Abstract
Background
A proportion of glioblastoma stemlike cells (GSCs) expressing endothelial cell marker CDH5 (vascular-endothelial–cadherin or CD144) can transdifferentiate into endothelial cells and form blood vessels. However, the implications of CDH5 expression in gliomas and how it is regulated in GSCs remain to be clarified.
Methods
The mRNA and protein levels of CDH5 were detected in glioma samples and cultured cell lines, and the prognostic value of the CDH5 expression level for GBM patients was evaluated. Bioinformatics analysis was performed to reveal the potential functional roles of CDH5 in glioblastoma multiforme. Gene knockdown induced by short hairpin RNA, chromatin immunoprecipitation analysis, and a vasculogenic tube formation assay were performed to investigate the relationships among hypoxia, CDH5 expression level, and angiogenesis.
Results
CDH5 was overexpressed in gliomas, correlated with tumor grades, and was an independent adverse prognostic predictor for glioblastoma multiforme patients. CDH5 was specifically activated in GSCs but not in non-GSCs or neural stem cells, and CDH5+ cells could produce xenografts in immunocompromised mice. Bioinformatics analysis demonstrated that CDH5 might interact directly with hypoxia-inducible factor (HIF)2α. CDH5 expression was significantly upregulated in GSCs, but not in non-GSCs or normal neural stem cells, under a 1% O2 condition. Both HIF1α and HIF2α positively regulated CDH5 level in GSCs and could bind to the promoter of CDH5. Furthermore, CDH5 contributed to the vasculogenic mimicry of GSCs, especially under hypoxic conditions.
Conclusions
The specific expression of CDH5 in GSCs may contribute to GSC-derived neovasculogenesis in glioblastoma multiforme, especially under hypoxic conditions, revealing novel tumorigenic mechanisms contributed by GSCs.
Keywords: bioinformatics, brain tumor stem cells, cancer stem cell, hypoxia, VE-cadherin
Glioblastoma multiforme (GBM), a World Health Organization (WHO) grade IV glioma that is the most common primary malignant brain tumor in adulthood, is among the most aggressive and lethal cancers in humans.1 Despite advances in neurosurgery and adjuvant treatment, the median survival of patients with GBM is only about 1 year.2 In recent years, one of the most promising advances in understanding the mechanism underlying the tumorigenesis of GBM has been promoted by the concept of cancer stem cells.3–6 A subpopulation of glioblastoma stemlike cells (GSCs)7–11 in GBM not only retain multipotency and an extraordinary potential to initiate tumors, but also contribute to the chemo- and radioresistance of GBM.4,5,12–14 In addition, GSCs are located in a perivascular niche and are regulated by hypoxia. These observations suggest that GSCs may be important therapeutic targets for GBM treatment.
Recently, new surprising features of GSCs have been reported, showing that GSCs even have the potential to transdifferentiate into endothelial cells (ECs),15–18 thus contributing to angiogenesis in GBM to meet the nutritional and oxygen requirements caused by overgrowth of the tumor, even where pre-existing blood vessels are lacking. This feature of GSCs is particularly important because GBM comprises highly vascularized tumors that are therefore attractive targets for anti-angiogenic therapies.19,20 The transdifferentiation of GSCs into ECs provides new insights into the angiogenic mechanisms in GBM and possible novel strategies for anti-angiogenic treatment. However, the mechanisms underlying such transdifferentiation remain to be clarified. Intriguingly, it was recently reported that a population of CD133+ GSCs express CDH5 (vascular-endothelial [VE]–cadherin/CD144), a specific marker for ECs, and GSCs may transdifferentiate into CDH5+ ECs.15–17 Considering that CDH5 plays a central role in EC biology21,22 and is an important target for angiogenesis in GBM,23,24 we focused on the roles of CDH5 in GBM, and particularly in GSCs. We hypothesized that CDH5 expressed in GBM may play a role in the process of GSC transdifferentiation into ECs. Here, we demonstrate that CDH5 is overexpressed in GBM tumor cells, and high expression of CDH5 predicts poor prognosis. In addition, CDH5 is specifically activated in GSCs but not in normal neural stem cells (NSCs) and non-GSCs. Interestingly, CDH5 expression in GSCs is intensively regulated by hypoxia through hypoxia-inducible factors (HIFs, including HIF1α and HIF2α), central transcriptional factors that mediate hypoxia-induced responses. Our results revealed promising mechanisms controlling transdifferentiation of GSCs into ECs, which are regulated by hypoxia, and provide new insights into the mechanisms of hypoxia-induced angiogenesis in GBM.
Materials and Methods
Glioma Samples
The following brain tumor samples used for real-time quantitative (q)PCR and Western blot were provided by the Department of Neurosurgery, Xijing Hospital, the Fourth Military Medical University: 31 GBM (M/F ratio: 19/12, median age: 54.0 y [range, 24–68]), 14 grade II astrocytomas (M/F ratio: 6/8, median age: 44.0 y [27–57]), 15 grade III anaplastic astrocytomas (M/F ratio: 9/6, median age: 47.8 y [24–65]), and 5 normal brain samples derived from brain lobectomy from patients with cerebral trauma. Tumors were histopathologically classified according to WHO classification. The 5 normal samples and additional archived 24 GBM samples were used to detect CDH5 expression by immunohistochemistry. Informed consent was obtained from each patient, and experiments were approved by the local ethics committee.
Culture of Primary GSCs and NSCs
GSCs were cultured as described previously.25,26 The study protocol was approved by the institutional review board of Xijing Hospital of the Fourth Military Medical University, and written informed consent was obtained from patients. Briefly, samples were dissociated to a single-cell suspension using a fire-polished Pasteur pipette and cultured in serum-free medium consisting of Dulbecco's modified Eagle's–F12 medium, 20 ng/mL epidermal growth factor (Invitrogen), 20 ng/mL basic fibroblast growth factor (Invitrogen), and B27 (1:50; Invitrogen) or in 10% serum-containing medium. Two human fetal cortical NSCs were isolated from spontaneous aborted fetuses (8–12 wk), which were dissociated into single cells as described and then cultured in serum-free medium.
In order to induce hypoxia, cells were cultured in a sealed modular incubator chamber (Billups-Rothenberg) flushed with 1% O2, 5% CO2, and 94% N2 and incubated at 37°C for 24 h.
Quantitative Real-time PCR
RNA was extracted from cultured cells and brain tumor tissues using Trizol Reagent (Invitrogen). The extracted RNA was then reverse transcribed into cDNA, and qPCR analysis was performed on an ABI 7700 instrument using SYBR Green PCR Core Reagents in 20 μL volume (Applied Biosystems). The primers used for qPCR analysis were as follows: CDH5 forward: 5′-TCACCTTCTGCGAGGATATGG-3′, reverse: 5′-GAGTTGAGCACCGACACATC-3′; HIF1α forward: 5′-TTCCAGTTACGTTCCTTCGATCA-3′, reverse: 5′-TTTGAGGACTTGCGCTTTCA-3′; HIF2α forward: 5′-GTGCTCCCACGGCCTGTA-3′, reverse: 5′-TTGTCACACCTATGGCATATCACA-3′; and β-actin forward: 5′-CCCAGCACAATGAAGATCAA-3′, reverse: 5′-GATCCACACGGAGTACTTG-3′. Quantitative PCR using water instead of template was used for negative control. All samples were assayed in triplicate, and the relative amount of target transcripts were normalized to the number of human β-actin transcripts found in the same sample. Specificity was verified by melting curve analysis and agarose gel electrophoresis. Relative fold changes were calculated using the ΔΔCt method with threshold cycle values of each sample.
Western Blotting
Cultured cells were lysed in sodium dodecyl sulfate (SDS) sample buffer, and 30 μg of the proteins were run on 6% SDS–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. Blots were blocked in phosphate buffered saline (PBS) containing 5% nonfat dry milk powder and incubated overnight at 4°C with primary antibody of CDH5 (1:1000; Cell Signaling Technology), HIF1α (1:1000; BD Biosciences), HIF2α (1:500; Novus Biologicals), or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1:500 000; Abcam). Blots were then washed with PBS containing 0.1% Tween 20 (PBST) and incubated in secondary antibodies coupled to peroxidase. After washing in PBST, blots were developed with enhanced chemiluminescence according to the manufacturer's instructions (Amersham Biosciences). All Western blot analyses were done in duplicates. The blot results were scanned and quantified by ImageJ software, and the relative levels of target proteins were normalized to nicotinamide adenine dinucleotide phosphate-oxidase.
Immunohistochemistry
Paraffin-embedded, 1-μm formalin fixed tissue sections were mounted on microscope slides and processed as previously described.25 Immunohistochemical staining of anti-human CDH5 (1:100, F-8; Santa Cruz Biotechnology) or human-specific nestin (1:100, 10C2, mouse monoclonal; Abcam) was performed on tissue sections. The sections were treated with a heat-induced epitope retrieval technique using a citrate buffer at pH 6.0. The sections were then blocked for endogenous peroxidase and biotin before incubation with primary antibodies for 3 h at room temperature. The Elite Vector Stain ABC System (Vector Laboratories) was used as the detection system and diaminobenzidine as the chromogen. Nuclei were counterstained with hematoxylin.
Flow Cytometry and Cell Sorting
Cells were counted by a hemocytometer, and 50 000 cells were placed in 200 μL PBS with 20% fetal bovine serum and allowed to incubate with fluorescein isothiocyanate (FITC)–conjugated anti-CDH5 (1:20; Abcam) or a nonspecific FITC-conjugated immunoglobulin G1 isotype control antibody (Miltenyi Biotec) for 1 h at 4°C. Cells were then washed, resuspended in PBS plus 20% fetal bovine serum, and analyzed by fluorescence activated cell sorting on a FACSCalibur (Becton Dickinson) flow cytometer. CDH5+ cells were isolated by positive magnetic affinity cell sorting selection using the FITC-conjugated CDH5 antibodies and anti-FITC magnetic beads (Miltenyi Biotec). All flow cytometry evaluations were performed in triplicate and analyzed with WinMDI software.
Intracranial Xenograft Tumors
Intracranial xenograft experiments were performed as described previously.25 Briefly, 105 isolated CDH5+ cells were resuspended in 10 μL PBS and stereotactically injected into the right striatum of the brains of nude mice (6–8 wk old; n = 6 each; Center of Experimental Animals, Fourth Military Medical University), following administration of general anesthesia. Coordinates for stereotactic injections into the adult mice were 2 mm to the right of the midline, 0.5 mm anterior to the coronal suture, and 3 mm deep. The mice were killed 2 months later and examined for tumor formation in the brains. All animal handling during the experiments was in strict accordance with the Animal Experiments guidelines in force at the Fourth Military Medical University.
Network Reconstruction and Informatics Analysis
The Algorithm for the Reconstruction of Accurate Cellular Networks (ARACNe), an information-theoretic algorithm for inferring transcriptional interactions,27 was used to identify a repertoire of candidate transcriptional regulators of interesting genes. Expression profiles used in the analysis were from 4 datasets: The Cancer Genome Atlas,28,29 a unified validation dataset,29 a high-grade glioma dataset from Gravendeel et al,30 and a GBM dataset from Lee et al.31 First, candidate interactions between a transcription factor (x) and its potential target (y) were identified by computing pairwise mutual information, MI[x; y], using a Gaussian kernel estimator and by thresholding the mutual information based on the null hypothesis of statistical independence (P < .05, Bonferroni corrected for the number of tested pairs). Then, indirect interactions were removed using a data processing inequality with a tolerance of 20%, a well-known property of the mutual information.
Short Hairpin RNA Infection
Short hairpin (sh)RNA lentivirus particles targeting HIF1α, HIF2α, CDH5, and scrambled nontargeting shRNA were purchased from Sigma. Cell lines were infected with the shRNA lentivirus according to the manufacturer’s protocol. Briefly, GSCs and NSCs growing as neurospheres and U87 cells were dissociated into single cells with Accutase and gentle trituration and then incubated with the lentivirus for 24 h. After ∼48 h in culture, 2 μg/mL puromycin was used to select infected cells.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described.32 Cultured cell lysates were precleared with Protein A/G beads (Santa Cruz) and incubated at 4°C overnight with 1 μg of polyclonal antibody specific for HIF1α (Santa Cruz), HIF2α (Novus), or normal rabbit immunoglobulins (Santa Cruz). DNA was eluted in 200 μL water, and 1 μL was analyzed by PCR. The primers used for ChIP PCR analysis were as follows: hypoxia response element (HRE)1,2-CDH5 forward: 5′-CCTCCAAAGACGGTCGGC-3′, reverse: 5′-GCCCTTGGCACTACCTCT.-3′; HRE3-CDH5 forward: 5′-CTTGGTTCTTCTGGGCTCTG-3′, reverse: 5′-GTCATCCTGGAGCCACAGTT-3′; HRE4,5-CDH5 forward: 5′-GGACTGTTCTCCTTCCAGCA-3′, reverse: 5′-GGGCTAGAGAAAGGGGAGAA-3′; HRE6-CDH5 forward: 5′-GAGACCCAGCAGGAAGCA-3′, reverse: 5′-CAACAGCCGATTGTGGAA-3′.
Vasculogenic Tube Formation Assay
Vasculogenic tube formation was tested using a commercial Matrigel assay kit (BD Biosciences). Twenty-four–well tissue culture plates were coated with Matrigel matrix (0.1 mL/well; BD Biosciences) and allowed to solidify at 37°C for 30 min. GSC cells were dissociated into single cells and resuspended at 6 × 104 cells/mL in endothelial basal medium containing 2% fetal calf serum. The cells in each group were then plated at 0.5 mL/well onto the surface of Matrigel and incubated at 37°C. Three random fields from each well were photographed at ×200 magnification (Olympus BX-51). Tube networks were quantified as the total number of pixels in thresholded images, using Image Pro Plus 6.0 software.
Statistical Analysis
Statistical analyses were performed using Student's t-tests and 1-way ANOVAs with least-squared-difference post-hoc tests, as appropriate. All P-values are 2-tailed, and P < .05 was considered statistically significant. Statistical analyses were performed with SPSS v.13.0.0.
Results
CDH5 Expression Levels Are Associated With Glioma Grades
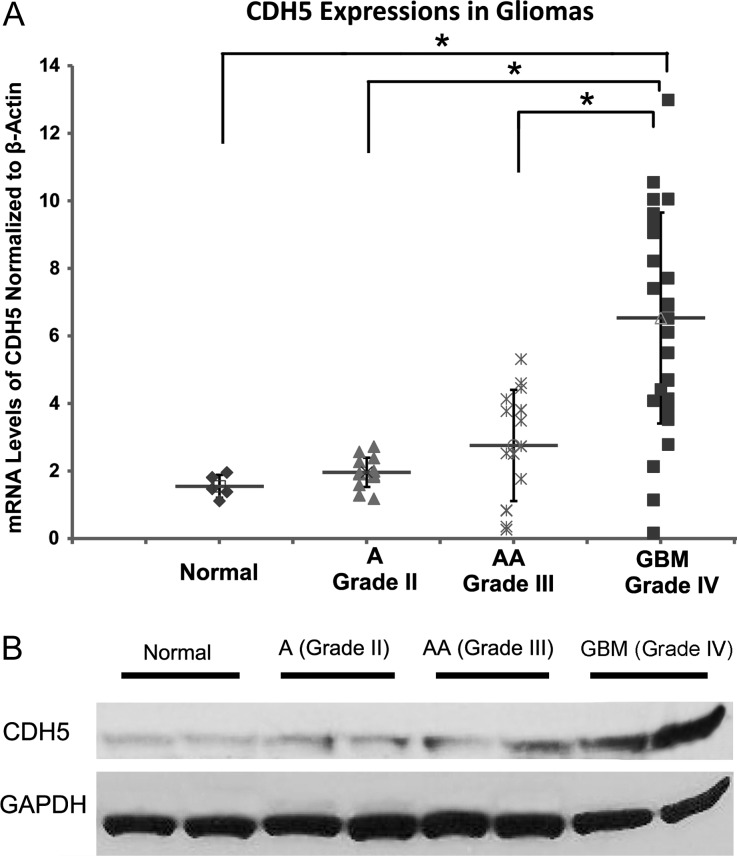
To examine whether CDH5 affects the tumor biology of gliomas, we first investigated CDH5 expression in glioma tissues by qPCR analysis of a cohort of normal and glioma samples, including 5 normal brain samples, 14 astrocytomas (WHO grade II), 15 anaplastic astrocytomas (WHO grade III), and 31 GBM tumors. CDH5 mRNA levels increased along with the tumor grades (P < .01; Fig. 1A). Compared with normal tissues, the fold increases of CDH5 mRNA levels in grades II and III astrocytomas and GBM were 1.17 ± 0.61, 1.51 ± 0.76, and 2.80 ± 1.86, respectively. Notably, CDH5 was more significantly overexpressed in GBM than in normal tissues and low-grade gliomas (P < .05, Tukey honestly significant difference [HSD] ANOVA), whereas grade II and grade III astrocytomas showed slightly, but not significantly, increased CDH5 levels compared with normal tissues (Fig. 1A), indicating that CDH5 is specifically activated in GBM. To further confirm the increased expression of CDH5 in gliomas, proteins were extracted from 2 normal brain samples, as well as 5 grade II astrocytomas, 5 grade III astrocytomas, and 5 GBM samples. Western blot analysis demonstrated that CDH5 protein was expressed in normal and glioma tissues but was more highly expressed in GBM (Fig. 1B). In addition, we performed a multivariate analysis of possible factors that may influence CDH5 expression, including age at diagnosis, sex, tumor grade, and KPS, by using the Repository of Molecular Brain Neoplasia Data (REMBRANDT) database of the National Cancer Institute (http://caintegrator-info.nci.nih.gov/rembrandt). In this analysis, tumor grade was the only independent factor that associated with CDH5 expression (P = .001), corroborating the conclusion that CDH5 is more highly expressed in GBM (Supplementary Table S1).
Fig. 1.
CDH5 expression in gliomas compared with normal brain tissue. (A) Quantitative PCR analysis of CDH5 expression in astrocytomas (A), anaplastic astrocytomas (AA), and GBM reveals increased expression in all types of gliomas, especially GBM, compared with normal brain tissue. GBM expressed significantly higher levels of CDH5 than normal tissue and low-grade gliomas (A and AA; P < .05). (B) Representative Western blots showing that CDH5 is more highly expressed in GBM than in normal and low-grade glioma tissues. β-actin and GAPDH were used as loading controls for qPCR and Western blotting, respectively.
Overexpression of CDH5 Predicts a Poor Prognosis for GBM Patients
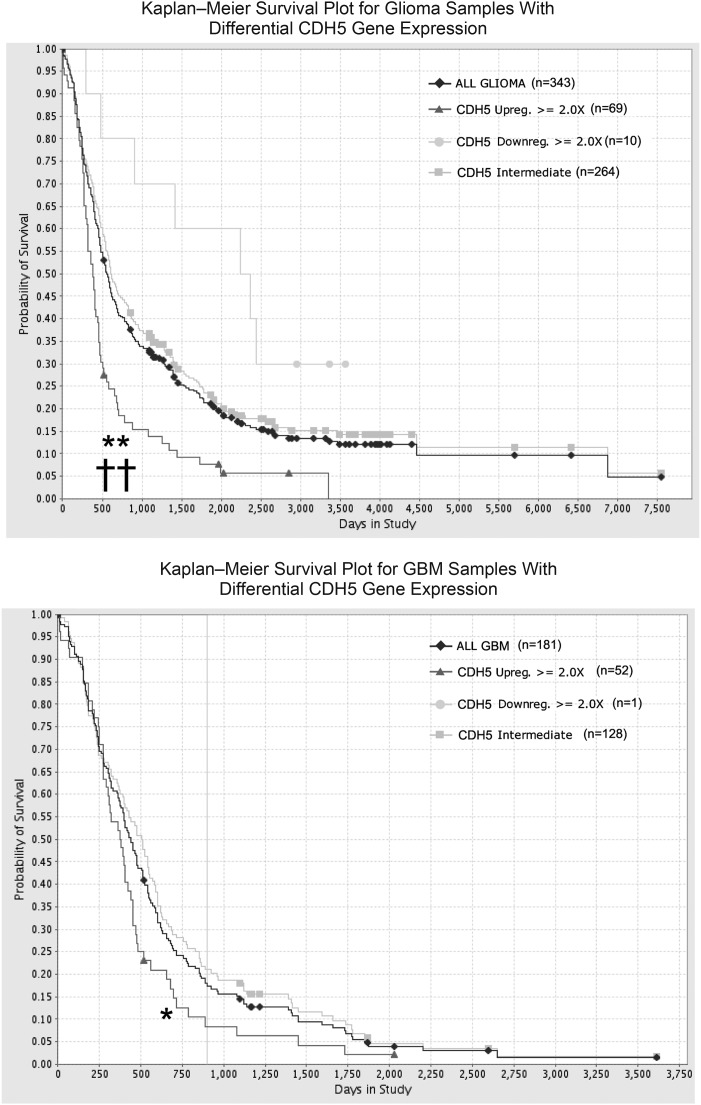
CDH5 is overexpressed in gliomas, especially in GBM, implying that it may influence clinical outcomes for glioma patients. We next used the REMBRANDT database to investigate whether targeting CDH5 might have a therapeutic benefit for glioma patients. The data were analyzed to determine the survival of glioma patients with intermediate, low, or high expression of CDH5. There were only 10 patients with >2-fold CDH5 downregulation, and we found a significant decrease in the probability of survival of patients with elevated CDH5 (P < .001) compared with samples with low or intermediate CDH5 levels (Fig. 2A). Because the amount of blood vessels may increase significantly along with glioma grade,33,34 and CDH5 is specifically overexpressed in GBM, to exclude that the prognostic value of CDH5 reflects only the amount of blood vessels and is increased with tumor grade, we further examined the association of CDH5 levels and the survival of GBM patients. The results revealed a statistically significant correlation between CDH5 expression and decreased survival of GBM patients (P < .05; Fig. 2B). Furthermore, we performed a multivariate Cox regression analysis of prognostic factors that included CDH5 expression, age at diagnosis, gender, KPS, extent of resection, and chemotherapy with temozolomide. For the glioma population stratified by tumor grade, CDH5 level was an independent predictive variable for shortened survival (P = .041; Supplementary Table S2). CDH5 also independently associated with shortened survival for GBM patients (P = .016, Supplementary Table S3). These data indicate that CDH5 expression is a potential independent prognostic factor for adverse survival of GBM patients.
Fig. 2.
CDH5 levels correlate with patient survival (REMBRANDT database) for all glioma patients (A) or GBM patients (B). There were only 4 patients with >2-fold ZNF217 downregulation in gliomas. *P < .05, **P < .001 with CDH5 high vs intermediate; ††P < .001 with CDH5 high vs low.
CDH5 Is Expressed in GBM Tumor Cells in Addition to ECs
Consistent with previous studies,15,16 we found that CDH5 was expressed in GBM tumor cells in addition to ECs. Immunohistochemical staining revealed that CDH5 was exclusively detected in vascular structures in normal brain tissues (Supplementary Fig. S1A), whereas it was detected in tumor cells (Supplementary Fig. S1C and D) in addition to vascular structures in GBM tissues. The positive tumor cells in GBM primarily exhibited membrane staining of CDH5 (Supplementary Fig. S1C and D). About 62.5% (15/24) of GBM samples displayed CDH5 immunoreactivity in tumor cells besides vascular structures. The patterns of CDH5 immunoreactivity in GBM tumor cells included scattered and clustered CDH5+ tumor cells. These results suggest that CDH5 is also activated in a proportion of GBM tumor cells, which may gain the potential to transdifferentiate into ECs and contribute to neovascularization in GBM.
CDH5 Is Specifically Activated in GSCs
Because it has been reported that GSCs may give rise to ECs, together with our finding that CDH5 is also expressed in tumor cells, we further explored the expression of CDH5 in GSCs, normal NSCs, and non-GSCs (serum-cultured primary and traditional cell lines). It has been suggested that glioma is driven by a subpopulation of GSCs that share some similar properties with normal NSCs and have characteristics that more closely resemble primary tumors than serum-cultured non-GSCs.35 Neurosphere cell lines derived from gliomas cultured in serum-free medium containing epidermal growth factor and basal fibroblast growth factor are widely used to enrich for GSCs,25,35,36 and serum-cultured primary glioma cells and traditional glioma cell lines have been reported to contain few or no GSCs.35 Therefore, 7 neurosphere cell lines derived from GBM tissues cultured in serum-free medium were used as GSCs. Primary glioma cells and GBM cell lines, including U87, U251, and A172, cultured in 10% serum medium were used as non-GSCs. Two neurosphere cell lines derived from human fetal brains cultured in serum-free medium were used as normal NSCs.26 The features of GSC, non-GSC, and NSC cell lines used here have been described in our previous studies,25,26 which demonstrated that GSC cells express the stem cell markers nestin, CD15, and CD133 and can differentiate into neuronal and glial lineages.
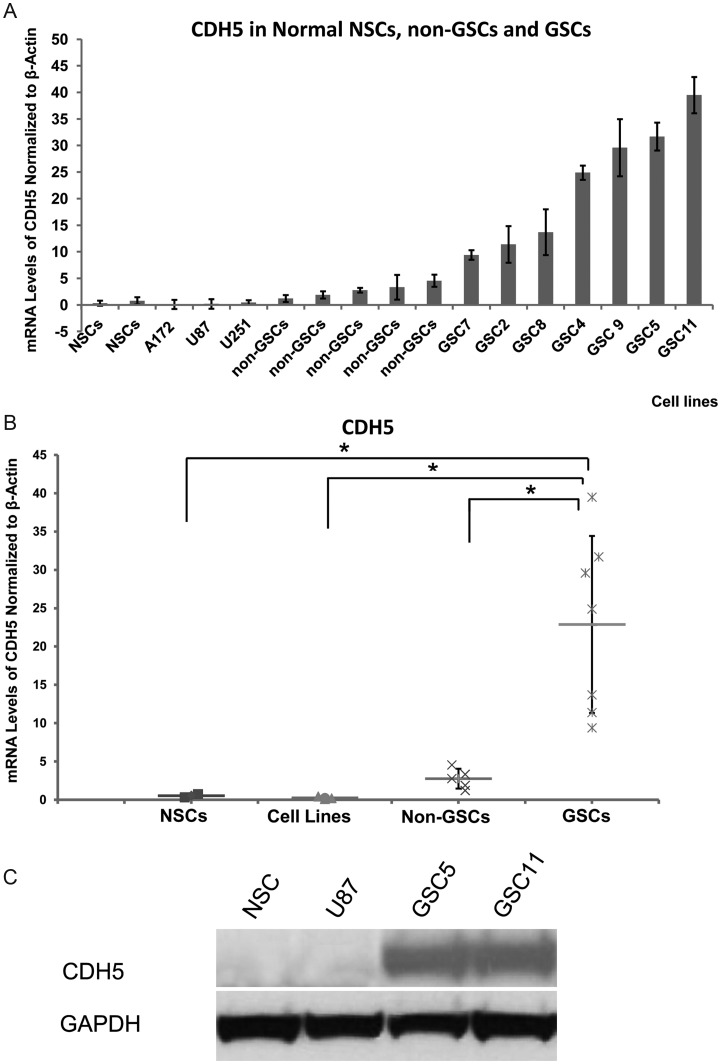
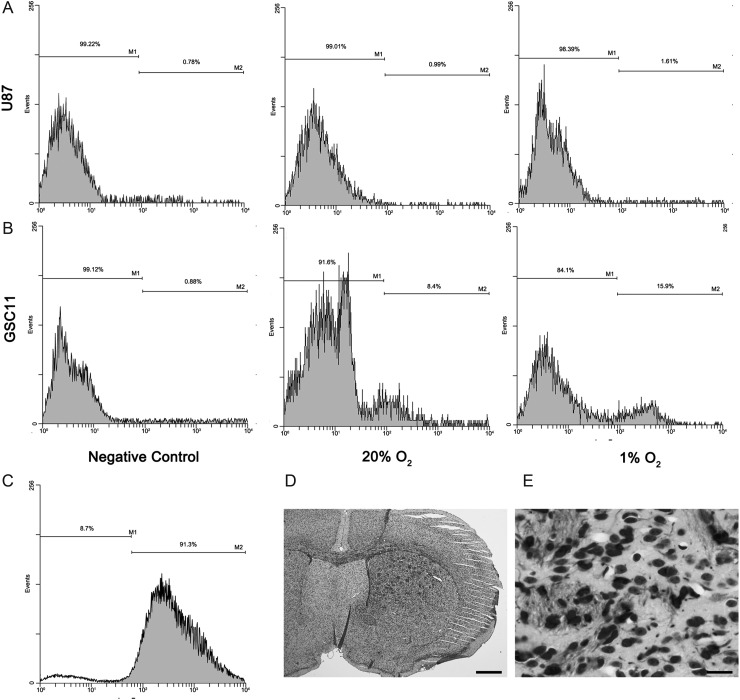
Notably, qPCR showed that CDH5 was more highly expressed in GSCs than in non-GSCs and normal NSCs (P < .05; Fig. 3A and B). Western blot analysis was performed to determine the CDH5 protein levels, and the results corroborated CDH5 expression in GSCs, but not in non-GSCs and NSCs (Fig. 3C). Because CDH5 is a membrane molecule, flow cytometric analysis revealed that 8.4% and 11.5% of CDH5+ cells were detected among GSC5 and GSC11 cells, respectively, whereas only 1.0% and 0.8% of CDH5+ cells were detected among U87 and NSCs (Fig. 4A and B).
Fig. 3.
Differential expression of CDH5 in GSCs, non-GSCs, and normal NSCs. (A, B) CDH5 expression in normal NSCs, serum-cultured GBM cell lines, non-GSCs, and GSCs. CDH5 is specifically expressed in GSCs compared with normal NSCs and non-GSCs. (C) Representative Western blots showing that CDH5 is detected in GSCs but not in NSCs and U87. *P < .05.
Fig. 4.
Flow cytometric analysis revealed that CDH5 was detected in GSC lines (A) (data of GSC11 are shown as representative), but not in non-GSC cells (B) (data of U87 are shown as representative). In addition, the percentage of CDH5+ cells was increased under hypoxic conditions in GSC11 (A) but not in U87 cells (B). (C–E) isolated CDH5+ cells produced xenograft tumors in the brains of nude mice (data from GSC5 are shown as representative). (C) The purity of isolated CDH5+ GSC5 cells revealed by flow cytometry. (D and E) Human-specific nestin staining revealed that CDH5+ cells produced tumors when injected into the brains of nude mice. (D) 40×; (E) 400×; bar = 500 μm in (D) and 50 μm in (E).
To further confirm that CDH5+ cells detected in the GSCs were tumor cells but not contaminated ECs, CDH5+ cells were purified from GSC5 and GSC11 (Fig. 4C, showing the purification of isolated CDH5+ cells) and injected into the brains of nude mice as orthotopic xenografts. The mice were sacrificed 2 months later to detect tumor formation in the brains. Human-specific nestin staining in the xenografts revealed that human-derived tumors were formed in the mouse brains (Fig. 4D and E). As a result, 80% (4/5) of the mice injected with GSC5 and 40% (2/5) of the mice injected with GSC11 CDH5+ cells developed tumors resembling those induced by their parent cell lines. The tumorigenic potential of the CDH5+ cells indicates that they are tumor cells rather than contaminated ECs.
Together, these data suggest that CDH5 is specifically activated in GSCs, implying that GSCs may acquire the potential to transdifferentiate into ECs or initiate the program to adopt an EC fate under suitable conditions.
CDH5 Closely Associates With EC Markers and Particularly HIF2α
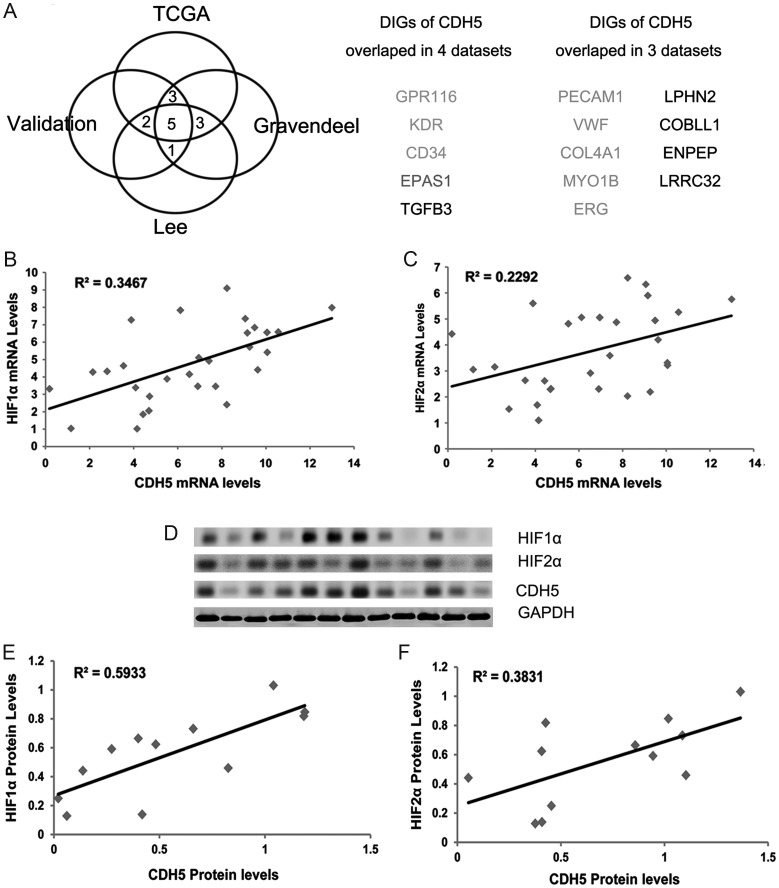
We have shown that CDH5 is overexpressed in GBM and is specifically activated in GSCs. However, its biological role in GBM is unclear. To address this issue, we used the ARACNe algorithm to assemble a genome-wide repertoire of CDH5-specific transcriptional interactions based on the mutual information between pairwise genes.27,37,38 As noted earlier, to exclude biases that may arise from using only 1 dataset, we used 4 datasets from independent groups to deduce candidate genes interacting with CDH5: The Cancer Genome Atlas,28,29 the unified validation database from Verhaak et al,29 a high-grade glioma dataset from Gravendeel et al,30 and a GBM dataset from Lee et al.31 A threshold of 10−7 was used to obtain candidate genes that interacted with CDH5 directly or indirectly. Next, we performed a data processing inequality with a tolerance of 20% to screen directly interacting genes (DIGs) of CDH5 (ie, genes that potentially interacted with CDH5 directly) by removing indirectly interacting genes.27 As a result, 4 sets of DIGs of CDH5 were obtained from the 4 datasets. We then identified 5 genes that overlapped in all 4 DIG sets, including G protein-coupled receptor (GPR)116, kinase insert domain receptor (KDR; vascular endothelial growth factor receptor [VEGFR]2), CD34, transforming growth factor–β3, and endothelial Per-Arnt-Sim domain-containing protein 1 (EPAS1; HIF2α), and 9 genes that overlapped in 3 of the DIG sets, such as platelet endothelial cell adhesion molecule–1 (PECAM1; CD31) and von Willebrand factor (VWF; Fig. 5A). These genes were the most reliable of those that may interact directly with CDH5 and regulate or be regulated by CDH5. Strikingly, most of these genes were EC markers or involved in angiogenesis, including CD34, CD31, KDR, VWF, collagen type IV alpha 1,39 GPR116, myosin-1B,40 erythroblast transformation-specific related gene (ERG),41,42 and EPAS1 (HIF2α),43 corroborating the results of the bioinformatics analysis (Fig. 5A). Notably, HIF2α genes, which are critical for hypoxia-induced phenotypes and important for the tumorigenic potential of GSCs,10 were among the most reliable DIGs. In fact, it has been reported that HIF2α is also selectively expressed in ECs,43 further confirming the results of our bioinformatics analysis. Moreover, in mouse cell lines, it has been reported that HIF2α regulates CDH5 expression directly by binding to the promoter of CDH5.44 However, it remains to be clarified whether HIF2α regulates CDH5 in human glioma cells. Considering the close relationship between hypoxia and angiogenesis, these results imply that hypoxia regulates CDH5 directly during the process of hypoxia-induced angiogenesis in human GBM.
Fig. 5.
Bioinformatics analysis from 4 datasets revealed putative DIGs for CDH5. (A) There are 5 DIGs overlapped in all of the 4 datasets and 9 in 3 of the datasets, representing the most reliable DIGs that may interact directly with CDH5 in GBMs. (B and C) Correlation analysis of CDH5, HIF1α, and HIF2α expression in GBM samples by qPCR demonstrated that CDH5 is significantly correlated with HIF1α and HIF2α (P < .05). (D) Immunoblots of 12 human primary GBM lysates probed with antibodies to HIF1α, HIF2α, CDH5, and GAPDH (loading control). (E and F) The intensities of the HIF1α, HIF2α, and CDH5 signals relative to GAPDH were quantified by densitometry, and correlation analysis revealed that CDH5 is significantly correlated with HIF1α and HIF2α in protein level (P < .05).
To further confirm the relationship between CDH5 and HIFs, we detected HIF1α and HIF2α by qPCR analysis in the cohort of GBM samples. Correlation analysis revealed that CDH5 was significantly correlated with both HIF1α (R2 = 0.35, P < .05) and HIF2α (R2 = 0.23, P < .05; Fig. 5B and C). Furthermore, quantification of Western-blot results from 12 GBM samples revealed that the protein levels of CDH5 were significantly correlated with those of both HIF1α (R2 = 0.59, P < .05) and HIF2α (R2 = 0.38, P < .05; Fig. 5D–F). These results imply potential interactions between CDH5 and HIFs.
CDH5 Is Upregulated in GSCs Under Hypoxic Conditions
Bioinformatics analysis and correlation results imply direct interaction between CDH5 and HIFs. In addition, hypoxia is an important factor for the GSC niche and can promote the tumorigenic capacity and clonogenicity of GSCs.10,11,45 Therefore, we next investigated whether CDH5 is regulated by hypoxia and contributes to hypoxia-induced angiogenesis.
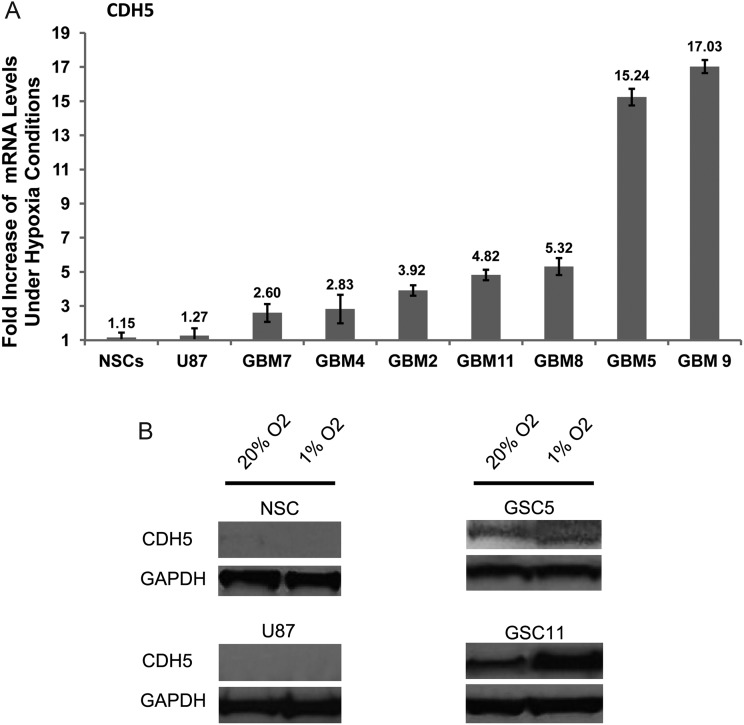
Two GSC lines, 2 non-GSC lines (a serum-cultured primary cell line and the U87 cell line), and normal NSCs were cultured under hypoxic (1% O2) or normoxic (20% O2) conditions for 24 h. Interestingly, under hypoxic conditions, CDH5 mRNA showed a 2.6- to 17.0-fold increase in GSCs (7.4 ± 6.1) (Fig. 6A), whereas a perceivable increase of CDH5 was not observed in NSCs and serum-cultured U87 cells. Western blot and flow cytometric analysis further confirmed that CDH5 was increased in GSCs during hypoxia but was not detected in non-GSCs or NSCs under either normoxic or hypoxic conditions (Figs 4 and 6B). These results suggest that CDH5 is specifically activated in GSCs and increased during hypoxia. In addition, although hypoxia increased CDH5 expression in GSCs, it had undetectable effects on non-GSCs and NSCs that expressed low or undetectable levels of CDH5 under normoxic conditions.
Fig. 6.
CDH5 is increased under hypoxia in GSCs. (A) Fold-increase of CDH5 mRNA levels in NSCs, U87, and GSCs after exposure to hypoxia for 24 h. (B) Representative Western blots exhibiting CDH5 were increased under 1% O2 hypoxic conditions for 24 h in GSC cells, but not in NSCs and U87.
CDH5 Is Regulated by HIF1α and HIF2α
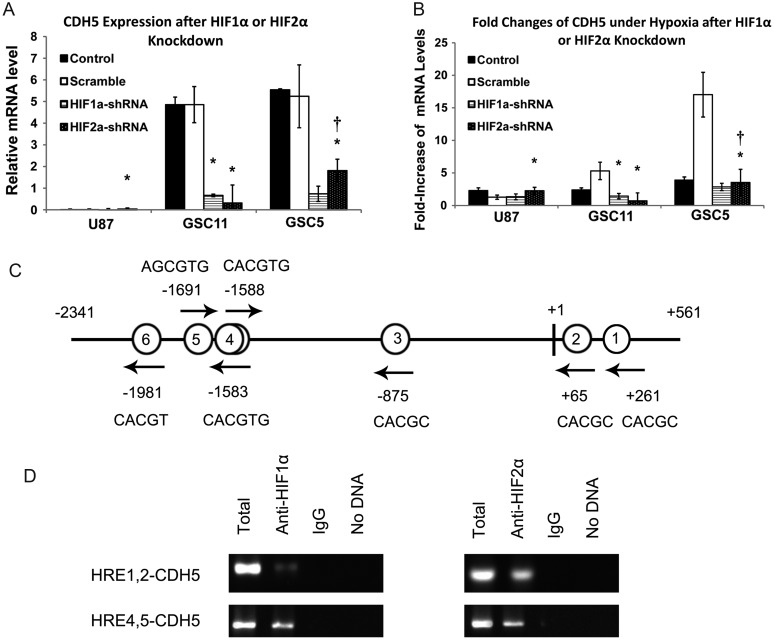
Because HIF1α and HIF2α play a central role in the hypoxia-induced responses of GSCs,10,45–47 we explored whether the increased CDH5 level under a hypoxic condition is regulated by HIFs. HIF1α and HIF2α were knocked down by lentivirus shRNA in U87 and 2 GSC cell lines, followed by selection with puromycin. The efficiencies of HIF1α and HIF2α knockdowns were detected by qPCR and are shown in Supplementary Fig. S2A and B. Quantitative PCR analysis indicated that knockdown of either HIF1α or HIF2α almost completely abolished CDH5 expression in the 2 GSC lines (CDH5 levels were repressed by 86%–93%; Fig. 7A). However, no significant change in CDH5 expression was detected in U87 cells, possibly due to the low expression of CDH5 in the parent cell line (Fig. 7A). To further confirm the regulation of CDH5 by HIFs, GSCs transfected with HIF1α, HIF2α, or nontargeting control shRNA were cultured under 20% or 1% O2 conditions for 24 h, and their changes of CDH5 expression were detected by qPCR. Compared with nontargeting controls, knockdown of HIF1α or HIF2α in GSCs significantly inhibited the increase of CDH5 expression under hypoxic conditions (Fig. 7B). Consistently, HIF1α or HIF2α knockdown had undetectable effects on CDH5 expression in U87 cells under hypoxic conditions (Fig. 7B). These data indicate that HIFs play an important role in the regulation of CDH5 expression in GSCs under hypoxic conditions.
Fig. 7.
CDH5 expression is regulated by HIFs in GSCs. (A) CDH5 expression was reduced in GSCs, while not significantly altered in U87, after HIF1α or HIF2α knockdown. *P < .05 compared with nontargeting scramble control; †P < .05 compared with HIF1α knockdown. (B) Fold-increase of CDH5 under hypoxia demonstrated that CDH5 upregulation was compromised in U87 and GSCs under hypoxia after HIF1α or HIF2α were knocked down in these cells. *P < .05, compared with the nontargeting scramble control; †P < .05 compared with HIF1α knockdown. Quantitative PCR data were normalized to β-actin; “Control” indicates parent cell line; “scramble” indicates GSCs treated with nontargeting scrambled shRNA. (C) Localization of putative HREs in the −2341/+561 CDH5 gene promoter. Schematic representation of the −2341/+561 region of the CDH5 gene. Putative HREs are represented by circled numbers and their sequence is displayed. (D) ChIP using HIF1α or HIF2α antibody and 2 different primer sets spanning different regions in the CDH5 promoter. IgG, immunoglobulin G.
HIFs Directly Interact With the CDH5 Promoter in GSCs
It has been reported that HIF2α binds to CDH5 promoter in mice.44 We also showed that CDH5 is regulated by HIFs. Together with the bioinformatics results, it is possible that HIFs directly regulate CDH5 in GSCs. The −2341/+561 promoter region of the CDH5 gene contains several putative HREs (5′-RCGTG-3′),48,49 including 5 single sites (HRE6, −1981; HRE5, −1691; HRE3, −875; HRE2, +65; HRE1, +261; Fig. 7C) and 1 head-to-head tandem (HRE4, −1583/−1588; Fig. 7C), suggesting that the gene may be regulated by HIFs. To confirm this hypothesis, we performed ChIP to investigate whether HIFs directly interact with the CDH5 promoter in GSCs. Primers were designed across the putative HRE sequence in 4 CDH5 promoter regions spanning the first and second HREs (HRE1,2-CDH5), the third HRE (HRE3-CDH5), the fourth and fifth HREs (HRE4,5-CDH5), and the sixth HRE (HRE6-CDH5). Then, PCR was performed with nuclear extracts from GSC5 cells after immunoprecipitation with anti-HIF antibodies. As a result, HIF2α bound to HRE4,5-CDH5 and HRE1,2-CDH5 strongly but did not bind to HRE3-CDH5 or HRE6-CDH5 (Fig. 7D). HIF1α bound to HRE4,5-CDH5 and weakly bound to HRE1,2-CDH5, while no signals were detected in the other regions (Fig. 7D). These results demonstrate that both HIF1α and HIF2α interact with the human CDH5 promoter, which is at least partly attributed to regulation of CDH5 expression by HIFs in GSCs.
CDH5 Contributes to Hypoxia-Induced Tubular Vasculogenic Mimicry of GSCs
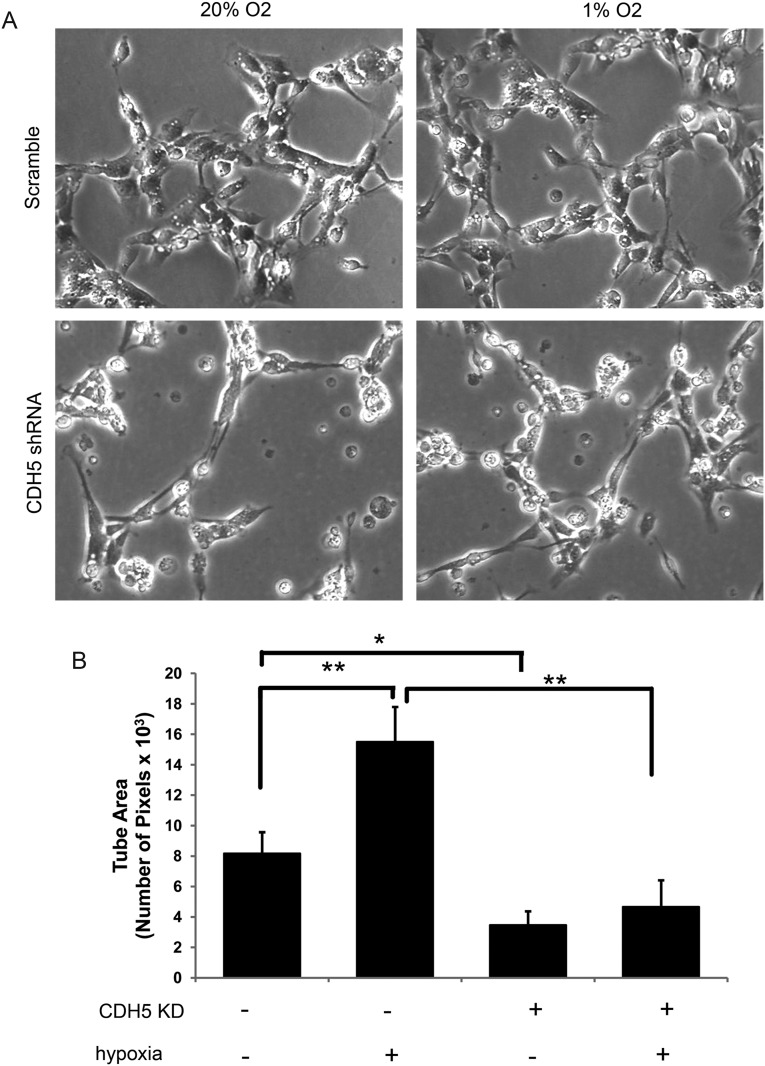
Because CDH5 is a critical regulator of vascular formation, we analyzed whether CDH5 is involved in GSC-derived vasculogenic mimicry. In addition, considering that hypoxia regulates both the expression of CDH5 and the vasculogenic mimicry of GSCs, we examined the effect of CDH5 on the vasculogenic mimicry of GSCs under a hypoxic condition in parallel. Three-dimensional Matrigel tube formation assays were performed, and the tube area, expressed as the total number of pixels, was measured to determine the vasculogenic capacity of GSCs. First, 3D Matrigel tube formation assays using the GSC lines revealed that GSC5, GSC9, and GSC11 cells exhibited various degrees of tube formation in vitro. Then, CDH5 was knocked down by shRNA in GSC5 cells to further explore the effects of CDH5 on vasculogenic mimicry. The efficiency of CDH5 knockdown in GSC5 cells detected by qPCR was about 83%. GSC5 cells treated with CDH5-shRNA or nonspecific scrambled shRNA were cultured in the Matrigel tube formation system under 20% or 1% O2 conditions for 24 h. Under the normoxic condition, the tube area per field in control GSC5 cells treated with nonspecific scrambled shRNA was 8.2 ± 1.4 × 103, which was significantly decreased after CDH5 knockdown (3.5 ± 0.9 × 103, P = .026, Tukey HSD; Fig. 8A and B). The tube area per field of GSC5 cells was increased under hypoxic conditions (16.3 ± 2.3 × 103, P = .001), consistent with previous reports.16 Interestingly, the tube area per field of GSC5 cells treated with CDH5-shRNA during hypoxia was 4.8 ± 1.3 × 103, less than that of control GSC5 cells under either normoxic (P = .111) or hypoxic conditions (P < .001; Fig. 8A and B). In addition, the fold increase of the tube area per field for control GSCs during hypoxia was 2.0 ± 0.3, which was decreased to 1.4 ± 0.4 after CDH5 knockdown (P = .083). These data suggest that CDH5 contributes to vasculogenic mimicry of GSCs, especially under hypoxic conditions.
Fig. 8.
CDH5 contributes to vasculogenic mimicry of GSC under hypoxia. (A) Effects of CDH5 knockdown and hypoxia on the capacity of vasculogenic mimicry of GSC5. (B) Tube area, taken from 3 random fields and expressed as total number of pixels, was used to compare the capacity of vasculogenic mimicry of GSC5 after CDH5 knockdown or under hypoxia conditions. *P < .05, **P < .001.
Discussion
GBM consists of highly vascularized tumors, and endothelial proliferation is one of the major hallmarks of this disease. In addition to providing oxygen and nutrition for tumor growth, the vasculature in GBM tumors also forms a special vascular niche for GSCs. Therefore, the use of anti-angiogenic agents is an important therapeutic strategy for tumor treatment. A major stimulator of angiogenesis in tumors is hypoxia. It has been shown that hypoxia induces angiogenesis via multiple mechanisms. Hypoxia regulates multiple steps involved in angiogenesis, including vascular permeability, endothelial sprouting, degradation of the extracellular matrix, and EC proliferation and migration, by regulating a cohort of molecules important for angiogenesis, including VEGF, Flt-1, KDR, angiopoietin-2, and matrix metalloproteinase 2.50 Nevertheless, the mechanisms underlying hypoxia-induced angiogenesis in GBM are not fully understood.
In recent years, GSCs have been reported to be the primary driving force of GBM growth and relapse. It has been well established that GSCs are enriched in vascular and perinecrotic/hypoxic niches in which hypoxia is a critical component of the microenvironment.8–11 In fact, hypoxia may promote cell reprogramming into more primitive cell types for many kinds of cells, including NSCs51 and, importantly, cancer stem cells.52,53 In addition, hypoxia contributes to the aggressiveness and therapeutic resistances of cancers.54,55 Hypoxia influences many key processes of cells by coordinated regulation of the expression of a large number of genes. HIF transcription factors, including HIF1α and HIF2α, which are regulated by oxygen levels, play central roles in the hypoxia-induced responses of these cells.46,56,57
Despite the importance of GSCs in GBM tumorigenesis, the intrinsic biological nature of GSCs is largely unknown. The importance of hypoxia to both angiogenesis and GSC maintenance indicates a potential relationship between GSCs and angiogenesis. In fact, recent studies have demonstrated that a proportion of CD133+ GSCs have the potential to transdifferentiate into vascular cells, and the process is regulated by hypoxia. However, the detailed relationships between GSCs and ECs and the mechanisms of transdifferentiation of GSCs into ECs are still unclear.
One of the specific markers for ECs is CDH5 (VE-cadherin/CD144). CDH5 plays a pivotal role in vascular integrity and permeability and controls EC assembly into tubular structures.21,22 CDH5 promotes cell survival and inhibits proliferation by interacting with β-catenin, phosphoinositide-3 kinase, and VEGFR-2 and modulating their signaling. Inactivation of the CDH5 gene is embryonically lethal in mice because of impaired vascular remodeling and maturation.21 CDH5 is also important for tumor angiogenesis, and blocking the CDH5 function with monoclonal antibodies in mouse tumor models leads to inhibition of tumor angiogenesis and growth.23,24 Interestingly, CDH5 is expressed in tumor cells in addition to ECs and promotes tumor progression via the transforming growth factor–β signaling pathway,58 implicating CDH5 in important roles for tumor cells.
Here, we first demonstrated that CDH5, a specific EC marker, is expressed in GBM tumor cells in addition to ECs. These results indicate that during transformation, a proportion of GBM cells also acquire a differentiation potential for cell types other than neural cells. Interestingly, we showed that CDH5 is specifically activated in GSCs but not in normal NSCs and non-GSCs. Therefore, it appears that the potential for transdifferentiation into ECs is specifically activated in GSCs, and this potential is intensively regulated by hypoxia conditions. Because GSCs reside in a hypoxic niche, it is reasonable to assume that hypoxia not only maintains the GSC state, but also elicits their transdifferentiation potentials. We further demonstrated that hypoxia-induced transdifferentiation of GSCs requires CDH5 expression that is regulated by HIFs. Surprisingly, knockdown of either HIF1α or HIF2α downregulated about 90% of CDH5 levels in GSCs, implying that both HIF1α and HIF2α are needed for CDH5 expression in GSCs. This notion is consistent with the fact that CDH5 is weakly or not expressed in NSCs and non-GSCs because it has been reported that HIF2α is specifically expressed in GSCs rather than NSCs and non-GSCs.10 Importantly, CDH5 expression is a potential independent prognostic factor for GBM, in which CDH5 expression may reflect the process of angiogenesis in GBM involving CDH5 induction or the existence of CDH5+ GSCs that acquire the potential to transdifferentiate into ECs.
It has been reported that normal NSCs can also transdifferentiate into ECs under certain conditions.59 However, CDH5 was weakly or not expressed in normal NSCs in the present study, implying that the capacity of NSCs to transdifferentiate into ECs is conditionally limited and can be activated only under certain conditions. However, this capacity is enforced in GSCs even in a “normal” tumor environment. One explanation is that CDH5 expression depends highly on both HIF1α and HIF2α, but HIF2α is weakly or not expressed in NSCs. The low expression of CDH5 in non-GSCs possibly reflects the diminished multipotency of these cells under differentiation conditions in cultures containing serum medium. Because complicated genetic and epigenetic alterations affect the states of GSCs, it is possible that the culture system for GSCs, which contains epidermal growth factor and basic fibroblast growth factor, may have a major effect on CDH5 expression. We found that both HIF2α and HIF1α bind to the CDH5 promoter, which is inconsistent with previous results, possibly due to the different cofactors of HIFs and the epigenetic contexts in these 2 different cell types. Different cofactors and epigenetic states may influence the affinity of transcriptional factors for their target genes.60,61 The dependence of CDH5 expression on both HIFs in GSCs implies that a transcriptional complex regulating CDH5 involves both HIF1α and HIF2α. It is possible that, like tumor stem cells, GSCs acquire an aberrant genetic and epigenetic background in which the program for EC differentiation can be activated and involves HIF2α and CDH5 expression. Such a differentiation program is compatible with the transformed states of GSCs, which confer upon them the ability to function as both tumor cells and ECs. Nonetheless, the detailed mechanisms controlling CDH5 expression in GSCs remain to be further explored.
An important result in this study was obtained in the bioinformatics analysis. Using 4 independent glioma datasets, we identified the most reliable (but not all) genes that may interact directly with CDH5, including several genes, such as ERG and EPAS1 (HIF2α), that have been confirmed by other studies. Notably, most of the remaining genes were EC markers or closely related to vasculogenesis. We also analyzed the other genes and found that all of them are closely related to ECs (data not shown). These results corroborate the results of the bioinformatics analysis, especially using more than 1 independent dataset. If the genes that interact directly with CDH5 and are closely related to CDH5 are treated as useful genes, then the reliable rate is almost 100% when considering the overlapped genes in all of the datasets.
We revealed novel features of GSCs that specifically express the EC marker CDH5 that is regulated by HIFs under hypoxic conditions. Therefore, neoangiogenesis may occur in glioma cells where tumor cells are overgrowing but blood vessels are lacking. Given the critical role of CDH5 in angiogenesis, it is possible that induction of CDH5 may be an important process during hypoxia-induced angiogenesis. However, the mechanisms of GSC transdifferentiation into ECs and how CDH5 expression is activated in GSCs are unclear. In addition, the exact roles of CDH5 during transdifferentiation of GSCs into ECs remain to be elucidated. Further study of these issues will provide a better understanding of the regulation mechanisms of the GSC state and thus provide potential therapeutic strategies to target GSCs.
Supplementary Material
Funding
This study was partially supported by the National Natural Science Foundation of China (81171087).
Supplementary Material
Acknowledgments
We thank Dr. Fucheng Ma for confirming the histopathological diagnosis of the samples used in this research. We also thank Juan Li, Yufen Shi, Xiaoyan Chen, and Junli Huo for technical and administrative support.
Conflict of interest statement. None declared.
References
- 1.Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY. Primary brain tumours in adults. Lancet. 2003;361(9354):323–331. doi: 10.1016/S0140-6736(03)12328-8. doi:10.1016/S0140-6736(03)12328-8. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. doi:10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. doi:10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Vescovi AL, Galli R, Reynolds BA. Brain tumour stem cells. Nat Rev Cancer. 2006;6(6):425–436. doi: 10.1038/nrc1889. doi:10.1038/nrc1889. [DOI] [PubMed] [Google Scholar]
- 5.Mao XG, Zhang X, Zhen HN. Progress on potential strategies to target brain tumor stem cells. Cell Mol Neurobiol. 2009;29(2):141–155. doi: 10.1007/s10571-008-9310-1. doi:10.1007/s10571-008-9310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. doi:10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 7.Charles N, Ozawa T, Squatrito M, et al. Perivascular nitric oxide activates Notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6(2):141–152. doi: 10.1016/j.stem.2010.01.001. doi:10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidel S, Garvalov BK, Wirta V, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain. 2010;133(Pt 4):983–995. doi: 10.1093/brain/awq042. doi:10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 9.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. doi:10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Bao S, Wu Q, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–513. doi: 10.1016/j.ccr.2009.03.018. doi:10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar EE, Lin A, Mahairaki V, Matsui W, Eberhart CG. Hypoxia increases the expression of stem-cell markers and promotes clonogenicity in glioblastoma neurospheres. Am J Pathol. 2010;177(3):1491–1502. doi: 10.2353/ajpath.2010.091021. doi:10.2353/ajpath.2010.091021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. doi:10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 13.Lu C, Shervington A. Chemoresistance in gliomas. Mol Cell Biochem. 2008;312(1–2):71–80. doi: 10.1007/s11010-008-9722-8. doi:10.1007/s11010-008-9722-8. [DOI] [PubMed] [Google Scholar]
- 14.Eramo A, Ricci-Vitiani L, Zeuner A, et al. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13(7):1238–1241. doi: 10.1038/sj.cdd.4401872. doi:10.1038/sj.cdd.4401872. [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Chadalavada K, Wilshire J, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–833. doi: 10.1038/nature09624. doi:10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 16.Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–828. doi: 10.1038/nature09557. doi:10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 17.Soda Y, Marumoto T, Friedmann-Morvinski D, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108(11):4274–4280. doi: 10.1073/pnas.1016030108. doi:10.1073/pnas.1016030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Hallani S, Boisselier B, Peglion F, et al. A new alternative mechanism in glioblastoma vascularization: tubular vasculogenic mimicry. Brain. 2010;133(Pt 4):973–982. doi: 10.1093/brain/awq044. doi:10.1093/brain/awq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paez-Ribes M, Allen E, Hudock J, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15(3):220–231. doi: 10.1016/j.ccr.2009.01.027. doi:10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keunen O, Johansson M, Oudin A, et al. Anti-VEGF treatment reduces blood supply and increases tumor cell invasion in glioblastoma. Proc Natl Acad Sci U S A. 2011;108(9):3749–3754. doi: 10.1073/pnas.1014480108. doi:10.1073/pnas.1014480108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmeliet P, Lampugnani MG, Moons L, et al. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98(2):147–157. doi: 10.1016/s0092-8674(00)81010-7. doi:10.1016/S0092-8674(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 22.Bach TL, Barsigian C, Chalupowicz DG, et al. VE-cadherin mediates endothelial cell capillary tube formation in fibrin and collagen gels. Exp Cell Res. 1998;238(2):324–334. doi: 10.1006/excr.1997.3844. doi:10.1006/excr.1997.3844. [DOI] [PubMed] [Google Scholar]
- 23.Corada M, Zanetta L, Orsenigo F, et al. A monoclonal antibody to vascular endothelial–cadherin inhibits tumor angiogenesis without side effects on endothelial permeability. Blood. 2002;100(3):905–911. doi: 10.1182/blood.v100.3.905. doi:10.1182/blood.V100.3.905. [DOI] [PubMed] [Google Scholar]
- 24.Liao F, Li Y, O'Connor W, et al. Monoclonal antibody to vascular endothelial–cadherin is a potent inhibitor of angiogenesis, tumor growth, and metastasis. Cancer Res. 2000;60(24):6805–6810. [PubMed] [Google Scholar]
- 25.Mao XG, Zhang X, Xue XY, et al. Brain tumor stem-like cells identified by neural stem cell marker CD15. Transl Oncol. 2009;2(4):247–257. doi: 10.1593/tlo.09136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao XG, Yan M, Xue XY, et al. Overexpression of ZNF217 in glioblastoma contributes to the maintenance of glioma stem cells regulated by hypoxia-inducible factors. Lab Invest. 2011;91(7):1068–1078. doi: 10.1038/labinvest.2011.56. doi:10.1038/labinvest.2011.56. [DOI] [PubMed] [Google Scholar]
- 27.Margolin AA, Nemenman I, Basso K, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. doi:10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.TCGA. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. doi:10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. doi:10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. doi:10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 31.Lee Y, Scheck AC, Cloughesy TF, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. doi:10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carro MS, Lim WK, Alvarez MJ, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463(7279):318–325. doi: 10.1038/nature08712. doi:10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma S, Sharma MC, Gupta DK, Sarkar C. Angiogenic patterns and their quantitation in high grade astrocytic tumors. J Neurooncol. 2006;79(1):19–30. doi: 10.1007/s11060-006-9120-6. doi:10.1007/s11060-006-9120-6. [DOI] [PubMed] [Google Scholar]
- 34.Folkerth RD. Descriptive analysis and quantification of angiogenesis in human brain tumors. J Neurooncol. 2000;50(1–2):165–172. doi: 10.1023/a:1006499824379. doi:10.1023/A:1006499824379. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. doi:10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 36.Mao XG, Guo G, Wang P, et al. Maintenance of critical properties of brain tumor stem like cells after cryopreservation. Cell Mol Neurobiol. 2010;30(5):775–786. doi: 10.1007/s10571-010-9505-0. doi:10.1007/s10571-010-9505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margolin AA, Wang K, Lim WK, Kustagi M, Nemenman I, Califano A. Reverse engineering cellular networks. Nat Protoc. 2006;1(2):662–671. doi: 10.1038/nprot.2006.106. doi:10.1038/nprot.2006.106. [DOI] [PubMed] [Google Scholar]
- 38.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37(4):382–390. doi: 10.1038/ng1532. doi:10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Carson-Walter EB, Cooper A, Winans BN, Johnson MD, Walter KA. Vascular gene expression patterns are conserved in primary and metastatic brain tumors. J Neurooncol. 2010;99(1):13–24. doi: 10.1007/s11060-009-0105-0. doi:10.1007/s11060-009-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallgard E, Larsson E, He L, et al. Identification of a core set of 58 gene transcripts with broad and specific expression in the microvasculature. Arterioscler Thromb Vasc Biol. 2008;28(8):1469–1476. doi: 10.1161/ATVBAHA.108.165738. doi:10.1161/ATVBAHA.108.165738. [DOI] [PubMed] [Google Scholar]
- 41.Yuan L, Sacharidou A, Stratman AN, et al. RhoJ is an endothelial cell-restricted Rho GTPase that mediates vascular morphogenesis and is regulated by the transcription factor ERG. Blood. 2011;118(4):1145–1153. doi: 10.1182/blood-2010-10-315275. doi:10.1182/blood-2010-10-315275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birdsey GM, Dryden NH, Amsellem V, et al. Transcription factor Erg regulates angiogenesis and endothelial apoptosis through VE-cadherin. Blood. 2008;111(7):3498–3506. doi: 10.1182/blood-2007-08-105346. doi:10.1182/blood-2007-08-105346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11(1):72–82. doi: 10.1101/gad.11.1.72. doi:10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 44.Le Bras A, Lionneton F, Mattot V, et al. HIF-2alpha specifically activates the VE-cadherin promoter independently of hypoxia and in synergy with Ets-1 through two essential ETS-binding sites. Oncogene. 2007;26(53):7480–7489. doi: 10.1038/sj.onc.1210566. doi:10.1038/sj.onc.1210566. [DOI] [PubMed] [Google Scholar]
- 45.Soeda A, Park M, Lee D, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28(45):3949–3959. doi: 10.1038/onc.2009.252. doi:10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 46.Mendez O, Zavadil J, Esencay M, et al. Knock down of HIF-1alpha in glioma cells reduces migration in vitro and invasion in vivo and impairs their ability to form tumor spheres. Mol Cancer. 2010;9:133. doi: 10.1186/1476-4598-9-133. doi:10.1186/1476-4598-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8(20):3274–3284. doi: 10.4161/cc.8.20.9701. doi:10.4161/cc.8.20.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Semenza GL, Jiang BH, Leung SW, et al. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. doi: 10.1074/jbc.271.51.32529. doi:10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 49.Mole DR, Blancher C, Copley RR, et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284(25):16767–16775. doi: 10.1074/jbc.M901790200. doi:10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9(6):677–684. doi: 10.1038/nm0603-677. doi:10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- 51.Mazumdar J, O'Brien WT, Johnson RS, et al. O(2) regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol. 2010;12(10):1007–1013. doi: 10.1038/ncb2102. doi:10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129(3):465–472. doi: 10.1016/j.cell.2007.04.019. doi:10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platet N, Liu SY, Atifi ME, et al. Influence of oxygen tension on CD133 phenotype in human glioma cell cultures. Cancer Lett. 2007;258(2):286–290. doi: 10.1016/j.canlet.2007.09.012. doi:10.1016/j.canlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 54.Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8(12):967–975. doi: 10.1038/nrc2540. doi:10.1038/nrc2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pouyssegur J, Dayan F, Mazure NM. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441(7092):437–443. doi: 10.1038/nature04871. doi:10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- 56.Semenza GL. Life with oxygen. Science. 2007;318(5847):62–64. doi: 10.1126/science.1147949. doi:10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- 57.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. doi:10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Labelle M, Schnittler HJ, Aust DE, et al. Vascular endothelial cadherin promotes breast cancer progression via transforming growth factor beta signaling. Cancer Res. 2008;68(5):1388–1397. doi: 10.1158/0008-5472.CAN-07-2706. doi:10.1158/0008-5472.CAN-07-2706. [DOI] [PubMed] [Google Scholar]
- 59.Wurmser AE, Nakashima K, Summers RG, et al. Cell fusion-independent differentiation of neural stem cells to the endothelial lineage. Nature. 2004;430(6997):350–356. doi: 10.1038/nature02604. doi:10.1038/nature02604. [DOI] [PubMed] [Google Scholar]
- 60.Xia X, Kung AL. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009;10(10):R113. doi: 10.1186/gb-2009-10-10-r113. doi:10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lou F, Chen X, Jalink M, et al. The opposing effect of hypoxia-inducible factor-2alpha on expression of telomerase reverse transcriptase. Mol Cancer Res. 2007;5(8):793–800. doi: 10.1158/1541-7786.MCR-07-0065. doi:10.1158/1541-7786.MCR-07-0065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.