Abstract
Coronavirus transcription leads to the synthesis of a nested set of mRNAs with a leader sequence derived from the 5′ end of the genome. The mRNAs are produced by a discontinuous transcription in which the leader is linked to the mRNA coding sequences. This process is regulated by transcription-regulating sequences (TRSs) preceding each mRNA, including a highly conserved core sequence (CS) with high identity to sequences present in the virus genome and at the 3′ end of the leader (TRS-L). The role of TRSs was analyzed by reverse genetics using a full-length infectious coronavirus cDNA and site-directed mutagenesis of the CS. The canonical CS-B was nonessential for the generation of subgenomic mRNAs (sgmRNAs), but its presence led to transcription levels at least 103-fold higher than those in its absence. The data obtained are compatible with a transcription mechanism including three steps: (i) formation of 5′-3′ complexes in the genomic RNA, (ii) base-pairing scanning of the nascent negative RNA strand by the TRS-L, and (iii) template switching during synthesis of the negative strand to complete the negative sgRNA. This template switch takes place after copying the CS sequence and was predicted in silico based on high base-pairing score between the nascent negative RNA strand and the TRS-L and minimum ΔG.
Transmissible gastroenteritis virus (TGEV) is a member of the Coronaviridae family, included in the Nidovirales order (10). TGEV is an enveloped virus with a single-stranded, positive-sense 28.5-kb RNA genome (28) for which infectious cDNA clones have been engineered (1, 12, 41). About the 5′ two-thirds of the entire RNA comprises open reading frames (ORFs) 1a and 1ab encoding the replicase (rep). The 3′ one-third of the genome includes the genes encoding the structural and nonstructural proteins, in the order 5′-S-3a-3b-E-M-N-7-3′ (9).
Coronavirus transcription is based on RNA-dependent RNA synthesis. The result of this process is the generation of a nested set of six to eight mRNAs of various sizes, depending on the coronavirus strain. These mRNAs are 5′- and 3′-coterminal with the genome. The largest mRNA is the genomic RNA (gRNA), which also serves as the mRNA for the rep1a and rep1b genes. A leader sequence of 93 nucleotides (nt), derived from the 5′ end of the genome, is fused to the 5′ end of the mRNA coding sequence (body) by a discontinuous transcription mechanism (18, 32).
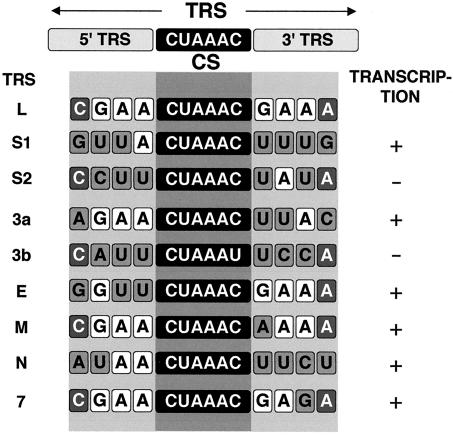
Sequences at the 5′ end of each gene represent signals that regulate the discontinuous transcription of subgenomic mRNAs (sgmRNAs). These are the transcription-regulating sequences (TRSs) that include a core sequence (CS; 5′-CUAAAC-3′), highly conserved in all TGEV genes, and the 5′ and 3′ flanking sequences (5′ TRS and 3′ TRS, respectively) that modulate transcription (2). Previous studies using TGEV minigenomes have shown that the CS was required for transcription and that the synthesis of sgmRNAs only proceeds when this CS is located in an appropriate sequence context (2).
Two major models have been proposed to explain the discontinuous transcription in coronavirus and arterivirus (18, 32). The discovery of transcriptionally active, subgenomic-size negative strands containing the antileader (cL) sequence and of transcription intermediates active in the synthesis of mRNAs (30, 31, 33, 34) favors the model of discontinuous transcription during the negative-strand synthesis (32). This concept was reinforced by demonstrating in arterivirus that the CS included in the sgmRNA was derived from the CS preceding each gene (CS-B) and not from the CS present at the 3′ end of the leader sequence (CS-L) (26, 38) (Fig. 1). According to this model of discontinuous sgRNA synthesis during production of the negative strand, the TRS-B acts as a slow-down and detaching signal for the transcription complex.
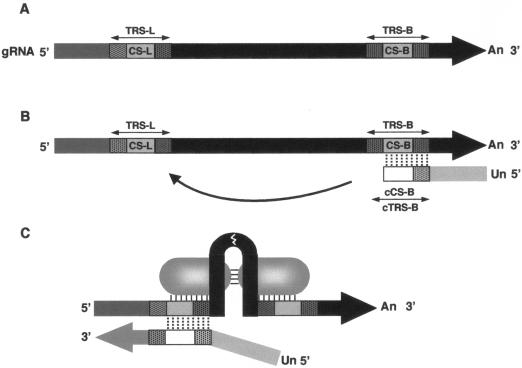
FIG. 1.
Diagram of the elements involved in coronavirus transcription. (A) The scheme represents all of the sequence elements probably involved in the discontinuous negative-strand synthesis model. CS-L, leader CS; CS-B, body CS. TRS-L and TRS-B, transcription-regulating sequences from the leader and body, respectively. An, poly(A). (B) Representation of the discontinuous transcription during negative-strand synthesis. cCS-B and cTRS-B represent the CS-B and TRS-B complementary sequences, respectively. Un, poly(U). (C) Leader and body sequences are probably located close to one another in higher-order structures maintained by RNA-protein and protein-protein interactions.
Transcription regulation is probably a multifactor process in which three factors may have a relevant role: (i) base pairing between the TRS-L and the nascent negative strand, (ii) proximity to the 3′ end of the genome, and (iii) RNA-protein and protein-protein interactions within TRSs.
The synthesis of a negative sgRNA is most probably mediated by a direct base-pairing interaction between the nascent negative body TRS (cTRS-B) and the 3′ end of the leader (TRS-L). The conserved sequence of this TRS, the CS-L, is probably exposed in a stem-loop at the 5′ end of the viral genome both in TGEV (S. Alonso, I. Sola, S. Zúñiga, and L. Enjuanes, unpublished) and in equine arteritis virus (EAV) (26, 38), although this RNA structure has not been experimentally proven.
Proximity to the 3′ end of the genome probably influences the relative amount of sgmRNAs, because the polymerase complex finds less slow-down and detaching signals during small negative sgRNA synthesis. Therefore, in principle, these RNAs could be the most abundant. Although this is the case in the order Mononegavirales (15, 39) and, in general, in coronaviruses, the relative amounts of coronavirus mRNAs are not strictly related to their proximity to the viral 3′ end (28, 37). Therefore, other factors may also regulate coronavirus transcription.
The interaction of RNA with viral and cellular proteins is probably involved in coronavirus transcription. The discontinuous synthesis of the negative RNA strand resembles a high-frequency copy-choice RNA recombination (3, 21, 26), in which the TRS-B (donor) and TRS-L (acceptor) sequences, located in distal domains in the RNA primary structure, are probably brought into physical proximity by RNA-protein and protein-protein interactions (Fig. 1C).
In arterivirus, base pairing between the leader CS and the negative-sense body CS (cCS-B) has been implicated in transcription, although the roles of other factors, such as relative TRS position in the genome and secondary structure, have led to less clear conclusions (25-27).
In this report, the role of CS sequences in coronavirus transcription is analyzed for the first time by using TGEV full-length genomes constructed with an infectious cDNA clone (1). The role of each nucleotide within the leader and body CSs has been studied by introducing point mutations in these sequences. A key strategy in these studies has been analysis of gene 3a transcription, because this gene is nonessential for TGEV replication (36). Therefore, infectious virus was rescued for all gene 3a CS-B mutants, allowing subsequent analysis. We show in the studies reported here that the presence of the highly conserved CS was associated with sgmRNA production and high virus titers, but that this sequence was not essential for sgmRNA synthesis when the TRS-L to cTRS-B duplex formation involved a high release of free energy (ΔG). In fact, the genome positions in which a negative sgRNA most frequently fused to the leader could be predicted in silico by determining the identity between the TRS-L and sequence domains of the genome. To this end, a computer-based program has been developed to assess the strength of base pairing between body and leader TRS that successfully predicts the authentic products as well as novel, mutant-derived sgmRNAs. In addition, it has been shown that nucleotide substitutions in the canonical CS led to the use of alternative noncanonical CSs, providing that sequences flanking the CS-L were also flanking the CS-B, leading to a favorable ΔG in duplex formation between TRS-L and cTRS-B. It has also been shown that during the synthesis of TGEV negative sgRNAs, template switching always took place after copying the canonical or noncanonical CS sequence, supporting the finding that coronavirus RNA discontinuous synthesis takes place during production of the negative strand. A three-step mechanism has been proposed as a working model for coronavirus mRNA transcription.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney cells (BHK-21) stably transformed with the gene coding for the porcine aminopeptidase N (BHK-pAPN) (6) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) and G418 (1.5 mg/ml) as a selection agent. Viruses were grown in swine testis (ST) cells (20).
Plasmid constructs.
TGEV cDNAs with point mutations in the leader and body CS were generated by overlapping PCR. To get leader CS mutants, the plasmid pBAC-TGEV(SrfI-NheI), which bears nt 1 to 15062 from the TGEV genome (GenBank accession no. AJ271965) except a ClaI-ClaI fragment (nt 4417 to 9615) (1), was used as template. Overlapping PCR fragments with point mutations were amplified by using the oligonucleotides described in Table 1. The final PCR product (2,415 bp), amplified with outer oligonucleotides Oli 5′I and Oli 3′D, was digested with SfiI and ApaLI and cloned into the same restriction sites of plasmid pBAC-TGEV(SrfI-NheI). To introduce mutations in the TGEV infectious cDNA, SfiI-ClaI fragment (5,277 bp) from pBAC-TGEV(SrfI-NheI) with the corresponding mutation was cloned into the same sites of pBAC-TGEVΔCla; after that, the toxic ClaI-ClaI fragment (5,198 bp) was introduced as previously described (1).
TABLE 1.
Oligonucleotides used for site-directed mutagenesis
| Mutant or othera | Oligonucleotideb | Oligonucleotide sequence 5′ → 3′c |
|---|---|---|
| L-mutant oligonucleotide | Oli 5′I | CGCGAATTCGATGATAAGCTGTCAAAC |
| Oli 3′D | CGCGAATTCCTCTACTACTTTCCAAGCGTC | |
| L-C1G | MutC94G-VS | CAACTCGAAGTAAACGAAATATT |
| MutC94G-RS | AATATTTCGTTTACTTCGAGTTG | |
| L-U2G | MutU95G-VS | CAACTCGAACGAAACGAAATATT |
| MutU95G-RS | AATATTTCGTTTCGTTCGAGTTG | |
| L-A3C | MutA96C-VS | CAACTCGAACTCAACGAAATATT |
| MutA96C-RS | AATATTTCGTTGAGTTCGAGTTG | |
| L-A4C | MutA97C-VS | CAACTCGAACTACACGAAATATT |
| MutA97C-RS | AATATTTCGTGTAGTTCGAGTTG | |
| L-A5C | MutA98C-VS | CAACTCGAACTAACCGAAATATT |
| MutA98C-RS | AATATTTCGGTTAGTTCGAGTTG | |
| L-C6G | MutC99G-VS | CAACTCGAACTAAAGGAAATATT |
| MutC99G-RS | AATATTTCCTTTAGTTCGAGTTG | |
| L-C1U | MutC94U-VS | CAACTCGAATTAAACGAAATATT |
| MutC94U-RS | AATATTTCGTTTAATTCGAGTTG | |
| L-A3U | MutA96U-VS | CAACTCGAACTTAACGAAATATT |
| MutA96U-RS | AATATTTCGTTAAGTTCGAGTTG | |
| L-A4U | MutA97U-VS | CAACTCGAACTATACGAAATATT |
| MutA97U-RS | AATATTTCGTATAGTTCGAGTTG | |
| L-A5U | MutA98U-VS | CAACTCGAACTAATCGAAATATT |
| MutA98U-RS | AATATTTCGATTAGTTCGAGTTG | |
| L-C6U | MutC99U-VS | CAACTCGAACTAAATGAAATATT |
| MutC99U-RS | AATATTTCATTTAGTTCGAGTTG | |
| B-mutant oligonucleotide | S-3839-VS | GTTGCAACTAGTTCTGACT |
| 3a-169-RS | CAATAATGGAGAGACCAAG | |
| B-C1G | MutC24798G-VS | TTTAAGAAGTAAACTTACGAGTC |
| MutC24798G-RS | GACTCGTAAGTTTACTTCTTAAA | |
| B-U2G | MutU24799G-VS | TTTAAGAACGAAACTTACGAGTC |
| MutU24799G-RS | GACTCGTAAGTTTCGTTCTTAAA | |
| B-A3C | MutA24800C-VS | TTTAAGAACTCAACTTACGAGTC |
| MutA24800C-RS | GACTCGTAAGTTGAGTTCTTAAA | |
| B-A4C | MutA24801C-VS | TTTAAGAACTACACTTACGAGTC |
| MutA24801C-RS | GACTCGTAAGTGTAGTTCTTAAA | |
| B-A5C | MutA24802C-VS | TTTAAGAACTAACCTTACGAGTC |
| MutA24802C-RS | GACTCGTAAGGTTAGTTCTTAAA | |
| B-C6G | MutC24803G-VS | TTTAAGAACTAAAGTTACGAGTC |
| MutC24803G-RS | GACTCGTAACTTTAGTTCTTAAA |
Virus names were derived from leader CS mutants (L) or CS-3a mutants (B) and indicate the nucleotide substitution and its position in the CS.
Oligonucleotides including the punctual mutations are named “Mut” and indicate the nucleotide substitution and its position at the TGEV genome. VS, virus sense; RS, reverse sense.
The mutated nucleotide is shown in boldface. Restriction endonuclease sites used for cloning are shown in italics (EcoRI, GAATTC; SpeI, ACTAGT). CS and cCS are underlined.
To generate CS-3a mutants, plasmid pSL(AvrII-AvrII), containing nt 22965 to 25865 from the TGEV genome, was used as a template for the overlapping PCR. Fragments were amplified with the oligonucleotides described in Table 1. The final PCR product (832 bp), amplified with outer oligonucleotides S-3839-VS and 3a-169-RS, was digested with SpeI and Tth111I and cloned in the same sites of pSL(AvrII-AvrII). To introduce mutations in the TGEV infectious cDNA, AvrII digestion product (2,900 bp) from pSL(AvrII-AvrII) with the corresponding mutation was cloned into the same sites of pBAC-TGEVΔC1a. To obtain the full-length TGEV cDNA, the toxic ClaI-ClaI fragment (5,198 bp) was introduced as previously described (1).
Double CS-L and CS-B mutants were obtained by introducing SfiI-ApaLI fragment from pBAC-TGEV(SrfI-NheI) plasmid with the leader mutation into the same restriction sites of pBAC-TGEVΔCla bearing the corresponding CS-3a mutation. The plasmid containing the full-length TGEV cDNA with point mutations was then generated as previously described.
All cloning steps were checked by sequencing the PCR-amplified fragments and cloning junctions.
Transfection and recovery of infectious TGEV from cDNA clones.
BHK-pAPN cells were grown to confluence in 35-mm-diameter plates and transfected with 4 μg of the appropriate full-length TGEV cDNA clone and 12 μl of Lipofectamine 2000 (Invitrogen) according to the manufacturer's specifications. The estimated transfection efficiency of the TGEV cDNA using this system was around 20% in all cases. Cells were incubated at 37°C for 6 h, and then the transfection medium was discarded, 200 μl of trypsin-EDTA was added, and trypsinized cells were plated over a confluent ST monolayer grown in a 35-mm-diameter plate. After a 2-day incubation period, the cell supernatants (referred to as passage 0) were harvested and stored. Virus from passage 0 supernatant was cloned by three plaque purification steps. Recombinant TGEV (rTGEV) viruses were grown and titrated as described previously (16).
RNA analysis by Northern blotting.
Total intracellular RNA was extracted at 18 to 24 h postinfection (hpi) from virus-infected ST cells by using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. RNAs were separated in denaturing 1% agarose-2.2 M formaldehyde gels and blotted onto positively charged nylon membranes (BrightStar-Plus; Ambion) as described previously (2). The 3′ untranslated region (UTR)-specific single-stranded DNA probe was complementary to nt 28300 to 28544 of the TGEV strain PUR46-MAD genome (28). Probe labeling was performed with the BrightStar psoralen-biotin nonisotopic labeling kit (Ambion), and Northern hybridizations were performed according to the manufacturer's instructions. Detection was done with the BrightStar BioDetect kit (Ambion).
RNA analysis by RT-PCR.
Analysis of mutant virus RNAs was performed by reverse transcription-PCR (RT-PCR). Total intracellular RNA was extracted at 18 hpi from ST cells infected with rTGEV viruses as previously described. cDNAs were synthesized at 42°C for 1 h with Moloney murine leukemia virus reverse transcriptase (Mo-MuLV-RT) (Ambion) and the antisense primers described in Table 2. The cDNAs generated were used as templates for specific PCR amplification using the reverse primers described in Table 2 and the forward primer SP (5′-GTGAGTGTAGCGTGGCTATATGTGT-3′), complementary to nt 15 to 39 of the TGEV leader sequence. RT-PCR products were separated by electrophoresis in 0.8% or 1.5% agarose gels, purified, and used for direct sequencing with the SP oligonucleotide and the same reverse primer used for PCR.
TABLE 2.
Reverse oligonucleotides used for RT-PCR analysis of RNA from rTGEV-infected cells
| sgmRNA | Primer | Sequence (5′→3′) | Amplicon size (bp) |
|---|---|---|---|
| Genomic | 1a-156-RS | TCCTTCGATCGCAATCAA | 473 |
| mRNA-S | S-449 | TAACCTGCACTCACTACCCC | 499 |
| mRNA-3a | 3a-169-RS | CAATAATGGAGAGACCAAG | 295 |
| mRNA-3b | X2B-112 | TTAACATACCAAAAGTATGC | 458 |
| mRNA-E | IGSM | CAGTCGACAGGCCTCGCCGGCGCGGCCGCGTTTAGTTCAAGC | 393 |
| mRNA-M | M.415RS | AGACCACCAAGAGTTAGTCC | 530 |
| mRNA-N | N-268RS | GGTCCGGTACCTAAGTAGTAGAAGAACC | 386 |
| mRNA-7 | 7(213)RS | TCTGTAGCAGCAAAATCC | 302 |
Real-time RT-PCR was used for quantitative analysis of gRNA (used as an endogenous standard) and mRNA 3a species. Oligonucleotides used for RT and PCRs, described in Table 3, were designed with Primer Express software. In the PCR step, SYBR Green PCR master mix (Applied Biosystems) was used according to the manufacturer's specifications. Detection was performed with an ABI PRISM 7000 sequence detection system (Applied Biosystems). Data were analyzed with ABI PRISM 7000 SDS version 1.0 software.
TABLE 3.
Oligonucleotides used for real-time RT-PCR analysis
| Amplicon | Size (bp) | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|---|
| Virus | 80 | TTCTTTTGACAAAACATACGGTGAA | CTAGGCAACTGGTTTGTAACATCTT |
| mRNA-3a.1 | 102 | CGGACACCAACTCGAACTAAACTTAC | ATCAAGTTCGTCAAGTACAGCATCTA |
| mRNA-3a.2 | 93 | CGTGGCTATATCTCTTCTTTTACTTTAACTAG | ATGGACGTGCACTTTTTCAATTG |
In silico analysis.
Free energy calculations were done using the two-state hybridization server (http://www.bioinfo.rpi.edu/applications/mfold/) (19). Potential base-pairing score calculations were done with the LALIGN program at the public ISREC LALIGN server (http://www.ch.embnet.org/). This is a local alignment tool that implements the algorithm of Huang and Miller (14). Briefly, the TGEV genome was divided into 500-nt pieces and compared with the leader TRS (nt 90 to 103 of TGEV genome) by using the LALIGN program. The alignment score and position data obtained from the LALIGN program were introduced in an Excel table to generate the graphical output. To automate this process, a PERL script was developed that fragments the complete TGEV genome sequence with the desired overlap (usually a 20-nt overlap was used), submits those fragments to LALIGN server automatically, and provides the results in a tabulated format ready to generate the graphical output with Excel.
The in silico analysis was performed with TRS-L sequences of different lengths and several coronavirus genomes: TGEV, human and bovine coronavirus (HCoV and BCoV, respectively). Since viral mRNAs always were generated from a TRS with a base-pairing score of ≥35, this value was selected as the threshold, although all of the values were taken into account. In these analyses, a score below 18 was never obtained, because the LALIGN program provides only the best local alignments. For the same reason, score values were discrete points in several positions distributed along the genome, but to facilitate data visualization, a continuous line representation was selected as the graphical output.
RESULTS
Relevance of base pairing between the CS-L and cCS-B in coronavirus transcription.
To study the relevance of the base pairing between CS-L and cCS-B, each of the 6 nt was substituted in the CS-L or in the gene 3a CS (CS-3a). Nucleotide changes within CS-3a, in principle, should only affect the synthesis of mRNA-3a. In contrast, nucleotide substitutions in the CS-L would have a pleiotropic effect on the synthesis of all mRNAs. Four groups of mutant TGEV infectious cDNA clones were generated (Fig. 2): (i) CS-B mutants, replacing each base of CS-3a by nucleotides that do not allow base pairing of the cCS-B with the CS-L (Fig. 2A); (ii) CS-L mutants with changes identical to those introduced in the CS-B mutants (Fig. 2B); (iii) CS-L mutants with changes allowing non-Watson-Crick base pairing with the cCS-B of all genes (Fig. 2C); and (iv) double mutants in which the complementarity between CS-L and cCS-3a was restored (Fig. 2D).
FIG. 2.
Mutations introduced in the TGEV full-length cDNA and virus recovery. Nucleotide substitutions were introduced in the 3a gene CS (CS-B mutants [A]), the leader CS (CS-L mutants [B]), in both the CS-L and CS-B (double mutants [D]), and leader CS mutants with changes allowing non-Watson-Crick base pairing with the body cCS (non-Watson-Crick mutants [C]). Virus titers (PFU per milliliter) obtained for the passage 0 supernatant are indicated in the figure.
Viruses were recovered from all CS-3a mutants, with titers similar to those obtained with the wild-type TGEV cDNA (Fig. 2A), as expected, since gene 3a is nonessential. In contrast, no virus was recovered from cDNAs when CS-L nt 1 to 3 were changed in the single or double mutants (Fig. 2B and D). Nucleotide substitutions in CS-L positions 4 to 6 led to the recovery of infectious recombinant TGEV (rTGEV) with titers up to 105-fold lower than the parental ones. Leader and double mutants showed the same behavior (Fig. 2B and D), as expected, since leader mutations affected the synthesis of all sgmRNAs.
Interestingly, infectious rTGEV was recovered from all non-Watson-Crick leader mutants, with titers ranging from wild-type levels, like those obtained for L-C6U mutant, to 105-fold lower for the L-C1U mutant (Fig. 2C). Overall, these data indicated the requirement of base pairing between CS-L and cCS-B for sgmRNA synthesis.
Relationship between CS-L and CS-3a sequences and sgmRNA levels.
It was postulated that synthesis of negative sgRNAs is mediated by direct base pairing between the TRS-L and the cTRS-B. This being the case, the CS-L and CS-3a sequences should modulate sgmRNA-3a levels. To determine whether this was the case, the pattern of sgmRNA synthesis produced by different rTGEVs with CS point mutations was analyzed by Northern blotting (Fig. 3). Nucleotide substitutions within the first 3 nt of CS-L led to no virus rescue, and it was not possible to analyze the sgmRNA pattern. To evaluate sgmRNA synthesis by Northern blot analysis, because mutations in CS-L sequence positions 4 to 6 considerably reduce sgmRNA production, the multiplicity of infection (MOI) and the amount of total RNA from the leader and double mutants loaded in the gel were increased in order to obtain similar levels of viral RNA (Fig. 3). The viral sgmRNA pattern for the wild-type virus was the expected one, but new bands were identified in all CS mutants (Fig. 3). Some of these unexpected bands were amplified by RT-PCR and sequenced, corresponding to alternative sgmRNAs for the S, 3a, and N genes. These data indicated that changes in the CS-L or CS-B opened new base-pairing possibilities throughout the genome, leading to the generation of alternative sgmRNAs.
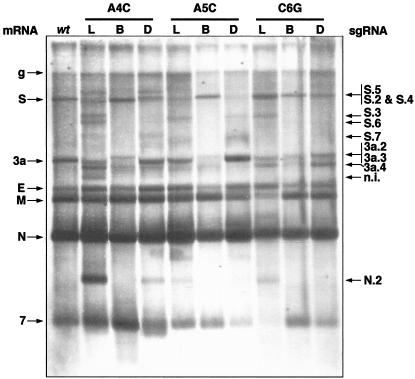
FIG. 3.
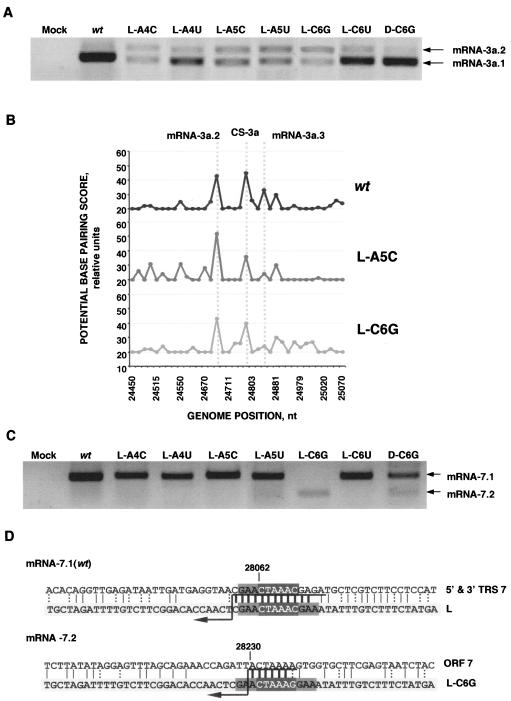
Northern blot analysis of rTGEVs. ST cells were infected with rTGEV at an MOI of 0.5 (for the wild type [wt] and CS-B mutants) or 1 (for CS-L and double mutants). Total RNA was extracted at 20 hpi and analyzed by Northern blotting with a probe complementary to the 3′ end of the gRNA. To normalize the amount of viral RNA in the gel, lanes L and D were loaded with three times the amount of the other lanes. L, CS-L mutant; B, CS-B mutant; D, double mutant. Viral mRNAs are indicated on the left side of the figure, and new sgmRNAs that have been clearly identified are indicated on the right (some of them correspond to the alternative sgRNAs analyzed in this work, indicated by the same number). n.i., still unidentified sgmRNAs.
The sgmRNA pattern for CS-3a mutants was also analyzed by RT-PCR. After ST cell infection with rTGEVs, total RNA was extracted, and genomic sequences from gene 3a were amplified by RT-PCR with oligonucleotides S-3839-VS and 3a-169-RS (Table 1 and Fig. 4A). Sequencing of these RT-PCR products showed that the nucleotide substitutions introduced within CS-3a were stably maintained during virus passage.
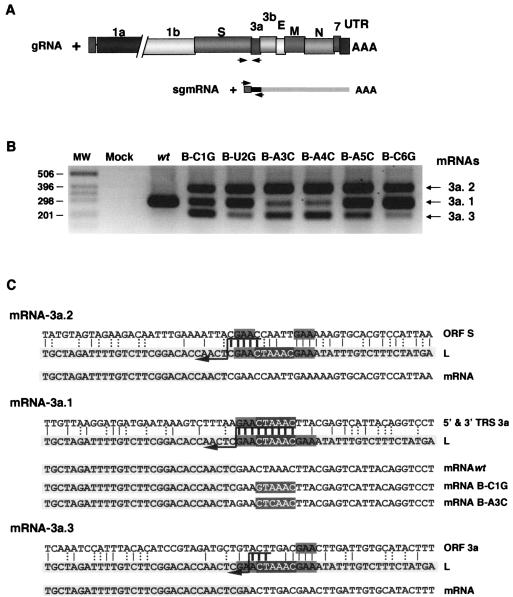
FIG. 4.
RT-PCR analysis of the CS-B mutants. (A) Scheme of the RT-PCR strategy for testing the gRNA and the mRNA-3a. Arrows indicate the approximate oligonucleotide position in the genome and sgmRNA. UTR, 3′ untranslated region. (B) mRNA-3a specific RT-PCR products were resolved in an agarose gel. mRNA-3a species were numbered 3a.1 (wild type [wt]), 3a.2, and 3a.3. MW, molecular weight markers. (C) Sequence analysis of the leader-body junction sites in the three mRNA-3a species. The sequence in the light-gray box corresponds to the leader (L) sequence. The CS appears as white letters in a dark-gray box in all cases. The sequence on top corresponds to the gRNA sequence in the fusion site; the sequence at the bottom is the mRNA sequence with nucleotides from the leader in a light-gray box. CS in white letters in a dark-gray box represents the mutated CS in each case; two examples of leader-to-body junction sites generating mRNA-3a.1 are presented: the B-C1G and B-A3C mutants. The GAA motif appears in a medium-gray box. Vertical bars represent the identity between the sequences, with thick bars at the possible fusion site. Dotted vertical bars represent the possible non-Watson-Crick interaction. Crossover should occur in any of the nucleotides above the arrow.
Using primers specific for mRNA-3a detection (SP and 3a-169-RS, Materials and Methods and Table 2) a single sgmRNA was detected in the wild-type virus, while in all CS-B mutants, three RT-PCR amplification bands were observed (Fig. 4B). This pattern was the same in the six mutants, although the relative band intensities were different. Moreover, sequencing of these cDNAs revealed that mRNA-3a.1 corresponded to the wild-type mRNA-3a, generated by a leader-to-body junction site within the CS-3a. The mRNA-3a.2 was generated in all CS-B mutants from a leader-to-body fusion site inside ORF S, 121 nt upstream of CS-3a. The third band (mRNA-3a.3) arose from a junction site 64 nt downstream of CS-3a, inside gene 3a.
Sequencing of the leader-to-body junction sites in the three sgmRNA-3a species showed that there was an extended identity between TRS-L and gRNA in sequence domains around the noncanonical CSs used (Fig. 4C). Interestingly, all of the mutations introduced in the CS-3a appeared in the mRNA-3a.1, including a substitution in the first CS-B nucleotide (B-C1G mutant), indicating that at least the whole-body CS was copied before template transfer. Nevertheless, because an extended upstream sequence identity is observed between the CS-L and CS-3a flanking sequences, the strand transfer point could not be accurately established. Even for the B-C1G mutation that remained in the mRNA-3a.1 sequence, strand transfer could happen in any of the 5′-GAA-3′ nucleotides upstream of CS-3a. However, in mutants B-A3C (Fig. 4C) and B-A5C (data not shown), template transfer had to occur at the A nucleotide, preceding GAA sequence upstream CS-3a, because the mRNA-3a.1 included the sequence 5′-AGAACUAAAC-3′ (Fig. 4C) derived from the gRNA sequence. The identity between leader and body sequences was frequently extended by including all or part of the sequence 5′-GAA-3′, at either the CS 5′ or 3′ end, or at both ends (Fig. 4C), suggesting that template switching during transcription required high complementarity between TRS-L and cTRS-B.
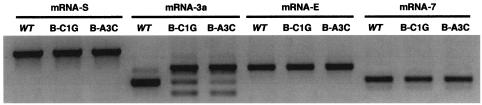
The transcription pattern in CS-3a mutants of proximal (gene E) or distal upstream (gene S) or downstream (gene 7) TGEV genes was analyzed by RT-PCR using specific oligonucleotides (Table 2), and no alteration was observed in the relative synthesis of these TGEV mRNAs (Fig. 5). These data suggested that the template switch was dependent on the nature of local sequences and was not influenced by sequences mapping 5′ or 3′ downstream.
FIG. 5.
Effect of CS-B mutations in the transcription of other TGEV mRNAs. mRNAs from genes S, 3a, E and 7 were analyzed by RT-PCR using specific oligonucleotides (Table 2). WT, wild-type virus; B-C1G and B-A3C, CS-3a mutants with mutation at positions 1 and 3, respectively.
Relationship between potential base pairing of the TRS-L with nascent negative RNA sequences and template transfer.
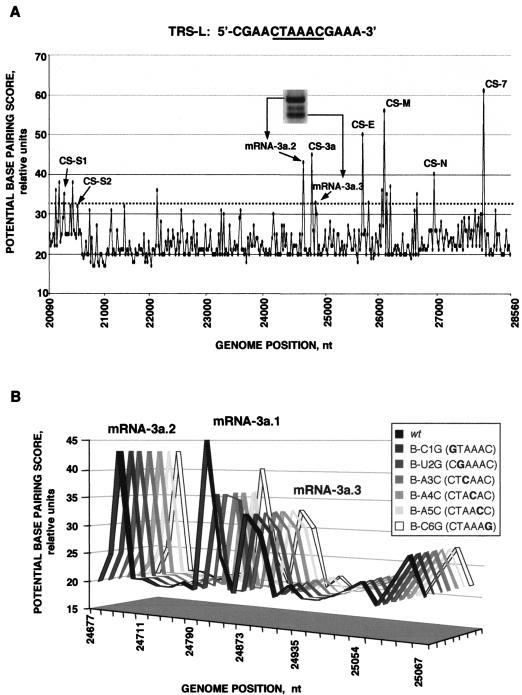
Termination of negative sgRNA synthesis seems to take place at sequence domains with high complementarity with the TRS-L. This complementarity would be the consequence of an identity between the TRS-L and sequences mapping throughout the genome. To determine whether a high identity score would promote template switching during the synthesis of viral negative RNA, an in silico approach was used that was based on a local alignment algorithm (14) that estimates the identity between the genomic RNA and the TRS-L, comprising the CS (5′-CUAAAC-3′) plus 3, 4, or 5 nt flanking the CS both at the 5′ and 3′ ends. In the case of 5 nt flanking the CS at both ends, the sequence considered was 5′-TCGAACTAAACGAAAT-3′ (the CS sequence is in boldface). In these three cases, the patterns of sequence domains with high identity were similar, differing only quantitatively.
Base-pairing scores throughout the 5′ two-thirds of the genome were very low (below a value of 35), except at the TRS-L, which obviously has the maximum base pairing score (a value of 70) (data not shown). Interestingly, potential base pairing throughout the one-third 3′ end of the genome, encoding the structural and nonstructural proteins, showed that the sequences with highest local identity correlated with template transfer sites leading to generation of the standard TGEV mRNAs (Fig. 6A). Intermediate values of local complementarity (between 32 and 40) were associated with the generation of sgmRNAs alternative to those generated by template transfer at positions of canonical CS-Bs. In contrast, no sgmRNAs were detected at sequence positions with a low potential base-pairing score (data not shown), suggesting a dominant role for the complementarity between TRS-L and cTRS-B in the control of sgmRNA levels.
FIG. 6.
In silico analysis of the identity between TRS-L and the TGEV genome. As indicated in Materials and Methods, a continuous line graph was selected to facilitate visualization of the data. (A) Graphical plot of the potential base-pairing score versus the genome position. All peaks assigned to the viral CSs are indicated as the peaks corresponding to the new 3a sgmRNA species. (B) Graphical plot of the potential base-pairing score versus the genome position around CS-3a. Each three-dimensional line represents either the wild-type (wt) situation or the body mutants. The peaks assigned to each 3a sgmRNA species are indicated.
Analysis of the potential base pairing between the TRS-L and sequences in the gRNA complementary to the fusion site of gene 3a showed that the three peaks of higher identity score surrounding the CS-3a corresponded to the canonical and noncanonical leader-to-body junction sites found in all CS-B mutants, generating mRNAs 3a.1, 3a.2, and 3a.3 (Fig. 6A). In silico analysis of the potential base pairing within this sequence domain showed that the potential base-pairing patterns were almost identical for all CS-3a mutants (Fig. 6B). The highest TRS-L to TRS-3a identity corresponded to the junction site 3a.1. In contrast, TRS-L to TRS-3a identity decreased in this sequence domain in all CS-3a mutants and was very close to the value at the sgRNA-3a.3 leader-to-body junction site. In these cases, the highest base-pairing value corresponded to the junction site upstream of the CS-3a sequence, within ORF S, generating sgRNA-3a.2. These results could explain the generation of the same new sgRNA species in all of the body mutants, despite the nucleotide change introduced, and suggested an important role for the GAA CS flanking sequence in junction site election, especially when a nucleotide substitution was introduced within the CS-3a.
Influence of CS-L to cCS-B duplex ΔG on sgRNA-3a levels.
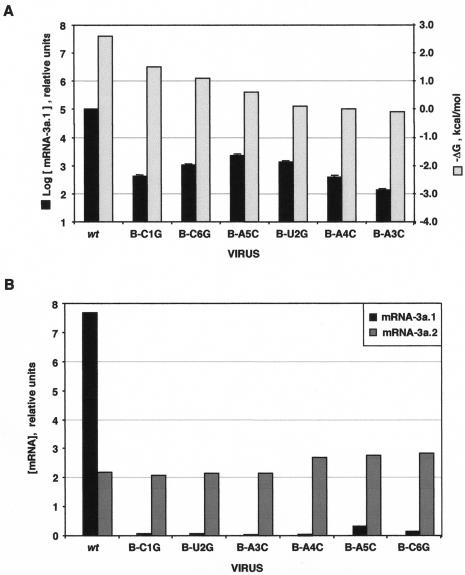
To study the influence of base pairing between the nascent negative sgRNA and the CS-L on sgmRNA synthesis, mRNA-3a.1 levels were quantified in all CS-B mutants by real-time RT-PCR using specific oligonucleotides (Table 3) and the gRNA as an internal standard for mRNA evaluation. The concentration of mRNA-3a.1 in CS-B mutant viruses was expressed in relation to that of the wild type. The results showed a significant decrease in mRNA-3a.1 levels of up to 103-fold and a good correlation between mRNA-3a.1 concentration and duplex ΔG except for nucleotide substitutions at both the 5′ and 3′ ends of the CS (B-C1G and B-C6G mutants) that had a higher effect than expected on sgmRNA levels (Fig. 7A). The additional decrease in the amount of mRNA-3a.1 in the B-C1G mutant could be due to the importance of this nucleotide to prime the synthesis of negative sgRNA after template switching. In addition, both the first and last CS nucleotides could play the extra role of stabilizing the formation of a duplex between the exposed CS-L and the cCS-B.
FIG. 7.
mRNA-3a quantification by real-time RT-PCR. (A) Amount of mRNA-3a.1, quantified by real-time RT-PCR, in the body mutants relative to the wild-type (wt) levels. Shown is a graphical representation of the ΔG (as −ΔG in kilocalories per mole) of the CS-L with cCS-B duplex and the relative amount of mRNA-3a.1 (represented as log [mRNA-3a.1] in relative units) for each virus. The data presented are the average of six independent experiments with duplicates in each case. Error bars represent the standard deviation in each case. (B) Graphical plot of the amounts of mRNA-3a.1 and mRNA-3a.2 relative to the level of gRNA, expressed as [mRNA] in relative units.
The mRNA-3a.1 levels were also quantified by real-time RT-PCR in the leader and double mutants with CS substitutions at positions 4 to 6 (data not shown). The amount of mRNA-3a.1 decreased in CS-L mutants at least 10-fold in relation to mRNA-3a.1 levels in the wild type. In mutants D-A4C and D-A5C, the amount of mRNA-3a.1 was similar to that obtained for the wild-type virus. However, mRNA-3a.1 levels were not restored in double mutant D-C6G and were at least 103-fold lower than that in the wild type (data not shown), reinforcing the possibility of an extra role for the nucleotide in the last position of the CS, such as the interaction of these sequences with regulatory proteins.
The amount of the alternative mRNA-3a species was also analyzed by real-time RT-PCR using specific oligonucleotides (Table 3). The level of mRNA-3a.2 in the CS-3a mutants did not change significantly when compared with that of the wild-type virus (Fig. 7B). The apparent discrepancy between the relative abundance of the mRNA-3a.2 bands (Fig. 4B) and the quantitative RT-PCR results for the wild-type virus (Fig. 7B) can be explained by primer sequestration by mRNA-3a.1, which was about 103-fold more abundant in the wild type than in the CS-3a mutants. As a consequence, the ratio of mRNA-3a.1 to mRNA-3a.2 was altered in all CS-B mutants. The alternative mRNA-3a.2 was also expressed in the wild-type virus as determined by real-time RT-PCR, although it was not detected by conventional RT-PCR due to the competition between the primers used. Unfortunately, real-time RT-PCR did not allow the quantification of mRNA-3a.3, since the design of specific oligonucleotides was not possible because a duplication of the sequence appears at the leader-to-body fusion site.
Effect of leader CS mutants on sgmRNA levels.
The introduction of nucleotide substitutions at CS-L could affect the potential base pairing between the TRS-L and cTRS-B of all TGEV genes, with the consequent reduction in sgmRNA and virus production. Alternatively, the decrease in virus titers could also be due to an effect of CS-L nucleotide substitutions in the TRS-L secondary structure. The transcription model proposed in this article, like the one proposed for arterivirus (26, 38), postulates exposure of the CS-L in a stem-loop within the TRS-L. In agreement with this model, virus production was only observed in TGEV mutants with a CS-L presented as a single-strand RNA according to secondary structure predictions (19; data not shown).
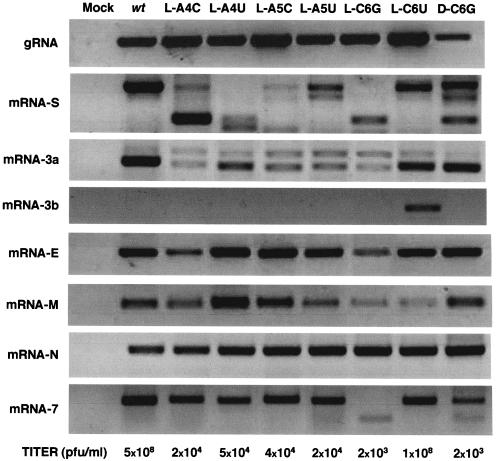
Construction of rTGEVs with nucleotide substitutions not allowing base pairing with cCS-B at each CS-L position led to the rescue of infectious viruses when these mutations were introduced within positions 4 to 6 of the CS, but not in positions 1 to 3. Therefore, the analysis of the sgmRNA generated after infection of cells was only possible in mutants with substitutions in positions 4 to 6. Total RNA from infected cells was analyzed by RT-PCR using specific oligonucleotides (Table 2) to amplify gRNA and mRNAs (Fig. 8). Nucleotide substitutions in CS-L positions 4 to 6 led to a reduction in virus titers higher than 104-fold in relation to wild-type virus (Fig. 8, bottom). rTGEV mRNAs could be clustered into two sets: one that in general led to a unique sgmRNA (genes E, M, and N) and another leading to alternative sgmRNAs (genes S, 3a, and 7). The sgmRNA corresponding to gene 3b was only produced when the mismatch in the sixth nucleotide of the CS-B present in the parental TGEV strain, considered in this report as the wild-type strain (2, 40), was compensated for by the mutation introduced within the CS-L (mutant L-C6U).
FIG. 8.
Analysis by RT-PCR of viral sgmRNAs generated by rTGEVs with CS-L substitutions. After ST cell infection with rTGEVs, total RNA was analyzed by RT-PCR with specific oligonucleotides to detect all viral mRNAs. Viruses with CS-L substitutions are indicated on top of the figure. The viral mRNA detected is shown to the left of the figure. The titer (PFU per milliliter) of each virus is shown at the bottom.
The pattern for sgRNAs S, 3a, and 7 in the virus mutants differed from that in the wild-type virus (Fig. 8). Different mutations in the same CS-L nucleotide position led to different sgmRNA species (for instance, compare sgmRNAs in mutants L-A4C to L-A4U or L-C6G and L-C6U). These results were expected, since changes in the CS-L were creating new possibilities of base pairing with alternative sequences in the nascent negative sgRNA, leading to the formation of new duplexes that could result in novel template switches during negative-strand synthesis and the production of new sgmRNA species.
All nucleotide substitutions introduced in the cDNA remained in the rescued virus genome (data not shown). Moreover, sequencing of 72 viral mRNA leader-to-body junction sites included in the sgmRNAs identified (Fig. 8) showed that nucleotide substitutions within the CS-L did not appear in the mRNA sequence, confirming that the CS sequence in the mRNA came from CS-B (data not shown). These results strongly suggest that the template switch was produced during negative sgRNA synthesis.
Synthesis of alternative sgmRNAs in viruses with nucleotide substitutions in CS-L.
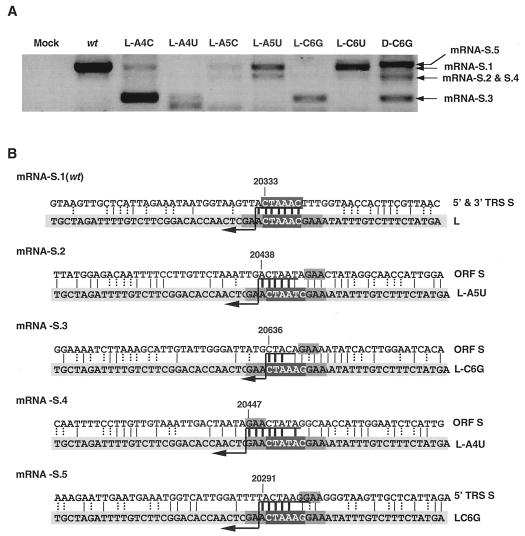
Mutations in CS-L led to the formation of at least five different sgRNA-S species, named mRNA-S.1 (wild type) to mRNA-S.5 (Fig. 9A). Some of these sgmRNA species, such as mRNA-S.2 and mRNA-S.4, were indistinguishable in agarose gel electrophoresis because of their similar size. RT-PCR amplification and sequencing of leader-to-body junction sites showed four new junction domains (extending nt 20291 to 20644 of the TGEV genome) leading to the synthesis of new sgmRNA species (Fig. 9B). Extended complementarity between leader and body sequences, mediated by the 5′-GAA-3′ sequence involved in the TRS-L with cTRS-B base pairing with noncanonical junction sites, was possible in all cases (Fig. 9B). Most likely, this extended complementarity leads to a higher base-pairing score between the nascent negative RNA strand and the TRS-L, promoting a template switch in these sequence positions and production of the corresponding sgmRNAs.
FIG. 9.
RT-PCR analysis of the S mRNA species present in leader mutants. (A) mRNA S detection by RT-PCR in leader and double mutants. sgmRNA species are named mRNA S.1, S.2, S.3, S.4, and S.5, as shown to the right of the panel. The oligonucleotides used for the analysis did not allow the detection of sgmRNAs S.6 and S.7. (B) Sequence analysis of the leader-to-body fusion site in all of the S gene sgmRNAs generated. The sequence in the light-gray box at the bottom represents the wild-type (wt) or mutated leader; the sequence on top is the gRNA sequence in the junction sites. CS is in white letters in a dark-gray box. The GAA motif is in a medium-gray box. Vertical bars represent the identity between the sequences; thick bars correspond to the possible fusion site, because crossover should occur in any nucleotide above the arrow. Dotted vertical bars represent the possible non-Watson-Crick interaction. Numbers indicate the position in the TGEV genome.
The TGEV sequence between nt 20438 and 20459 presented high identity to TRS-L (Fig. 6A), and different base pairings were possible, depending on the leader mutant. In fact, two sgmRNAs were generated (mRNA-S.2 or mRNA-S.4) (Fig. 9B). Interestingly, a potential mRNA that would use the canonical sequence CS-S2 located 121 nt upstream of the first S gene nucleotide was not detected, suggesting that sequences flanking this internal CS are instrumental in sgmRNA synthesis (2).
Nucleotide substitutions in the CS-L led to two gene 3a sgmRNA species, named mRNA-3a.1 (wild type) and mRNA-3a.2 (Fig. 10A). These sgmRNAs corresponded to the two larger gene 3a sgmRNA species found in the corresponding CS-3a mutants. The potential base-pairing score between the mutated TRS-L and the TRS-B, leading to mRNA-3a.3 synthesis, was smaller in the mutants than in the wild-type virus (Fig. 10B). Furthermore, the estimated ΔG indicated that base pairing in this junction site was energetically disfavored, providing a justification for the lack of template switch and production of mRNA-3a.3.
FIG. 10.
Analysis of 3a and 7 sgmRNAs present in leader mutants. (A) mRNA-3a detection by RT-PCR. sgmRNA species are named as mentioned before. (B) In silico analysis of the identity between the wild-type (wt) or mutated TRS-L and the TGEV genome surrounding the 3a gene CS. Data are graphically plotted as potential base-pairing score versus the genome position. (C) mRNA-7 detection by RT-PCR. The sgRNA species are named mRNA 7.1 and 7.2. (D) Sequence analysis of the leader-to-body junction sites in all of the 7 gene sgRNAs generated. The sequence at the bottom (light-gray box) represents the wild-type or mutated leader, and the one on top represents the gRNA in the fusion site context. CS is in white letters in a dark-gray box. Vertical bars show the identity between the sequences, and thick bars represent the possible fusion site, because strand transfer should occur in any of the nucleotides above the arrow. Dotted vertical bars represent the possible non-Watson-Crick interaction. Numbers indicate the position in the TGEV genome.
For TGEV gene 7, only one alternative sgmRNA was found in the L-C6G mutant and its double mutant, D-C6G (Fig. 10C). The new mRNA-7.2 was generated by a leader-to-body fusion near the ORF 7 3′ end, with high identity with the mutated TRS-L, as shown by RT-PCR amplification and sequencing of leader-to-body junction sites (Fig. 10D). Although this sgmRNA could not be translated, because there is no ATG codon, progeny virus was produced, since gene 7 is not essential (22).
Overall, analysis of the alternative sgRNAs produced in virus with nucleotide substitutions in the CS-L indicated that production of novel sgRNAs was associated with the possibility of duplex formation between the TRS-L and cTRS-B with a high base-pairing score.
DISCUSSION
In this report, the mechanism of coronavirus transcription has been studied by reverse genetics using full-length genomes for the first time. We have shown that discontinuous coronavirus transcription takes place during the synthesis of a negative sgRNA, and most template switch sites can be predicted by estimating the potential base-pairing score and free energy of the duplex between TRS-L and cTRS-B, using an adapted computer-based program to assess the strength of base pairing between body and leader TRS. In addition, it has been shown that the body canonical core sequence 5′-CUAAAC-3′, although nonessential for the generation of sgmRNA, promotes transcription levels by more than 103-fold over sgmRNAs associated with TRSs without a canonical CS-B. The modification of a canonical CS led to the synthesis of alternative sgmRNAs using noncanonical CSs. It has also been shown that the core sequence and flanking regions in the TGEV genome play a role in discontinuous transcription and modulate sgmRNA levels principally by the extent of base pairing. The data obtained were compatible with a three-step mechanism for coronavirus transcription postulating first the formation of a complex between the TRS-L and the 3′ end of the genome and, in a second step, a base-pairing scanning to determine the complementarity between the nascent negative RNA chain and the TRS-L. If duplex formation is favored, in a third step, a template switch is produced, leading to generation of a negative sgRNA.
A key strategy in our study of transcription regulation was the selection of gene 3a, a nonessential gene for TGEV growth both in vitro and in vivo, since modification of this gene did not affect the recovery of mutant viruses (36).
The requirement of complementarity between the CS-L and the cCS-B for the synthesis of a negative sgRNA was reinforced by showing a reduction in the sgmRNA synthesis associated with point mutations reducing complementarity between CS-L and cCS-B, and by demonstrating that, in general, sgmRNA synthesis was partially or completely restored by the introduction of nucleotide substitutions allowing formation of non-Watson-Crick or Watson-Crick base pairs.
The extent of sgmRNA synthesis was related to the free energy of the duplex between CS-L and cCS-B. The potential base-pairing score of sequence domains complementary to genomic RNA with the TRS-L ranged between 15 and 70. According to this score, the local sequence domains could be classified into domains with low (<35), and high (>35) base-pairing potential. In general, sequences with local base pairing of <35 led to no significant production of sgmRNA; in contrast, local base pairing higher than 35 led to the synthesis of standard viral mRNAs. These findings validated the in silico analysis method. This method was also found reliable for the prediction of most sgmRNAs synthesized by TGEV leader mutants and by other coronaviruses, such as HCoV 229E and BCoV (data not shown). The presence of a canonical CS within the TRS promoted higher sgmRNA levels. Nevertheless, the presence of a canonical CS within the TGEV genome did not guarantee the synthesis of an sgmRNA. The requirement of an appropriate sequence context was confirmed by showing that a 5′-CUAAAC-3′ sequence present 121 nt downstream of the gene S initiation codon (CS-S2) did not lead to synthesis of the corresponding sgmRNA as a consequence of the 5′ and 3′ flanking TRS sequences (2). The lack of sgmRNA synthesis could be explained by the relatively low potential base-pairing score and ΔG values (32 and −3.0 kcal/mol, respectively) between the corresponding TRS-L and cTRS-B, values lower than those estimated for the canonical CS-S1 used (35 and −4.3 kcal/mol, respectively).
The presence of TRS-L complementary sequences flanking noncanonical cCS-Bs led to the use of alternative TRS-B sequences. These sgmRNAs, such as mRNAs 3a.3 and 7.2, although produced in significant amounts, were generally not translated into truncated proteins, because there was no initiation codon in their sequence. Nevertheless, in a minority of cases, new sgmRNAs could encode essential proteins, and the use of alternative noncanonical CSs could be a safeguard mechanism for virus survival. In fact, this could be the case of gene S alternative sgmRNAs that could lead to the production of truncated S proteins, similar to that found in field variants of TGEV, such as the porcine respiratory coronavirus (PRCV) (4, 29). The synthesis of alternative sgmRNAs using noncanonical CS as part of the viral life cycle has also been reported for other coronavirus (17, 23), in arterivirus (24), and in nidovirus-derived expression systems (7, 11, 36).
Nucleotide substitutions, not allowing base pairing with cCS-B, within the first 3 nt of the CS-L led to the inhibition of sgmRNA synthesis and, consequently, to a failure in infectious virus production. In contrast, mutation of CS-L nt 4 to 6 promoted infectious virus recovery, although virus production was reduced by more than 104-fold. The higher restriction within the first 3 CS-L nt during template switch fits the discontinuous negative-strand synthesis model (18, 32), because extension of the negative sgRNA body sequences to add the cL sequence could not proceed in the absence of a complementarity between the 3′ end of the growing negative strand and the TRS-L. However, alternative explanations, such as the effect of these nucleotide substitutions on key CS-L structural motifs, cannot be discarded.
Our data indicate that template switching during synthesis of the negative strand takes place after termination of the complementarity between the 3′ end of the nascent RNA strand (negative polarity) and the TRS-L. In this process, mismatches may be tolerated, providing that several complementary nucleotides between the TRS-L and cTRS-B upstream of the CS-B would be present. This conclusion is based on the sequence of the leader-to-body junctions in a collection of 72 sgmRNAs generated after the introduction of point mutations within CS-L and CS-3a. For instance, nucleotide substitutions introduced in the first CS nucleotide (B-C1G) of gene 3a were transferred to the sgmRNA, probably because the identity between the TRS-B and the TRS-L was extended 3 nt upstream of the CS (Fig. 11). Similarly, nucleotide substitutions in the first CS-L nucleotide (L-C1U) never appeared in the sgmRNA sequence—except in the mRNA-E sequence, since in this case there was no immediately adjacent upstream complementarity (Fig. 11) (data not shown)—reinforcing the concept that template switching takes place at the end of the complementarity between the TRS-L with the cTRS-B.
FIG. 11.
CS adjacent flanking sequences identity. Identity between the TRS-L sequence and TRS-Bs for all TGEV sgmRNAs is shown in the figure. The CS sequence is in white letters in a black box. White boxes highlight the identity in the sequences immediately flanking CS both at the 5′ and 3′ ends.
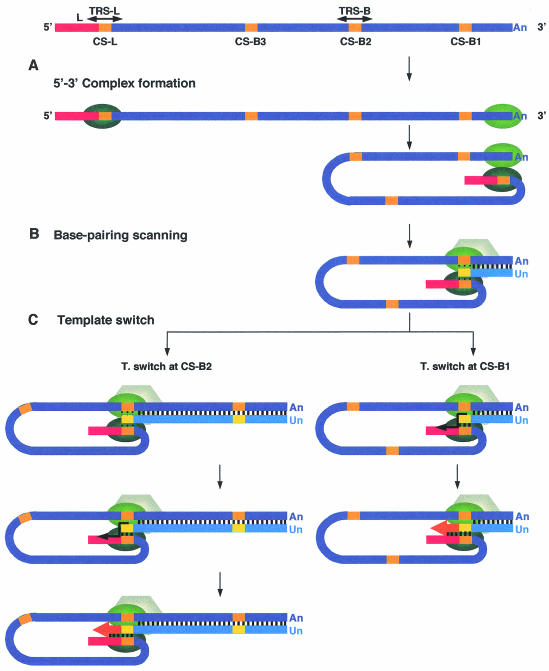
The data presented are compatible with the working model of coronavirus transcription shown in Fig. 12. This model includes three steps in the selection of the gRNA sequence in which template switching takes place. The first step would involve the formation of 5′-end-3′-end complexes mediated by protein-RNA and protein-protein interactions, by which the TRS-L would be located in close proximity to sequences located at the 3′ end of genomic RNA (Fig. 12A). In the mouse hepatitis virus (MHV), it has been shown that members of the heterogeneous nuclear ribonucleoprotein (hnRNP) family, like hnRNP A1 and the polypyrimidine track binding protein (PTB), could interact either with the 5′ or 3′ ends of the viral genome and between them (5, 13, 35), suggesting that viral ends could be in close proximity during virus transcription and replication. This step is a requirement for the viability of the next step.
FIG. 12.
Three-step working model of coronavirus transcription. (A) The 5′-3′ complex formation step. Proteins binding the 5′- and 3′-end TGEV sequences are represented by the green ovals. The leader sequence is red, and CS sequences are yellow. An, poly(A) tail. (B) Base-pairing scanning step. Negative-strand RNA is in a lighter color than positive-strand RNA. The transcription complex is represented by the hexagon. Vertical dotted bars represent the base-pairing scanning by the TRS-L sequence in the transcription process. Vertical solid bars indicate complementarity between gRNA and the nascent negative strand. Un, poly(U) tail. (C) Template switch step. The thick arrow indicates the switch in the template made by the transcription complex to complete the synthesis of negative sgRNA.
In the second base-pairing scanning step, the TRS-L would scan the nascent negative chain, looking for complementary sequence domains (Fig. 12B) leading to a high potential base-pairing score, associated with a favorable ΔG. This scanning has been postulated based on the observed relationship between the presence of a high potential base-pairing score between the TRS-B and the TRS-L and synthesis of sgmRNA. This correlation implies that during the synthesis of the negative RNA, the nascent chain has to be screened by the TRS-L, probably partially exposed at the top of a stem-loop. If the complementarity is above a certain threshold, the third template switch step takes place (Fig. 12C) in a proportion of the nascent chains, and a copy of the gRNA leader is made, leading to termination of a negative sgRNA that will be used to generate an sgmRNA with the same length. The existence of this step is required to explain the primary structure of a high number (>60) of sgmRNAs described in this article. The transcription model postulated in this article reinforces and extends a previously proposed model for coronavirus and arterivirus (26, 27, 30, 32, 38).
The proposed coronavirus transcription mechanism implies a close interaction between TRS-L and each cTRS-B present in the gRNA. Therefore, there should be a restriction in the evolution of these TRSs, because changes in a given TRS-B would affect synthesis of the specific sgmRNA. More importantly, changes within the TRS-L should have a pleiotropic effect on the synthesis of all viral mRNAs. Therefore, the degree of freedom in the evolution of these TRSs should be limited, particularly for essential genes. Nucleotide substitutions within a TRS should only be fixed if the sequences flanking a canonical or noncanonical CS-B compensate the decrease in the base pairing between cCS-B and CS-L. Therefore, the probability of a nucleotide substitution within a TRS-B should be lower than that of an average nucleotide substitution within RNA virus genomes (i.e., <1 × 10−4) (8). Furthermore, the fixation of a nucleotide substitution within the TRS-L, particularly within the first 3 CS-L nt, would require the simultaneous incorporation of complementary mutations within the CS-B of at least the four essential TGEV genes encoding proteins S, E, M, and N. Therefore, this event should have an even lower probability (<1 × 10−20 in TGEV). These theoretical considerations are supported in TGEV by the lack of CS-L evolution throughout more than 50 years of virus replication.
Multiple factors seem to regulate the transcription process (2, 26, 27). These factors would probably imply protein-RNA and protein-protein recognition, including viral and host cell components that will be the subject of future studies.
Acknowledgments
We thank D. Sawicki, F. Almazán, J. Ortego, and D. Escors for critically reading the manuscript and helpful discussions. We are also grateful to J. C. Oliveros for the PERL script used in the in silico analysis and D. Dorado and V. Hernández for technical assistance.
This work was supported by grants from the Comisión Interminis-terial de Ciencia y Tecnología (CICYT), La Consejería de Educación y Cultura de la Comunidad de Madrid, Fort Dodge Veterinaria, and the European Communities (Frame V, Key Action 2, Control of Infectious Disease Projects). I.S., S.A., and S.Z. received postdoctoral fellowships from the Community of Madrid and the European Union (Frame V, Key Action 2, Control of Infectious Disease Projects: QLRT-1999-00002, QLRT-1999-30739, and QLRT-2000-00874).
REFERENCES
- 1.Almazán, F., J. M. González, Z. Pénzes, A. Izeta, E. Calvo, J. Plana-Durán, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso, S., A. Izeta, I. Sola, and L. Enjuanes. 2002. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J. Virol. 76:1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brian, D. A., and W. J. M. Spaan. 1997. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 8:101-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callebaut, P., I. Correa, M. Pensaert, G. Jiménez, and L. Enjuanes. 1988. Antigenic differentiation between transmissible gastroenteritis virus of swine and a related porcine respiratory coronavirus. J. Gen. Virol. 69:1725-1730. [DOI] [PubMed] [Google Scholar]
- 5.Choi, K. S., P.-Y. Huang, and M. C. C. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. [DOI] [PubMed] [Google Scholar]
- 6.Delmas, B., J. Gelfi, H. Sjöström, O. Noren, and H. Laude. 1994. Further characterization of aminopeptidase-N as a receptor for coronaviruses. Adv. Exp. Med. Biol. 342:293-298. [DOI] [PubMed] [Google Scholar]
- 7.de Vries, A. A. F., A. L. Glaser, M. J. B. Raamsman, and P. J. M. Rottier. 2001. Recombinant equine arteritis virus as an expression vector. Virology 284:259-276. [DOI] [PubMed] [Google Scholar]
- 8.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 9.Enjuanes, L., D. Brian, D. Cavanagh, K. Holmes, M. M. C. Lai, H. Laude, P. Masters, P. Rottier, S. G. Siddell, W. J. M. Spaan, F. Taguchi, and P. Talbot. 2000. Coronaviridae, p. 835-849. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 10.Enjuanes, L., W. Spaan, E. Snijder, and D. Cavanagh. 2000. Nidovirales, p. 827-834. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, D. J. McGeoch, J. Maniloff, M. A. Mayo, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Classification and nomenclature of viruses. Academic Press, San Diego, Calif.
- 11.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González, J. M., Z. Pénzes, F. Almazán, E. Calvo, and L. Enjuanes. 2002. Stabilization of a full-length infectious cDNA clone of transmissible gastroenteritis coronavirus by insertion of an intron. J. Virol. 76:4655-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, P., and M. M. C. Lai. 2001. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, X., and W. Miller. 1991. A time-efficient, linear-space local-similarity algorithm. Adv. Appl. Math. 12:337-357. [Google Scholar]
- 15.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477-484. [DOI] [PubMed] [Google Scholar]
- 16.Jiménez, G., I. Correa, M. P. Melgosa, M. J. Bullido, and L. Enjuanes. 1986. Critical epitopes in transmissible gastroenteritis virus neutralization. J. Virol. 60:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 20.McClurkin, A. W., and J. O. Norman. 1966. Studies on transmissible gastroenteritis of swine. II. Selected characteristics of a cytopathogenic virus common to five isolates from transmissible gastroenteritis. Can. J. Comp. Med. Vet. Sci. 30:190-198. [PMC free article] [PubMed] [Google Scholar]
- 21.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 22.Ortego, J., I. Sola, F. Almazan, J. E. Ceriani, C. Riquelme, M. Balasch, J. Plana-Durán, and L. Enjuanes. 2003. Transmissible gastroenteritis coronavirus gene 7 is not essential but influences in vivo virus replication and virulence. Virology 308:13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pasternak, A. O., A. P. Gultyaev, W. J. M. Spaan, and E. J. Snijder. 2000. Genetic manipulation of arterivirus alternative mRNA leader-body junction sites reveals tight regulation of structural protein expression. J. Virol. 74:11642-11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pasternak, A. O., W. J. M. Spaan, and E. J. Snijder. Regulation of relative abundance of arterivirus subgenomic mRNAs. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 26.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasternak, A. O., E. van den Born, W. J. M. Spaan, and E. J. Snijder. 2003. The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis. J. Virol. 77:1175-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Penzes, Z., J. M. González, E. Calvo, A. Izeta, C. Smerdou, A. Mendez, C. M. Sánchez, I. Sola, F. Almazán, and L. Enjuanes. 2001. Complete genome sequence of transmissible gastroenteritis coronavirus PUR46-MAD clone and evolution of the Purdue virus cluster. Virus Genes 23:105-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sánchez, C. M., G. Jiménez, M. D. Laviada, I. Correa, C. Suñé, M. J. Bullido, F. Gebauer, C. Smerdou, P. Callebaut, J. M. Escribano, and L. Enjuanes. 1990. Antigenic homology among coronaviruses related to transmissible gastroenteritis virus. Virology 174:410-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawicki, D. L., T. Wang, and S. G. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:386-396. [DOI] [PubMed] [Google Scholar]
- 31.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 440:215-220. [DOI] [PubMed] [Google Scholar]
- 33.Schaad, M. C., and R. S. Baric. 1994. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J. Virol. 68:8169-8179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethna, P. B., S.-L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, S. T., P. Huang, H.-P. Li, and M. M. C. Lai. 2000. Heterogeneous nuclear ribonucleoprotein A1 regulates RNA synthesis of a cytoplasmic virus. EMBO J. 19:4701-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sola, I., S. Alonso, S. Zúñiga, M. Balach, J. Plana-Durán, and L. Enjuanes. 2003. Engineering transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity. J. Virol. 77:4357-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription, and RNA recombination, p. 11-31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 38.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. M. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sc. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wesley, R. D., A. K. Cheung, D. M. Michael, and R. D. Woods. 1989. Nucleotide sequence of coronavirus TGEV genomic RNA: evidence of 3 mRNA species between the peplomer and matrix protein genes. Virus Res. 13:87-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]