Abstract
Background
The Cancer Genome Atlas (TCGA) project is a large-scale effort with the goal of identifying novel molecular aberrations in glioblastoma (GBM).
Methods
Here, we describe an in-depth analysis of gene expression data and copy number aberration (CNA) data to classify GBMs into prognostic groups to determine correlates of subtypes that may be biologically significant.
Results
To identify predictive survival models, we searched TCGA in 173 patients and identified 42 probe sets (P = .0005) that could be used to divide the tumor samples into 3 groups and showed a significantly (P = .0006) improved overall survival. Kaplan-Meier plots showed that the median survival of group 3 was markedly longer (127 weeks) than that of groups 1 and 2 (47 and 52 weeks, respectively). We then validated the 42 probe sets to stratify the patients according to survival in other public GBM gene expression datasets (eg, GSE4290 dataset). An overall analysis of the gene expression and copy number aberration using a multivariate Cox regression model showed that the 42 probe sets had a significant (P < .018) prognostic value independent of other variables.
Conclusions
By integrating multidimensional genomic data from TCGA, we identified a specific survival model in a new prognostic group of GBM and suggest that molecular stratification of patients with GBM into homogeneous subgroups may provide opportunities for the development of new treatment modalities.
Keywords: comparative genomic hybridization, EMT, gene expression, glioblastoma, prognostic marker, TCGA
Glioblastomas (GBMs) are aggressive brain tumors for which there are few prognostic markers and predictors of therapeutic response.1–3 Although clinical and pathological subtype study of GBM has been increasing in recent years,4–6 identification of the prognostication and targeting of treatment has been increasing only recently. Gene expression studies have identified several distinct GBM subtypes based on an intrinsic gene list that differentiate GBMs into 4 separate groups: proneural, neural, classical and mesenchymal.1,7 The proneural class was described as the concomitant overexpressing the p53 and IDH1 mutation. This subtype was significantly younger. The neural subtype did not show significantly higher or lower rates of mutations. This normal-like subgroup was characterized by the expression of several gene types of the brain's noncancerous nerve cells, or neurons. The classical subtype expresses abnormally high levels of epidermal growth factor receptor (EGFR) and EGFR vIII mutation, whereas TP53 is not mutated in this classical GBM tumors. The mesenchymal subtype was reflected by the most frequent mutations in the NF1 tumor suppressor gene and an epithelial-to-mesenchymal transition (EMT). However, there was no association of GBM subtype with a trend toward longer survival among patients with a signature. Although several etiologic factors have been established as important events for each subtype of GBM, these subtype analyses show no favorable patient outcomes and overall survivals were not even favorable for neural and proneural subtypes.7 Because subtypes and clinical correlations have been commonly used, questions remain as to whether these groups are clinically relevant. Thus, studies have been needed to associate prognostic and etiologic importance with clinical features, such as disease subtype, and clinical outcome.
In this study, we systematically evaluated the biologically interesting features of these molecular subtypes and their relationship with GBM survivorship from The Cancer Genome Atlas (TCGA)4 and validated our results using other independent glioma datasets (eg, GSE4290 dataset8). Finally, we related our results to distinct patterns of EMT activation in different subtypes and characterized their prognostic effects. Therefore, the expression of the genes in this study clearly revealed the evidence that elucidates their functional significance between the molecular environments. Our findings also have important implications for glioma biology and heterogeneity and possible therapeutic intervention for patients with GBM.
Materials and Method
Sample Description
We used public TCGA (http://cancergenome.nih.gov/) data repositories as our primary source of samples. To analyze the data generated by TCGA, we directly accessed the input data (gene expression of the Genechip Human Genome HT-HG-U133A of TCGA and CNA of Agilent Human Genome CGH Microarray 244A). In total, 254 tumors having clinical data (date of download: September 2009) were profiled for class discovery and survival analysis. Survival was defined as the time interval from surgery until the date of death. For validation, we used REMBRANDT (http://caintegrator-info.nci.nih.gov/rembrandt) (HG-U133plus 2.0 of GSE4290).
Statistics for Classification
The CEL files were reprocessed using the R statistical computing platform and packages from Biconductor bioinformatics software project,9 and a robust multiarray average intensity on a log-squared scale was generated for each probe set. We identified genes that were differentially expressed between the 2 classes with use of a random-variance t test.10 This method is incorporated into BRB-ArrayTools, version 3.1.11 Genes were considered to have statistically significant differences in expression if P < .001. We performed hierarchical clustering based on the most variably expressed genes using the Euclidean distance as the similarity metric and the complete linkage method as the between-cluster distance metric. To build an aggregate CGH profile based on the frequency of a particular copy number aberration segment in the population of samples, we directly accessed the dataset (CNA of Agilent Human Genome CGH Microarray 244A) from the TCGA database and imported the normalized data into the Nexus 4.0 copy number analysis program (BioDiscovery Inc., El Segundo, CA), and CNA regions were called using BioDiscovery's rank segmentation algorithm.12
IPA and Multivariate Analysis
We analyzed the gene ontology, canonical pathways, and functional networks with use of tools from the Ingenuity Pathways Analysis tools (Ingenuity Systems, Mountain View, CA). In addition, we used Fisher's exact test to determine the significance of the frequency differences. Kaplan-Meier survival analysis was performed to estimate the survival distributions and the log-rank test to assess the statistical significance of the differences between the stratified survival groups using GraphPad Prism (version 5, GraphPad Software Inc., San Diego, CA).13 Logistic regression using SPSS software, version 11.0 (SPSS, Chicago, IL), was applied for multivariate analysis of significant variables for predicting overall survival patterns.14 The differences between covariates were tested using log-rank analyses. The joint effect of different covariates was assessed using multivariate Cox's regression. Differences were considered to be statistically significant when P < .05.
Results
A Model Building Phase Using TCGA Dataset
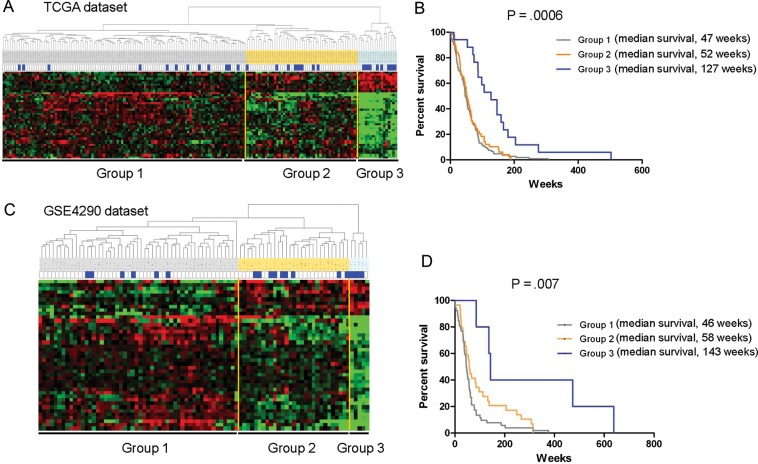
Of the 173 patients with GBM in the TCGA dataset, 142 were considered to be short-term survivors (<2 years) and 31 were considered to be long-term survivors (≥2 years). We first sought to find probe sets that are differentially expressed in 2 groups. Genes with an expression ratio that differed by a factor of at least 1.5-fold were selected (8015 gene features). We generated 42 probe sets by applying the 2-sample t test (P = .0005) (Table 1). Hierarchical clustering analysis of the expression data from the 173 patients samples revealed 3 distinctive types of gene expression patterns (Fig. 1A). Of note, 17 GBMs could tightly group on the basis of these expression patterns, whereas the remaining GBMs had wide variations, which led us to classify them in a second group. The 42 probe sets indicated a significantly improved overall survival (P = .0006 by log-rank test). Kaplan-Meier plots and log-rank survival analyses (Fig. 1B) showed that the median overall survival time of group 3 was markedly longer (127 weeks) than that of groups 1 and 2 (47 and 52 weeks, respectively; ie, the molecular differences between groups 1 and 2 and group 3 were associated with differences in clinical outcomes) (Supplementary Table S1).
Table 1.
Forty-two probe sets and their frequency difference between 2 survivor populations in TCGA dataset
| Gene Symbol | Description | Frequency difference by CAN (Group 3 – Group 1 and 2) |
|
|---|---|---|---|
| Gain (%) | Loss (%) | ||
| DEPDC6 | DEP domain containing 6 | 34.87 | −0.95 |
| RPRM | reprimo, TP53 dependent G2 arrest mediator candidate | −1.97 | 0 |
| NET1 | neuroepithelial cell transforming gene 1 | 18.09 | −72.04 |
| NET1 | neuroepithelial cell transforming gene 1 | – | – |
| WAC | WW domain containing adaptor with coiled-coil | 24.34 | −79.61 |
| March8 | membrane-associated ring finger (C3HC4) 8 | 6.25 | −82.89 |
| AI054381 | Transcribed locus | – | – |
| REPS2 | RALBP1 associated Eps domain containing 2 | −1.32 | 11.84 |
| ZNF609 | zinc finger protein 609 | 4.28 | −7.89 |
| KLF13 | Kruppel-like factor 13 | 0 | 19.41 |
| IL8 | interleukin 8 | 4.28 | 6.25 |
| ADM | adrenomedullin | −0.66 | 44.74 |
| PDPN | podoplanin | 0.33 | 4.61 |
| IGFBP2 | insulin-like growth factor binding protein 2, 36 kDa | −2.63 | −1.32 |
| MDK | midkine (neurite growth-promoting factor 2) | 5.59 | 12.17 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 0 | 5.59 |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | −4.93 | 1.97 |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | – | – |
| ACOX2 | acyl-Coenzyme A oxidase 2, branched chain | −3.29 | 9.87 |
| TAGLN2 | transgelin 2 | 2.63 | 0 |
| SLC43A3 | solute carrier family 43, member 3 | −1.32 | 5.26 |
| LGALS8 | lectin, galactoside-binding, soluble, 8 (galectin 8) | −1.64 | −1.97 |
| LGALS8 | lectin, galactoside-binding, soluble, 8 (galectin 8) | – | – |
| DYNLT3 | dynein, light chain, Tctex-type 3 | −0.66 | 11.84 |
| KIAA0323 | KIAA0323 | −0.66 | −16.78 |
| TFRC | transferrin receptor (p90, CD71) | −5.92 | 0.99 |
| KIAA0495 | KIAA0495 | −3.95 | 12.5 |
| FBXO17 | F-box protein 17 | −28.29 | 14.8 |
| TMEM22 | transmembrane protein 22 | 0.99 | –3.95 |
| LOC390940 | similar to R28379_1 | – | – |
| MT1E | metallothionein 1E | 19.74 | –9.21 |
| DCTD | dCMP deaminase | 3.62 | 14.14 |
| FLJ11286 | hypothetical protein FLJ11286 | – | – |
| C13orf18 | chromosome 13 open reading frame 18 | −0.66 | 15.79 |
| C13orf18 | chromosome 13 open reading frame 18 | – | – |
| HOMER1 | homer homolog 1 (Drosophila) | −4.61 | 3.62 |
| FAM3C | family with sequence similarity 3, member C | −61.51 | −0.66 |
| CASP3 | caspase 3, apoptosis-related cysteine peptidase | 3.62 | 13.49 |
| NSUN5 | NOL1/NOP2/Sun domain family, member 5 | −70.22 | −0.66 |
| NSUN5 | NOL1/NOP2/Sun domain family, member 5 | – | – |
| PDLIM3 | PDZ and LIM domain 3 | 3.62 | 13.49 |
| MT1M | metallothionein 1M | 19.74 | −9.21 |
Fig. 1.
Hierarchical clustering analysis. (A) Thirty-one patients with GBM in the TCGA dataset were classified as long-term survivors (≥2 years), and the remaining 142 were classified as short-term survivors (<2 years). Forty-two probe sets were generated using a 2-sample t-test (P = .0005) between the 2 survivor populations. The 42 probe sets are presented using hierarchical clustering in matrix format, where rows represent individual genes and columns represent each tissue. Each cell in the matrix represents the expression level of a gene in an individual tissue. Red and green cells reflect high and low expression levels, respectively. Each blue bar represents a ≥2-year survival among the patients with GBM. (B) Kaplan-Meier plot of overall survival among patients with GBM who were grouped on the basis of expression of the 42 probe sets. The difference between the groups was significant, with group 3 having the longest survival times (P = .0006, log-rank test). (C) Hierarchical clustering analysis was applied to gene expression data from the GSE4290 dataset based on the 42 probe sets. Nineteen patients with GBM were classified as long-term survivors (≥2 years), and the remaining 67 were classified as short-term survivors (<2 years). (D) Kaplan-Meier plot of overall survival among patients with GBM in the GSE4290 dataset who were grouped together on the basis of expression of the 42 probe sets.
Validation Models Using GSE4290 and the TCGA Second Batch Dataset
To establish the reproducibility of the 42 probe sets, we assessed the generality of the 42 probe sets' expression signatures with other independent public GBM gene expression datasets (eg, GSE4290 dataset15). In the GSE4290 dataset (67 short-term survivors and 19 long-term survivors) (Supplementary Table S2), the expression levels of the 42 probe sets were highly consistent with the TCGA dataset (Fig. 1C and D). The difference between the groups was significant, with group 3 having the longest survival times (P = .007, log-rank test). Of interest, in the GSE4290 datasets for astrocytomas (II and III) and oligodendrogliomas (II and III), no correlations were identified with the 42 probe sets (Supplementary Fig. S1 and Table S3). It is posssible that these tumors do not have the more aggressive group 1 or group 2 classifications and all could be classified as group 3. Thus, we clustered the astrocytomas and oligodendrogliomas with the GBMs to see whether the lower grade gliomas cluster with group 3 (Supplementary Fig. S2). The comparisons showed that they are closer to group 3 than the other groups, suggesting that the 42 probe set signature was specific for lower grade gliomas and group 3 GBMs.
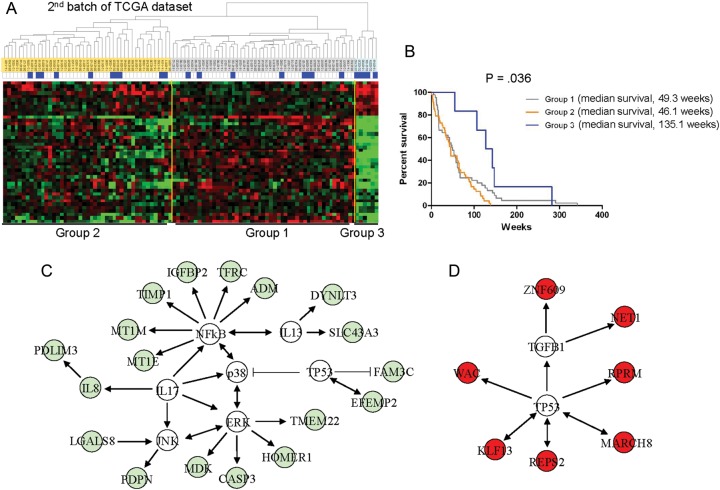
Since our initial analysis (173 samples), TCGA data set has expanded; thus, we validated our model by analyzing a second batch of 100 patients. Twenty-three patients were classified as long-term survivors (≥2 years), and the remaining 77 as short-term survivors (<2 years). It showed a similar clustering pattern, and the difference between the groups was significant (P = .036, log-rank test), confirming that the 42 probe sets could be used to segregate 3 distinctive types of gene expression patterns with use of a supervised hierarchical clustering (Fig. 2A and B, and Supplementary Table S4).
Fig. 2.
Hierarchical clustering analysis of the second batch of TCGA dataset. (A) The data are presented in matrix format, in which rows represent individual genes and columns represent each tissue. (B) Kaplan-Meier plot of overall survival among patients with GBM in the second batch of TCGA data grouped on the basis of gene expression profiling. (C) Deposition of the 42 probe sets into the Ingenuity knowledge database. Eighteen genes (green color) down-regulated in group 3 were mapped to highly significant networks of the NFkB, P38, ERK, JNK, and IL-17 signaling pathways. (D) Seven genes (red color) up-regulated in group 3 were mapped to p53 signaling pathways. Genes not included in the 42 probe sets are shown in white.
To compare our signatures with data on previously described GBM subtypes, we assessed the expression levels of the 42 probe sets with 2 GBM datasets.1,7 It appears that the expression signatures highly resemble proneural subtype observed in Phillips et al (Supplementary Fig. S3). However, there are 20 grade III gliomas of 30 patient samples in proneural subtype,1 not all of which are grade IV glioblastoma, showing that the 42 probe sets may have association of GBM subtype with a trend toward longer survival among patients. In this comparison, IL-8 was missed in the 42 probesets. On the other hand, the analysis of the TCGA data into biologically based subtypes was not prognostic.7 Although HOMER1 showed opposite expression into proneural, the expression signatures of the 42 probe sets are similar to proneural subtype observed in Verhaak et al (Supplementary Fig. S4A). Most (168 of 173) samples in the set are also part of the article by Verhaak et al. We used the data from our set to make the table for the 168 samples (Supplementary Fig. S4B). Group 3 is enriched for proneural, although not all of the proneural, but groups 1 and 2 are a mixture of the other Verhaak subtypes, suggesting that the group classification to identify survival-based subtypes could be different but similar with characteristics from the previously published subtypes.
In addition, to gain an integrated view of the biological insights into the differential expression of this probe set, we categorized the 42 survival-related probe sets according to the gene ontology (GO) database and analyzed the probe sets using the Ingenuity Pathways knowledge database (Fig. 2C and D). An initial analysis of the GO database revealed significant down-regulation of the genes involved in apoptosis (20.6%; P < .0000034), cellular movement (10.3%; P < .0005), and inflammatory disorder (24.1%; P < .011). In network analysis based on predetermined knowledge about individually modeled relationships between genes, we identified 2 highly significant, overlapping networks in the dataset. The top-scoring network built around NFkB, P38, ERK, JNK, and IL-17 displayed high-level functions in cell death and inflammatory response and included several interacting genes, such as IL-8 and TIMP1. This analysis also identified a highly interconnected network of aberrations with p53 pathway and suggested that these pathways might have important roles in long-term survival in group 3. In biologic pathway analysis, we found that the IL-17 canonical pathway was significantly associated with the molecular pathway (P < .0084). This analysis identified a highly interconnected network of aberrations, including NFkB, p38, ERK, JNK, IL-13, and IL-17, with the down-regulated probes in group 3,16 and an interconnection between TP53 and the up-regulated probes in group 3, which suggests that these interconnections might have important roles in long-term survival among patients in group 3.
We next used 5 statistical methods to determine whether gene expression patterns could predict the signature-based survival patterns in the 3 groups of patients with GBM: linear discriminator analysis, support vector machines, nearest centroid, nearest neighbor, and compound covariate predictor.17 Again, we identified the 42 probes using the 5 different algorithms. These genes were combined to form a series of classifiers that estimate the probability that a specific tumor belongs to a subgroup. To validate the statistical methods used in this study, the number of genes in the classifiers was optimized to minimize classification errors during the leave-one-out cross-validation of the tumors.11 Hierarchical clustering analyses with all 5 models showed consistent prediction patterns. These results showed that the GBM groups with the same survival rate could be identified by a 42 probe set expression signature (Supplementary Fig. S5).
Genomic Aberrations Using CNA Analysis
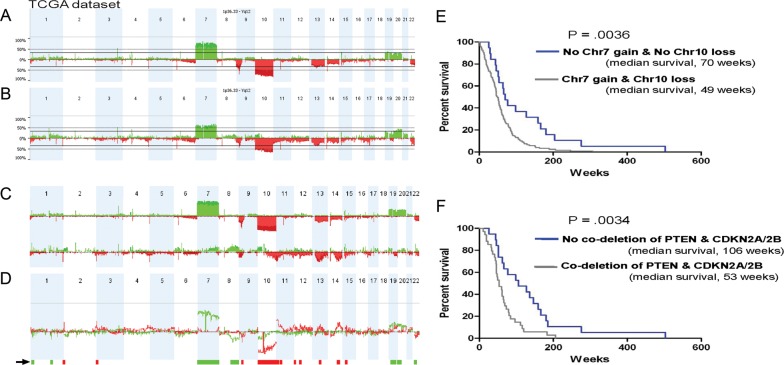
We performed a parallel CNA analysis of the Agilent 244K oligonucleotide array data on 170 matched GBM tumors (140 from short-term survivors and 30 from long-term survivors) (Supplementary Table S5). The CNA profiles showed well-correlated frequencies of several known chromosomal alterations, such as Chr7 gain and Chr10 loss (Fig. 3A and B).6,18 This analysis revealed 2 distinctive CNA profiles, one for groups 1 and 2 and a markedly different one for group 3 (Fig. 3C and D). Among the 42 probe sets, DEPDC6, NET1, WAC, MARCH8, ADM, FBXO17, FAM3C, and NSUN5 had >25% frequency difference in their CNA profiles, and these genes were associated with the survival characteristics in each group. Furthermore, most of the amplified and overexpressed genes (NET1, DEPDC6, and WAC) and deleted and down-regulated genes (ADM, FBXO17, FAM3C, and NSUN5) were found in group 3. Of interest, there was a strong correlation between the frequency differences defined by CNA and the gene expression predictors in each survival group (Table 1). Because frequent deletions and mutations of the PTEN lipid phosphatase tumor suppressor and deletion of the CDKN2A/B locus are a prominent signature in human GBM,19 we compared associations between survival and the status of Chr7 gain and Chr10 loss and the codeletion of PTEN and CDKN2A/2B. A survival analysis of the CNA data showed that the median survival in a group of patients with no Chr10 loss and no Chr7 gain was longer (70 weeks) than that in the other groups of patients with Chr10 loss and Chr7 gain (49 weeks) (Fig. 3E). The difference between the groups was significant (P = .0036, log-rank test). In addition, results divided patients into either a longer-term survival group (median, 106 weeks), those without codeletion of CDKN2A/2B and PTEN, or a shorter-term survival group (median, 53 weeks), those with codeletion of CDKN2A/2B and PTEN. The difference between groups was statistically significant (P = .0034, log-rank test) (Fig. 3F).
Fig. 3.
Genome-wide profile of representative TCGA GBM tumors; 30 were classified as long-term survivors (≥2 years) and the remaining 140 as short-term survivors (<2 years). (A) Genome-wide overview of CGH array data that graphically depicts changes in copy number for short-term survivors. On the x-axis, the numbers are organized along the whole chromosomes, and the y-axis depicts frequency of CNA. Green represents gain, and red represents loss. (B) Genome-wide overview of CGH array data for long-term survivors. (C) A genome-wide overview of the CGH array data that graphically depicts changes in copy number for groups 1 and 2 (upper panel) and group 3 (lower panel). On the x-axis, the numbers are organized along the 22 chromosomes, and the y-axis depicts the frequency of CNA. Green represents gain, and red represents loss. (D) Differences in the CNA profiles between groups 1 and 2 and group 3. Green represents gain differences, and red represents loss differences between 2 groups. The arrow indicates regions that are differentially altered between groups 1 and 2 and group 3 (P < .05; frequency >25%). (E) Kaplan-Meier plot of overall survival among patients with GBM grouped on the basis of Chr7 gain and Chr10 loss. F, Kaplan-Meier plot of overall survival among patients with GBM who were grouped on the basis of codeletion of PTEN and CDKN2A/2B.
EMT-Associated Changes
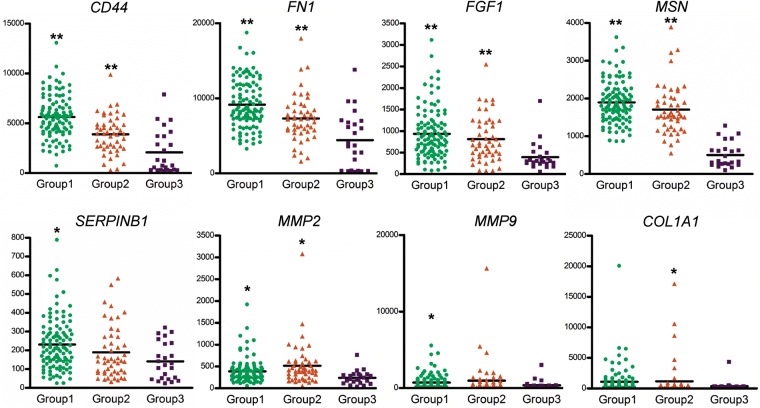
To explore the biology underlying the subgroup, we compared each group with several potentially relevant mesenchymal-associated genes.20 A molecular program of epithelial-mesenchymal transition (EMT) is frequently seen in malignant tumor with a highly invasive phenotype.21 On the basis of this information, we first investigated the expression of EMT-related genes in each group and generated 29 probe sets (15 genes) by applying the 2-sample t test (P = .05). As shown in Fig. 4 and Supplementary Fig. S6, the expression of various mesenchymal type genes was decreased in group 3, including CD44, FN1, FGF1, MSN, SERPINB1, MMP2, MMP9, and COL1A1. CD44, which is highly overexpresssed in glioblastoma and induced tumor cell growth,22 was significantly increased in group 1. Conversely, MMP2 and MMP9 were relatively overexpressed in group 2. Of interest, the differential expression of epithelial type genes, such as CDH1 and KRT18, was not found among these 3 groups. These results showed that the coexpression of these mesenchymal-associated genes suggest the presence of a molecular program of EMT in group 1 and 2.
Fig. 4.
Tumor subgroups are distinguished by the differential expression of various mesenchymal type genes. Horizontal bars denote mean values. Mesenchymal genes enriched in group 1 and group 2. Significant difference was detected by t test. *P < .05, and **P < .001, compared with the group 3 using GraphPad Prism (version 5, GraphPad Software Inc., San Diego, CA).
Statistical Modeling Using Multivariate Cox Analysis
On the basis of the independent gene expression signatures, we identified patients with clinical information and CGH data and observed a significant association with overall survival in group 3. On the other hand, group 3 only had 17 samples, not all of which had survival times >2 years. Of the long-term survivors (≥2 years), 10 (58.8%) were classified in group 3. Many of the GBMs in groups 1 and 2 had overall survival >2 years (21 of 156). We sought to find probe sets that are differentially expressed between the groups 1 and 2 long-term survival patients and the group 3 long-term survival patients and generated 1088 probe sets by applying the 2-sample t test (P = .0005). Hierarchical clustering analysis of the expression data from the 31 long-term survivors showed distinctive difference in gene expression patterns (Supplementary Fig. S7), suggesting the difference in survival between the groups 1 and 2 long-term survival patients and the group 3 long-term survival patients.
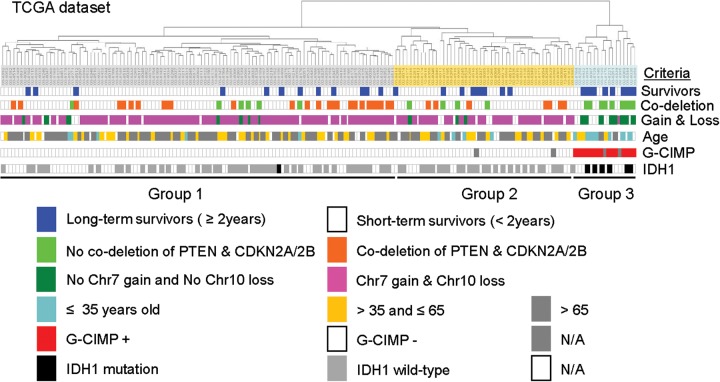
The 42 probe molecular signature and CNA data were the dominant characteristics that permitted the stratification of individuals into long-term and short-term survival groups (Fig. 5). Younger age appeared to be correlated with group 3. Eight younger age survivors, 3 intermediate age survivors, and 2 older age survivors were in group 3 (four survivors were not accessed). Although 50% of group 3 were younger age survivors, the median survival was not remarkable (Supplementary Fig. S8), not similar to either no codeletion of CDKN2A/B + PTEN or no Chr7 gain + No Chr10 loss cases. To identify the factors that were most associated with long-term survival in patients with GBM, we applied a logistic regression model for multivariate Cox hazards models 23 of the signatures with the known clinical parameters (age and sex) and molecular signature parameters (42 probe set, no Chr7 gain + no Chr10 loss, and codeletion of PTEN and CDKN2A/2B) (Table 2). All 9 covariates were included in the multivariate analysis, and we added the P values for those showing significance in multivariate analysis. Although that the difference was significant in univariate analysis with codeletion of CDNK2A/B + PTEN and Chr7 gain + Chr10 loss, we found that the 42 probe molecular signature gave additional information that was not captured by the other 2 significant covariates in predicting long-term survival (P < .018). A subset of samples in the TCGA dataset showed a glioma-CpG Island Methylator Phenotype (G-CIMP) phenotype and enriched for IDH1 mutations.24 We classified these 2 markers in the expression group (Fig. 5). The G-CIMP tumors represent 88% (15 of 17) of all group 3 and 86% (6 of 7) of IDH1 mutations. It appears that the G-CIMP tumors with IDH1 mutations had similar characteristics in group 3.
Fig. 5.
A dendrogram of the cluster analysis of the integrated GBMs and clinical factors of TCGA dataset. The expression pattern of the long-term survival group 3 is associated with several factors, compared with the short-term survival groups 1 and 2. The data are presented in matrix format, where columns represent individual genes and rows represent each tissue. Each colored bar represents each molecular signature and clinical factor.
Table 2.
Univariate and multivariate analyses for GBM among several survival factors (n = 173) in TCGA dataset
| Variable | Univariate log-rank test | Multivariate analysis | Hazard ratio |
|---|---|---|---|
| 42 probe sets | 0.0006 | 0.018 | 2.582 |
| Co-deletion of CDKN2A/2B + PTEN | 0.0034 | 0.632 | 1.081 |
| Deletion of CDKN2A/2B | 0.0457 | 0.583 | 1.068 |
| Deletion of PTEN | 0.0167 | 0.707 | 0.841 |
| Chr7 gain + No Chr10 loss | 0.0036 | 0.803 | 0.967 |
| Chr7 gain | 0.098 | 0.851 | 0.969 |
| Chr10 loss | 0.0051 | 0.883 | 0.876 |
| Age | 0.0196 | 0.031 | 1.190 |
| Sex | 0.1957 | 0.707 | 1.537 |
Discussion
Because each tumor type possesses an enormous degree of heterogeneity,25,26 different therapies depending on patients' underlying biology have been challenging. In defining and categorizing the subtype in terms of the gene expression pattern of their classifiers, there have been several studies to find a first step toward developing personalized rational therapy that targets the unique gene changes in each patient's individual tumor.27 Most recently, the 4 groups were named proneural, neural, classical, and mesenchymal, showing distinct, but overlapping, copy number alterations and gene expression,7 suggesting that the family of gliomas constitutes at least 6 major biological subtypes.27 The GBM subtypes, however, were found to show a lack of prognosis, insufficient tumor classification, limited sample size, and variation of the signature of the subtype.1,7,28,29 Through a comprehensive genomic-based classification of GBM, we identified the existence in specific tumors of a pattern of genomic alterations associated with tumor aggressiveness and a list of genes that are related with EMT process in GBM. The tumor subtypes identified in this study were different, compared with previously reported subtypes identified by expression profiling.1,7 At first, it appears that the gene expression and copy number highly resemble the proneural subtype observed in Phillips et al and Verhaak et al. However, the comparison showed that the group classification could be different but similar characteristics from the previously published subtypes and the 42 probe sets to identify survival-based subtypes may have association of GBM subtype with a trend toward longer survival among patients. Through the comparison of these 3 survival-derived subtypes with the previously published subtypes, our study shows a clear hypothesis to explain the divergence between the poor prognosis subtypes of different signaling pathways, which seem to progress through a different and independent mechanism. A subset of samples in the TCGA dataset showed a G-CIMP phenotype and enriched for IDH1 mutations.24 It had reduced copy number gains of Chr7 and fewer losses of Chr10, was a subset of the proneural mRNA subtype, and had significantly improved outcomes. Of the 24 G-CIMP tumors, 21 (87.5%) were classified in the proneural expression group. These G-CIMP tumors represent 21 (30%) of 71 of all proneural GBM tumors in Verhaak et al. In this study, it appears that the G-CIMP tumors with IDH1 mutations had similar characteristics in group 3. Thus, because the comparison between G-CIMP classification and the group 3 classification was highly overlapped, it could be very interesting to show that this potential subset of samples can be identified by both methylation subtypes and survival-based subtypes.
Our observations demonstrate the prognostic value of 42 probe sets in glioblastomas showing better outcome, compared with poor prognosis subtypes. Although this may support the notion that GBM subtypes follow different pathways to malignancy, this study shows that specific expression and CNA variation have an important role in signaling pathways implicated in gliomagenesis and predicting outcome of GBM cases. With use of 76 grade III or IV GBM samples, the 35-gene signature was identified that correlated with patient survival and was used to separate the tumors into 3 groups: proneural, mesenchymal, and proliferative.1 Later, the 38-gene set revealed consistent association with patient outcome.28 The 9-gene set was then validated and confirmed as an independent predictor of outcome. In general, the large-scale gene signature classification studies have demonstrated the heterogeneous nature of glial tumors. Thus, although these genes may indeed play a role in the biology of gliomas, their use as diagnostic markers is not yet clear, perhaps because of unrecognized molecular heterogeneity in the tumor groupings.30,31 Very recently, the heterogeneity between tumors and in individual tumors was noted to understanding the molecular aspects of tumorigenesis essential to finding effective therapies.32
By inspecting the lists of genes across the panel of data, we identified 42 unique transcripts that identify each specific tumor subtype. This gene list contains genes of known pathologic significance in GBM that were not previously recognized as targets. These included genes involved in nerve growth factor receptor signaling pathway (eg, neuroepithelial-transforming protein 1), histone H2B conserved C-terminal lysine ubiquitination (eg, WW domain containing adaptor with coiled-coil), and negative regulation of cell proliferation (eg, Kruppel-like factor 13). At present, it is unclear what the interpretation of the genes presented here should be. Understanding whether any of these targets are driver genes of aberrant tumor growth and survival in GBM is a next major challenge of our research. One subtype, which we term group 3, is distinguished by markedly better prognosis. Two poor prognoses subtypes showed more heterogeneity than the other group and activation of gene expression programs indicative of cell proliferation by expressing genes associated with EMT that has been associated with poor outcome in several tumor types.33 Recent observations demonstrate that mesenchymal transition that is associated with poor outcomes in GBM and is analogous to the epithelial-to-mesenchymal transition.28,34 Another example of aberrant gene expression that plays in the regulation of cellular properties of glioma is the phenomenon of mesenchymal drift. This means a change in transcription factor networks and epigenetic processes toward a mesenchymal signature and indicates a more aggressive phenotype.1,34 It has been noted that a series of mesenchymal-associated genes only overexpressed in a subset of primary glioblastomas but not in secondary glioblastoma, low-grade astrocytoma, or normal brain.35 Our data support a hypothesis in which overexpression of the genes may facilitate tumor aggressiveness by inducing EMT through an oncogene-mediated increase in MMP9, TGFB2, FGF1, and SERPINB1. Consistent with a recent report linking GBM to EMT,36 we also noted a strong correlation between 42 probe set expression and EMT signature. The patient population in the younger group had a uniformly better prognosis, which provides strong correlation with clinical outcome or expression information. It was shown that the older age of patients with mesenchymal subtype tumors can acquire the phenotype through accumulation of genetic or epigenetic abnormalities.37 The mesenchymal phenotype in GBM is also associated with a stem-like phenotype,38 showing its role in the regulation of stem cell pluripotency and differentiation.39 Therefore, these observations support the possibility of a fundamentally distinct mechanism, possibly facilitating EMT process in at least a subset of the poor subtypes, regardless of underlying mechanism for the biology of disease progression. Thus, drugs targeting EMT-transformed cancer stem–like cells have been promising therapies for patients with the poor prognosis molecular profile.28,40,41
Several studies have shown that losses on Chr10, loss of PTEN locus, CDKN2A homozygous deletions, and chromosome 7/7p amplification are the most frequently observed in primary glioblastomas that are associated with poor prognosis.6,42,43 Many mesenchymal-associated genes on chromosome 7 (MET, HIF-2, CAV1, CAV2, SERPINE1, PBEF, GPNMB, UPP1, MEOX2, EGFR, and SEC61G) were reported to be associated with Akt phosphorylation, antiapoptosis, hypoxia, angiogenesis, and EMT,1,35 whereas better survival groups did not have these alterations. In addition, a gain of chromosome 7 has been suggested to confer radiation resistance,44 and primary glioblastomas simultaneously express a host of transcripts typically expressed in mature mesenchymal lineage cell types.35 These findings are consistent with those of other studies that the cells undergoing a change from an epithelial to a mesenchymal phenotype after PTEN knockdown and further establishes a connection between EMT and the PI3K pathway.45,46 Downregulation of PTEN, however, is not sufficient to trigger EMT-like phenotype and metastatic properties, such as a complete loss of epithelial cell polarity and cell detachment.47 Therefore, inappropriate reactivation of the EMT process needs additional abnormalities in addition to the loss of PTEN to become fully invasive and metastastic.46,48 It was reported that, in combination with KRAS, p53, and SMAD4 mutations, loss of epithelial PTEN function leads to invasive lesions or even metastasis.49–51 Two separate studies identified the CDKN2A–CDKN2B locus as risk factors using genome-wide association studies.19,52 CDKN2A-deficient tumors yielded distinctive gene expression profiles that closely mirrored those typical of p53 deficiency, indicating that CDKN2A deficiency, like p53 deficiency, promotes EMT-associated relapse.53 In this study, the CNA data showed that the median survival in a group of patients without codeletion of CDKN2A/2B and PTEN was longer than that in the other groups of patients with codeletion of CDKN2A/2B and PTEN. Our study shows that the copy number aberration of poor subtype is strongly correlated with EMT process, which is an integrated genomic predictor model of each subtype. Thus, chromosomal deletion of the CDKN2A/2B and PTEN locus may be a better predictor of survival potency. Further confirmation of the biology of disease progression and glioma aggressiveness could offer an important insight into the ways tumor subtypes differ in their etiologies.
Supplementary Material
Funding
This study was supported by grants from the National Cancer Institute (CA 123304 to W.K.A.Y., P30 CA 016672 to MD Anderson, and P30 CA 127001 to W.K.A.Y.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest statement: None declared.
Supplementary Material
Acknowledgements
We thank Ann M. Sutton and Kathryn B. Carnes (Department of Scientific Publications, The University of Texas MD Anderson Cancer Center) for editing the manuscript. We are also indebted to Dr. Hyunsun Lim (Gangnam Severance Hospital Biomedical Research Center, Yonsei University College of Medicine) for her statistical assistance.
References
- 1.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Fukuda ME, Iwadate Y, Machida T, et al. Cathepsin D is a potential serum marker for poor prognosis in glioma patients. Cancer Res. 2005;65(12):5190–5194. doi: 10.1158/0008-5472.CAN-04-4134. [DOI] [PubMed] [Google Scholar]
- 3.Yamanaka R. Cell- and peptide-based immunotherapeutic approaches for glioma. Trends Mol Med. 2008;14(5):228–235. doi: 10.1016/j.molmed.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freire P, Vilela M, Deus H, et al. Exploratory analysis of the copy number alterations in glioblastoma multiforme. PLoS One. 2008;3(12):e4076. doi: 10.1371/journal.pone.0004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgson JG, Yeh RF, Ray A, et al. Comparative analyses of gene copy number and mRNA expression in GBM tumors and GBM xenografts. Neuro Oncol. 2009;11(5):477–487. doi: 10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Cancer Institute. REMBRANDT home page 2005. http://rembrandt.nci.nih.gov .
- 9.Zorn KK, Bonome T, Gangi L, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11(18):6422–6430. doi: 10.1158/1078-0432.CCR-05-0508. [DOI] [PubMed] [Google Scholar]
- 10.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19(18):2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 11.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-ArrayTools. Cancer informatics. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 12.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 13.Alonso MM, Fueyo J, Shay JW, et al. Expression of transcription factor E2F1 and telomerase in glioblastomas: mechanistic linkage and prognostic significance. J Natl Cancer Inst. 2005;97(21):1589–1600. doi: 10.1093/jnci/dji340. [DOI] [PubMed] [Google Scholar]
- 14.Kim SH, Kim H, Kim TS. Clinical, histological, and immunohistochemical features predicting 1p/19q loss of heterozygosity in oligodendroglial tumors. Acta Neuropathol. 2005;110(1):27–38. doi: 10.1007/s00401-005-1020-x. [DOI] [PubMed] [Google Scholar]
- 15.Li A, Walling J, Ahn S, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kehlen A, Thiele K, Riemann D, Rainov N, Langner J. Interleukin-17 stimulates the expression of IkappaB alpha mRNA and the secretion of IL-6 and IL-8 in glioblastoma cell lines. J Neuroimmunol. 1999;101(1):1–6. doi: 10.1016/s0165-5728(99)00111-3. [DOI] [PubMed] [Google Scholar]
- 17.Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9(3):505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 18.Maher EA, Brennan C, Wen PY, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66(23):11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 19.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi E, Nagano O, Ishimoto T, et al. Tumor necrosis factor-alpha regulates transforming growth factor-beta-dependent epithelial-mesenchymal transition by promoting hyaluronan-CD44-moesin interaction. J Biol Chem. 2010;285(6):4060–4073. doi: 10.1074/jbc.M109.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moody SE, Perez D, Pan TC, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8(3):197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 22.Weber GF, Ashkar S, Glimcher MJ, Cantor H. Receptor-ligand interaction between CD44 and osteopontin (Eta-1) Science. 1996;271(5248):509–512. doi: 10.1126/science.271.5248.509. [DOI] [PubMed] [Google Scholar]
- 23.Lamborn KR, Chang SM, Prados MD. Prognostic factors for survival of patients with glioblastoma: recursive partitioning analysis. Neuro Oncol. 2004;6(3):227–235. doi: 10.1215/S1152851703000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loeper S, Romeike BF, Heckmann N, et al. Frequent mitotic errors in tumor cells of genetically micro-heterogeneous glioblastomas. Cytogenetics and Cell Genetics. 2001;94(1–2):1–8. doi: 10.1159/000048773. [DOI] [PubMed] [Google Scholar]
- 26.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 27.Riddick G, Fine HA. Integration and analysis of genome-scale data from gliomas. Nature reviews. Neurology. 2011;7(8):439–450. doi: 10.1038/nrneurol.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colman H, Zhang L, Sulman EP, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan X, Ma L, Yi D, et al. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci USA. 2011;108(4):1591–1596. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nature Reviews. Neuroscience. 2011;12(9):495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]
- 31.Betensky RA, Louis DN, Cairncross JG. Influence of unrecognized molecular heterogeneity on randomized clinical trials. J Clin Oncol. 2002;20(10):2495–2499. doi: 10.1200/JCO.2002.06.140. [DOI] [PubMed] [Google Scholar]
- 32.Jones TS, Holland EC. Molecular pathogenesis of malignant glial tumors. Toxicologic pathology. 2011;39(1):158–166. doi: 10.1177/0192623310387617. [DOI] [PubMed] [Google Scholar]
- 33.Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi: 10.1038/nrc2620. [DOI] [PubMed] [Google Scholar]
- 34.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tso CL, Shintaku P, Chen J, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4(9):607–619. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 36.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krex D, Klink B, Hartmann C, et al. Long-term survival with glioblastoma multiforme. Brain: A Journal of Neurology. 2007;130(Pt 10):2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 38.Kong D, Li Y, Wang Z, Sarkar FH. Cancer Stem Cells and Epithelial-to-Mesenchymal Transition (EMT)-Phenotypic Cells: Are They Cousins or Twins? Cancers (Basel) 2011;3(1):716–729. doi: 10.3390/cancers30100716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kashyap V, Rezende NC, Scotland KB, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells and Development. 2009;18(7):1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed N, Abubaker K, Findlay J, Quinn M. Epithelial mesenchymal transition and cancer stem cell-like phenotypes facilitate chemoresistance in recurrent ovarian cancer. Current Cancer Drug Targets. 2010;10(3):268–278. doi: 10.2174/156800910791190175. [DOI] [PubMed] [Google Scholar]
- 41.Gallo D, Ferlini C, Scambia G. The epithelial-mesenchymal transition and the estrogen-signaling in ovarian cancer. Current Drug Targets. 2010;11(4):474–481. doi: 10.2174/138945010790980385. [DOI] [PubMed] [Google Scholar]
- 42.Arslantas A, Artan S, Oner U, et al. The importance of genomic copy number changes in the prognosis of glioblastoma multiforme. Neurosurgical Review. 2004;27(1):58–64. doi: 10.1007/s10143-003-0279-4. [DOI] [PubMed] [Google Scholar]
- 43.Chakravarti A, Zhai G, Suzuki Y, et al. The prognostic significance of phosphatidylinositol 3-kinase pathway activation in human gliomas. J Clin Oncol. 2004;22(10):1926–1933. doi: 10.1200/JCO.2004.07.193. [DOI] [PubMed] [Google Scholar]
- 44.Misra A, Pellarin M, Hu L, et al. Chromosome transfer experiments link regions on chromosome 7 to radiation resistance in human glioblastoma multiforme. Genes, Chromosomes and Cancer. 2006;45(1):20–30. doi: 10.1002/gcc.20257. [DOI] [PubMed] [Google Scholar]
- 45.Bowen KA, Doan HQ, Zhou BP, et al. PTEN loss induces epithelial–mesenchymal transition in human colon cancer cells. Anticancer Research. 2009;29(11):4439–4449. [PMC free article] [PubMed] [Google Scholar]
- 46.Langlois MJ, Bergeron S, Bernatchez G, et al. The PTEN phosphatase controls intestinal epithelial cell polarity and barrier function: role in colorectal cancer progression. PLoS One. 2010;5(12):e15742. doi: 10.1371/journal.pone.0015742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoo LI, Liu DW, Le Vu S, Bronson RT, Wu H, Yuan J. Pten deficiency activates distinct downstream signaling pathways in a tissue-specific manner. Cancer Res. 2006;66(4):1929–1939. doi: 10.1158/0008-5472.CAN-05-1986. [DOI] [PubMed] [Google Scholar]
- 48.Shorning BY, Griffiths D, Clarke AR. Lkb1 and Pten synergise to suppress mTOR-mediated tumorigenesis and epithelial-mesenchymal transition in the mouse bladder. PLoS One. 2011;6(1):0–e16209. doi: 10.1371/journal.pone.0016209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27(2):222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 50.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes and Development. 2009;23(6):675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin P, Liu YN, Pierce R, et al. Prostate epithelial Pten/TP53 loss leads to transformation of multipotential progenitors and epithelial to mesenchymal transition. The American Journal of Pathology. 2011;179(1):422–435. doi: 10.1016/j.ajpath.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debies MT, Gestl SA, Mathers JL, et al. Tumor escape in a Wnt1-dependent mouse breast cancer model is enabled by p19Arf/p53 pathway lesions but not p16 Ink4a loss. J Clin Invest. 2008;118(1):51–63. doi: 10.1172/JCI33320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.