropism of immunodeficiency-inducing variants. J. Virol. 75:8888-8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leverett, B., K. B. Farrell, M. V. Eiden, and C. A. Wilson. 1998. Amphotropic murine leukemia virus receptor function is altered by a combination of amino acid residues in the putative second extracellular domain. J. Virol. 72:4956-4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundorf, M. D., F. S. Pedersen, B. O'Hara, and L. Pedersen. 1999. Amphotropic murine leukemia virus entry is determined by specific combinations of residues from receptor loops 2 and 4. J. Virol. 73:3169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundorf, M. D., F. S. Pedersen, B. O'Hara, and L. Pedersen. 1998. Single amino acid insertion in loop 4 confers amphotropic murine leukemia virus receptor function upon murine PiT1. J. Virol. 72:4524-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marin, M., D. Lavillette, S. M. Kelly, and D. Kabat. 2003. N-linked glycosylation and sequence changes in a critical negative control region of the ASCT1 and ASCT2 neutral amino acid transporters determine their retroviral receptor function. J. Virol. 77:2934-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, D. G., and A. D. Miller. 1993. Inhibitors of retrovirus infection are secreted by several hamster cell lines and are also present in hamster sera. J. Virol. 67:5346-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, D. G., and A. D. Miller. 1992. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J. Virol. 66:78-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Hara, B., S. V. Johann, H. P. Klinger, D. G. Blair, H. Rubinson, K. J. Dunne, P. Sass, S. M. Vitek, and T. Robins. 1990. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1:119-127. [PubMed] [Google Scholar]
- 19.Olah, Z., C. Lehel, W. B. Anderson, M. V. Eiden, and C. A. Wilson. 1994. The cellular receptor for gibbon ape leukemia virus is a novel high affinity phosphate transporter. J. Biol. Chem. 269:25426-25431. [PubMed] [Google Scholar]
- 20.Overbaugh, J., A. D. Miller, and M. V. Eiden. 2001. Receptors and entry cofactors for retroviruses include single and multiple transmembrane spanning proteins as well as newly described GPI-anchored and secreted proteins. Mol. Biol. Rev. 65:371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pedersen, L., S. Johann, M. van Zeijl, F. Pedersen, and B. O'Hara. 1995. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J. Virol. 69:2401-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen, L., M. V. Zeijl, S. V. Johann, and B. O'Hara. 1997. Fungal phosphate transporter serves as a receptor backbone for gibbon ape leukemia virus. J. Virol. 71:7619-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quigley, J., C. Burns, M. Anderson, E. Lynch, K. Sabo, and J. Overbaugh. 2000. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood 95:1093-1099. [PubMed] [Google Scholar]
- 24.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64:354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salaün, C., E. Gyan, P. Rodrigues, and J. M. Heard. 2002. PiT2 assemblies at the cell surface are modulated by extracellular inorganic phosphate concentration. J. Virol. 76:4304-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salaün, C., P. Rodrigues, and J. M. Heard. 2001. Transmembrane topology of PiT-2, a phosphate transporter-retrovirus receptor. J. Virol. 75:5584-5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneiderman, R. D., K. B. Farrell, C. A. Wilson, and M. V. Eiden. 1996. The Japanese feral mouse PiT1 and PiT2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J. Virol. 70:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tailor, C., and D. Kabat. 1997. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J. Virol. 71:9383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tailor, C., B. J. Willett, and D. Kabat. 1999. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter superfamily. J. Virol. 73:6500-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tailor, C. S., A. Nouri, and D. Kabat. 2000. Cellular and species resistance to murine amphotropic, gibbon ape, and feline subgroup C leukemia viruses is strongly influenced by receptor expression levels and by receptor masking mechanisms. J. Virol. 74:9797-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozal, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tailor, C. S., Y. Takeuchi, B. O'Hara, S. V. Johann, R. A. Weiss, and M. K. Collins. 1993. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J. Virol. 67:6737-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ting, Y.-T., C. A. Wilson, K. B. Farrell, G. J. Chaudry, and M. V. Eiden. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite their bearing functional receptors. J. Virol. 72:9453-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Zeijl, M., S. V. Johann, E. Cross, J. Cunningham, R. Eddy, T. B. Shows, and B. O'Hara. 1994. An amphotropic virus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 91:1168-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, C., and M. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, C. A., M. V. Eiden, W. B. Anderson, C. Lehel, and Z. Olah. 1995. The dual-function hamster receptor for amphotropic murine leukemia virus, 10A1 MuLV and gibbon ape leukemia virus is a phosphate symporter. J. Virol. 69:534-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, Y., L. Guo, S. Xu, H. Ca, T. Kitamura, K. Hunter, and J. Cunningham. 1999. Receptors for polytropic and xenotropic mouse leukemia viruses encoded by a single gene at Rmc1. Nat. Genet. 21:216-219. [DOI] [PubMed] [Google Scholar]