Summary
Cell-cycle arrest, apoptosis, and senescence are widely accepted as major mechanisms by which p53 inhibits tumor formation. Nevertheless, it remains unclear whether they are rate-limiting steps in tumor suppression. Here, we have generated mice bearing lysine to arginine mutations at one (p53K117R) or three (p533KR; K117R+K161R+K162R) of the critical p53 acetylation sites. While p53K117R/K117R cells are competent for p53-mediated cell-cycle arrest and senescence, but not apoptosis, all three of these processes are ablated in p533KR/3KR cells. Surprisingly, unlike p53-null mice, which rapidly succumb to spontaneous thymic lymphomas, early-onset tumor formation does not occur in either p53K117R/K117R or p533KR/3KR animals. Notably, p533KR retains the ability to modulate energy metabolism and reactive oxygen species (ROS) production by regulating metabolic p53 target genes. These findings underscore the crucial role of acetylation in differentially modulating p53 responses and suggest that unconventional activities of p53, such as metabolic regulation and antioxidant function, are critical for suppression of early-onset spontaneous tumorigenesis.
Keywords: p53, Tip60, CBP/p300, K120, k164, Mdm2, Mdmx, acetylation, deacetylation, Growth arrest, apoptosis, p21, PUMA, DNA binding domain
Introduction
The p53 tumor suppressor regulates multiple signaling pathways triggered by diverse cellular stresses, including DNA damage, abnormal oncogenic events, loss of normal cell contacts, and hypoxia, as well as some normal cellular processes (Vousden and Prives, 2009; Feng and Levine, 2010; Kruse and Gu, 2009). Inactivation of p53 function is a pivotal aspect of tumor formation in a broad spectrum of human cancers. More than 50% of sporadic tumors harbor somatic mutations of the p53 gene, while Li-Fraumeni patients carry germline p53 mutations that confer a high familial risk of cancer (Vogelstein et al., 2000; Whibley et al., 2009). The principal functions of p53 are mediated through its ability to serve as a DNA-binding transcription factor, although transcription-independent activities of p53 have also been identified. The importance of its transcriptional activity is underscored by the fact that most tumor-associated p53 mutations occur within the central core domain responsible for sequence-specific DNA binding (Vogelstein et al., 2000). Although the molecular mechanism of p53-mediated tumor suppression is not completely understood, it is widely accepted that tumor cell growth can be inhibited by p53-mediated cell cycle arrest, apoptotic cell death, and/or cellular senescence.
p53 is regulated by an exquisite network of fine-tuning mechanisms that ensure proper responses to the various stress signals encountered by cells. In broad terms, p53 activities are controlled via protein levels, coactivator/corepressor recruitment, and a diverse array of post-translational modifications that include acetylation, ubiquitination, phosphorylation, methylation, sumoylation, and neddylation (Brooks and Gu, 2003; Kruse and Gu, 2009). The phenotypic outcome of p53 activation is apparently determined by promoter-specific induction of certain p53 transcription target genes, a number of which have already been implicated in specific p53-dependent processes such as cell cycle arrest (e.g., p21, 14-3-3σ), apoptotic cell death (PUMA, NOXA, Killer/DR5), and cellular senescence (p21, PAI-1, PML) (Vousden and Prives, 2009). Nevertheless, the precise mechanisms by which p53 achieves promoter specificity are complex and remain poorly understood.
Numerous studies demonstrate that non-histone protein acetylation is critically involved in regulating diverse cellular processes such as transcription, protein folding and cellular metabolism (Kim et al., 2006; Choudhary et al., 2009; Wang et al., 2010; Zhao et al., 2010). p53 was the first non-histone protein shown to be regulated functionally by acetylation and deacetylation (Gu and Roeder, 1997; Luo et al., 2000; Luo et al., 2001; Vaziri et al., 2001) and subsequent work has established that acetylation plays a key role in controlling promoter-specific activation of p53 targets during stress responses (Berger, 2010; Kruse and Gu, 2009; Mellert and McMahon, 2009; Loewer et al., 2010). The major acetylation sites of human p53 include two lysine residues (K120 and K164) within the DNA-binding domain and a cluster of six lysines with the C-terminal domain. We and other recently showed that p53 is acetylated at K120 by Tip60/MOF (Sykes et al., 2006; Tang et al., 2006; Li et al., 2009), while K164 is acetylated by CBP and p300, but not Tip60/MOF (Tang et al., 2008). Both sites are mutated in human tumors and well conserved in all species known to encode p53. Interestingly, K120 acetylation is crucial for p53-mediated apoptosis but has no obvious effect on cell cycle arrest, while simultaneous loss of acetylation at all major sites abolishes the ability of p53 to induce both cell cycle arrest and apoptosis (Tang et al., 2008), suggesting that acetylation is essential for both p53-mediated processes. However, the role of acetylation in the most critical aspect of p53 function, its ability to suppress tumor formation, has not been addressed.
To investigate whether p53 acetylation is important for tumor suppression, we generated p53-mutant mice (p53K117R/K117R) in which K117 (K120 in human) is replaced by arginine. In these animals, p53-mediated apoptosis is completely abrogated but p53-dependent cell cycle arrest and senescence remain intact. We also established mice (p533KR/3KR) in which the three acetylation sites of the DNA-binding domain (K117, K161, K162) were simultaneously replaced by arginine. Significantly, loss of acetylation at these three sites completely abolished the ability of p53 to mediate cell cycle arrest, apoptosis, and senescence in vivo. To evaluate whether these p53-dependent processes are required for tumor suppression, we monitored tumor formation in cohorts of p53 acetylation-deficient mice. Although p53-null mice rapidly develop spontaneous thymic lymphomas (Donehower et al., 1992; Jacks et al., 1994; Lozano, 2010), neither p53K117R/K117R nor p533KR/3KR mice are prone to early-onset tumorigenesis. Since tumor suppression can be mediated by a p53 polypeptide (e.g., p533KR) that lacks the ability to induce p53-dependent cell cycle arrest, apoptosis, and senescence, these results indicate that other aspects of p53 function are sufficient to suppress tumor formation. Strikingly, the p533KR mutant retains the capacity to inhibit glycolysis and reduce the levels of reactive oxygen species (ROS). These findings suggest that current views regarding the mechanism of p53-mediated tumor suppression should be reconsidered.
Results
Generation of p53K117R knock-in mice
To elucidate the physiological functions of p53 acetylation, we first used mass spectrometric (MS/MS) analysis of mouse p53 protein to confirm that lysine 117 (K117) (corresponding human lysine 120) is indeed acetylated (Figure S1). Mouse embryonic stem (ES) cells were then electroporated with a knockin targeting construct designed to replace lysine codon 117 with an arginine codon (K117R) (Figure S2A). Properly targeted G418-resistant clones were identified by Southern analysis (Figure S2B), and two independent clones were injected into C57BL/6 blastocysts to achieve germline transmission of the p53K117R allele. The heterozygous F1 mice were then bred with Rosa26-Cre transgenic mice to delete the LoxP-flanked Neomycin resistant gene (Neor) from the targeted allele. The resultant Neor-deleted heterozygous p53K117R/+ mice were intercrossed and their progeny genotyped by PCR (Figure S2C). Of note, homozygous p53K117R/K117R mice were obtained at the expected Mendelian ratio (Figure S2D) and displayed no apparent developmental abnormalities. Expression of the mutant gene product was confirmed by sequence analysis of p53 cDNAs from p53K117R/K117R cells (Figure S2E).
K117R mutation has no effect on p53 stabilization but completely abolishes p53-mediated apoptosis
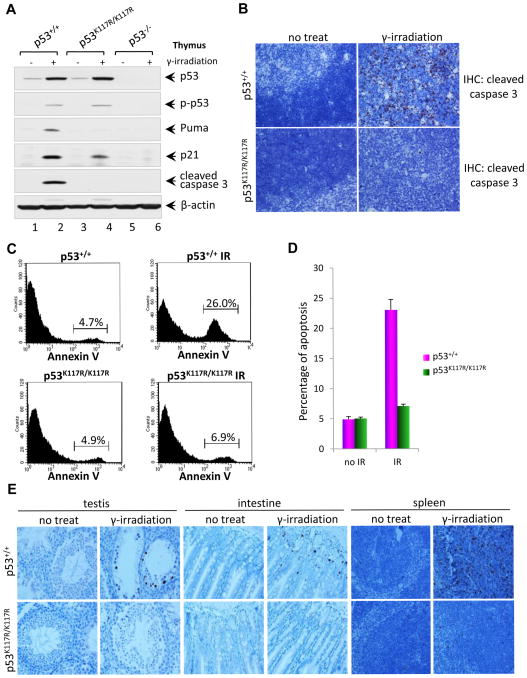
Previous studies in human cell lines indicate that K120 acetylation is crucial for apoptosis but has no obvious effect on p53 protein stability or p53-mediated cell cycle arrest (Sykes et al., 2006; Tang et al., 2006). To ascertain whether K117 acetylation regulates p53 apoptotic function similarly in mice, we analyzed p53-dependent apoptosis in p53K117R/K117R thymocytes subjected to ionizing radiation (IR). The levels of p53 protein were induced in a similar fashion by IR in p53K117R/K117R and wildtype thymocytes (Figure 1A), and IR-induced p53 phosphorylation (p-p53) remained intact. Although IR-induced transactivation of p21, a p53 target gene required for cell cycle arrest, was observed in p53K117R/K117R thymocytes, IR induction of PUMA, an apoptotic p53 target, was completely abrogated. Moreover, immunostaining for the active form of caspase-3 revealed that IR-induced apoptosis was also abolished in p53K117R/K117R thymocytes (Figure 1B). To corroborate these findings, thymocytes were isolated from both p53K117R/K117R and wildtype mice four hours after irradiation and stained with Annexin V-FITC. FACS analysis showed that IR-induced apoptosis was virtually abrogated in p53K117R/K117R thymocytes (Figure 1C and 1D). To examine whether these effects are cell type-specific, we also examined apoptotic responses in other tissues sensitive to DNA damage. As shown in Figure 1E, K117R mutation completely blocked the apoptotic response in testis, intestine, and spleen. Detailed analysis again showed that p53-mediated activation of proapoptotic target genes and p53-dependent apoptosis were ablated in p53K117R/K117R spleens (Figure S3). These data indicate that K117 acetylation is required for p53-dependent apoptosis in vivo.
Figure 1. p53K117R Activates p21 Normally but Completely Abrogates Activation of Propoptotic Target Genes.
(A) Western blot analysis of the thymus of p53+/+, p53K117R/K117R and p53−/− mice. Mice were either untreated or exposed to 12.5 Gy of γ-irradiation, four hours later; thymocytes were isolated and analyzed for the expression of p53, phospho-p53, and p21, Puma, cleaved caspase 3 and β-actin.
(B) Representative immunohistochemical staining of thymi from p53+/+ and p53K117R/K117R mice for cleaved caspase3. Eight-week-old mice were either untreated or exposed to 5 Gy of γ-irradiation. Four hours later, thymus from these mice were fixed overnight then processed, paraffin-embedded, sectioned and stained with anti-mouse cleaved caspase 3 antibodies according to the standard protocol.
(C) In vivo apoptotic analysis of p53+/+ and p53K117R/K117R mice after 5 Gy of γ-irradiation. Mice were treated as described in (B), Then single-cell suspension of thymocytes from mice were prepared and stained with Annexin V-FITC for FACS analysis.
(D) Quantification of apoptosis in vivo represented by Annexin-V positive thymocytes. Error bars represent averages ± SD from at least three mice for each genotype.
(E) Representative cleaved caspase 3 immunohistochemical staining images showing apoptotic cells in the spleen, testis and small intestine of p53+/+ and p53K117R/K117R mice. Eight-week-old mice were either untreated or exposed to 12.5 Gy of γ-irradiation. Then tissues were collected 4 hr later and processed as described in (B).
See also Figure S1, Figure S2 and Figure S3.
p53-dependent cell cycle arrest and senescence remain intact in p53K117R/K117R MEFs
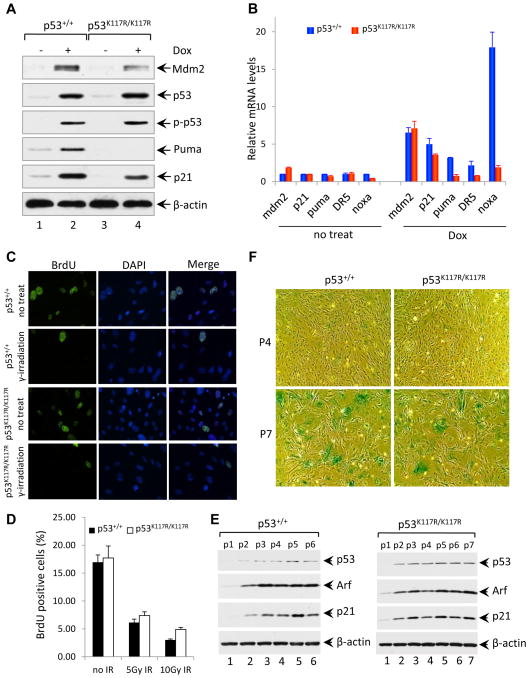
To determine whether the K117R mutation affects p53-dependent cell cycle arrest and senescence, we compared the response of p53K117R/K117R and wildtype mouse embryonic fibroblasts (MEFs) to treatment with the genotoxic agent doxorubicin. As shown in Figure 2A, stabilization and phosphorylation of p53K117R polypeptides occurred normally in response to doxorubicin. Consistent with results obtained in mouse tissues (Figures 1A), p53-mediated transactivation of PUMA, but not p21 or Mdm2, was completely abrogated in p53K117R/K117R MEFs (Figure 2A). Moreover, p53K117R/K117R MEFs were also defective for induction of other known proapoptotic p53 target genes such as NOXA and DR5 (Figure 2B). To examine whether the K117R mutation affects p53-mediated growth arrest, p53K117R/K117R and wildtype MEFs were pulse-labeled for BrdU, fixed in 70% ethanol, and double-stained with anti-BrdU antibody and DAPI. As shown in Figure 2C, IR treatment reduced the levels of BrdU incorporation to a comparable degree in both p53K117R/K117R and wildtype MEFs (Figure 2C and 2D). In addition, we compared the growth and senescence of wildtype and p53K117R/K117R MEFs following a standard 3T3 proliferation protocol. As expected, the expression levels of p53, p21, and Arf increased gradually upon passage of both wildtype and p53K117R MEFs (Figure 2E). Although p53K117R/K117R cells proliferated slightly faster than wildtype MEFs (Figure S4), both reached senescence similarly at passage 7 (Figure 2F). Together, these data suggest that the p53K117R mutant retains its ability to induce both cell cycle arrest and senescence in vivo.
Figure 2. P53K117R/K117R MEFs Retain Intact Cell Cycle Checkpoint and Cellular Senescence Function in Response to DNA Damage.
(A) Western blot analysis of Mdm2, p53, phosphorylated p53, Puma and p21 in p53+/+ and p53K117R/K117R MEFs either left untreated or treated with 1.0 μM Dox (doxorubicin) for 8 hours, β-actin was used as a loading control.
(B) qRT-PCR analysis of Mdm2, p21, Puma, Killer/DR5, and Noxa mRNA in p53+/+ and p53 K117R/K117R MEFs either untreated or treated with 1.0 μM Dox for 8 hours. Data shown are the relative amount of specific mRNA normalized first to β-actin and then to the untreated wildtype sample values from three independent experiments with different MEF lines. Results are reported as average ± standard error of the mean (SEM).
(C) Cell growth arrest analysis of p53+/+ and p53K117R/K117R MEFs either left untreated or exposed to 5 Gy of γ-irradiation. 23 hours after irradiation, MEFs were pulsed with 10 μM BrdU for 45 minutes, and then cells were fixed and processed for immunofluorescence analysis for BrdU (green). The nuclei were stained with DAPI (blue).
(D) Quantification of BrdU labeling of p53+/+ and p53K117R/K117R MEFs irradiated with 5 and 10 Gy. Error bars represent ±SD from three independent experiments.
(E) Western blot analysis of p53+/+ and p53K117R/K117R MEFs at different passages cultured according to a 3T3 protocol for the expression of p53, Arf and p21.
(F) Senescence-associated β-galactosidase (SA-β-gal) staining of p53+/+ and p53K117R/K117R MEFs cultured according to a 3T3 protocol. MEFs at indicated passages were fixed and stained for β-galactosidase activity as described in the Extended Experimental Procedures.
See also Figure S4.
Loss of acetylation at three sites (K117, K161 and K162) of mouse p53 abrogates transcriptional induction of both p21 and PUMA
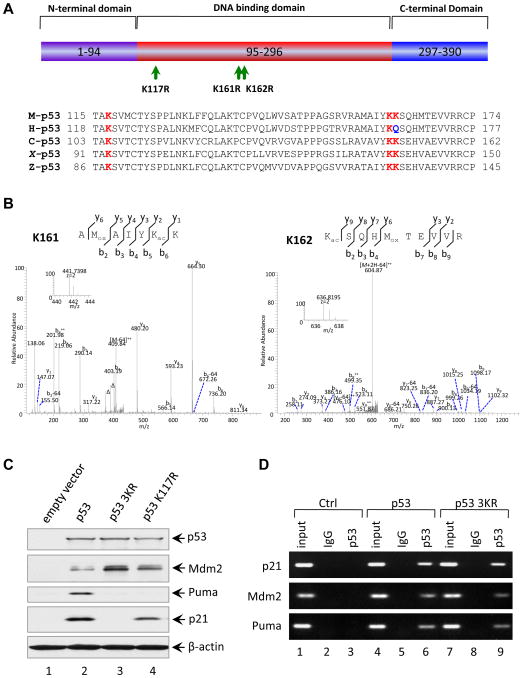
Although K117 acetylation is required for the p53-mediated apoptotic response in vivo (Figure 1), we observed no obvious differences between p53K117R/K117R and p53 wildtype cells with respect to either cell-cycle arrest or senescence. Notably, our recent study showed that elimination of multiple acetylation sites on human p53 abrogates both the growth arrest and apoptosis functions of p53 (Tang et al., 2008). To explore this possibility, we sought to generate a mutant mouse that would also be defective for p53-dependent cell-cycle arrest and senescence. Our previous studies indicated that K164 acetylation of human p53 (corresponding to K161 of mouse p53) is critical for p53-mediated cell cycle arrest (Tang et al., 2008). Interestingly, as shown in Figure 3A, both K161 and K162 of mouse p53 (corresponding to K164 and Q165 of human p53) are evolutionarily conserved in all species, with the exception of Q165 in human. Moreover, mass spectrometry analysis of mouse p53 revealed that both K161 and K162 are acetylated in vivo (Figure 3B). Therefore, we generated an expression plasmid encoding a mouse p53 mutant (p533KR) in which all three acetylation sites (K117, K161, and K162) are replaced by arginine. As expected, expression of wildtype mouse p53 in p53-null human H1299 cells induced transcription of both p21 and PUMA (lane 2, Figure 3C). Moreover, consistent with the data from p53K117R/K117R mice (Figure 2A), expression of p53K117R induced p21, but not PUMA, transcription (lane 4). Strikingly, loss of acetylation at all three sites (K117R, K161R and K162R: p533KR) completely abolished the induction of both p21 and PUMA (lane 3). Since the three mutated residues all reside within the DNA-binding domain, we examined the sequence-specific DNA-binding properties of p533KR in a chromatin immunoprecipitation assay (ChIP). As shown in Figure 3D, the DNA-binding activities of the wildtype and p533KR-mutant forms of mouse p53 were indistinguishable at a number of p53 target promoters, including p21, PUMA, and Mdm2. Moreover, p533KR readily induced expression of endogenous Mdm2 (lane 3, Figure 3C). Thus, we have identified a mouse p53 mutant (p533KR) that retains its ability to bind DNA and activate RNA transcription but is likely defective for both p53-dependent cell cycle arrest and apoptosis.
Figure 3. Loss of Acetylation Sites at K117, K161 and K162 of Mouse p53 Abolishes the Activation of p21 and PUMA.
(A) Schematic representation of the mouse p53 protein with three mutated acetylation sites indicated and alignment of the K117, K161 and K162 flanking regions of the mouse p53 with those of other species. The conserved lysines were highlighted in red and Q165 of human p53 highlighted in blue is evolutionarily reminiscent of acetylated K.
(B) Mass spectrometry analysis of tryptic mouse p53 peptides containing K161 and K162. The fragmentation spectrum of 156AMoxAIYKacK162 and 162 KacSQHMoxTEVVR171 revealed the presence of peptides with acetylation at K161 and K162, respectively. The protein was prepared as described in the Experimental Procedures. Inset shows the high-resolution precursor ion mass. The label “Δ” designates “b” or “y” ions with water and/or ammonia loss. “Kac” and “Mox” designate acetyllysine and oxidized methionine, respectively. Neutral loss of sulfenic acid from oxidized methionine was indicated as “-64”.
(C) Western blot analysis of p53, Mdm2, Puma and p21 in H1299 cells transfected with plasmids expressing mouse wildtype p53, p53-K117R and p53-3KR mutants. β-actin was used as a loading control.
(D) ChIP assay for the binding of p53 or p53-3KR mutant to the consensus sites in the p21, Puma and Mdm2 promoters. H1299 cells transfected with plasmids DNA expressing mouse p53 wildtype (WT) and p53-3KR mutant were treated with 1% formaldehyde for 10 min and processed for ChIP analysis. The occupancy of p53 or p53-3KR of the p21, Puma and Mdm2 promoters were detected by PCR-agarose gel electrophoresis. IgG antibody serves as a negative control.
p53-mediated cell cycle arrest and apoptosis are abrogated in p533KR/3KR mice
To elucidate the role of p53 acetylation in vivo, we used a similar knockin targeting strategy to introduce a p533KR allele into the germline of mice (Figures S5A and S5B). Upon intercrossing p533KR/+ heterozygotes, viable p533KR/3KR homozygous mice were obtained at normal Mendelian ratios (Figure S5C and S5D), and expression of the mutated p533KR allele was confirmed by sequence analysis of cDNA from p533KR/3KR cells (Figure S5E).
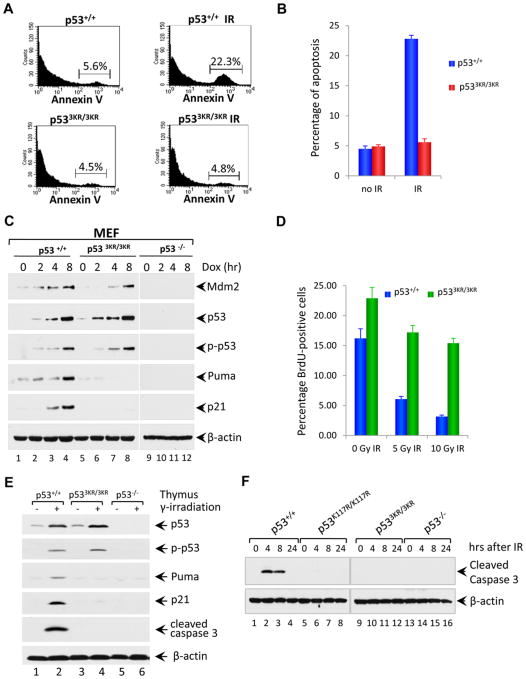
To examine p533KR-mediated function in vivo, we analyzed p53-dependent apoptosis in p533KR/3KR thymocytes subjected to ionizing radiation (IR). As expected, apoptosis was completely abrogated in p533KR/3KR thymocytes (Figures 4A and 4B). To test whether the p533KR mutation affects cell cycle arrest, MEFs from embryonic day 13.5 were treated with doxorubicin. As shown in Figure 4C, doxorubicin induced the stabilization and phosphorylation of p53 polypeptides to a comparable degree in p533KR/3KR and wildtype MEFs. In accordance with results obtained using exogenous p53 expression (Figures 3C and 3D), doxorubicin induced Mdm2 expression in both p533KR/3KR and wildtype cells (albeit to a slightly less degree in p533KR/3KR MEFs), whereas p21 and PUMA induction was specifically ablated in p533KR/KR MEFs (Figure 4C). Similar results were also obtained when p53-mediated responses were examined under different types of stress conditions such as etoposide, UV and oxidative stress (Figure S6A, S6B and S6C). Moreover, by using BrdU incorporation analysis, we found that p533KR/3KR cells were defective for IR-induced arrest of cell proliferation (Figure 4D and S6D). Consistent with these observations, p53 protein levels were induced to a similar degree in thymocytes from both wildtype and p533KR/3KR mice (Figure 4E), while IR induction of p21 and PUMA transcription was completely abrogated (Figure 4E). Moreover, to exclude the possibility that the mutant cells die with slower kinetics, we compared the levels of apoptosis induced by DNA damage in p53-wt, p53K117R/K117R, p533KR/3KR and p53-null mice. To this end, p53-wt, p53K117R/K117R, p533KR/3KR and p53-null mice were treated with 12.5 Gy of γ-radiation, and thymocytes isolated from these mice at different time points after irradiation (4hr, 8hr, and 24 hr) were analyzed for levels of apoptosis. As shown in Figure 4F, the active form of caspase-3 was readily detected in p53-wt mice at 4hr and 8hr after irradiation, indicating strong IR-induced apoptosis under these conditions. In contrast, significant apoptosis was not observed in thymocytes isolated from p53K117R/K117R, p533KR/3KR or p53-null mice at any time point after γ-radiation.
Figure 4. p533KR Mutation Severely Abrogates the p53-mediated Cell Cycle Arrest and Apoptotic Cell Death.
(A) Analysis of apoptosis in vivo for p53+/+ and p533KR/3KR mice. Mice were either untreated or exposed to 5 Gy of γ-irradiation, and then thymocytes were isolated and stained with Annexin V-FITC for FACS analysis.
(B) Annexin-V positive cells representing apoptotic thymocytes are quantified in (E). Data are shown as average ± SD from at least three mice for each genotype.
(C) Western blot analysis of Mdm2, p53, phosphorylated p53, Puma and p21 in p53+/+ and p533KR/3KR and p53−/− MEFs either untreated or treated with 0.2 μg/ml Dox for 2, 4 and 8 hours. β-actin serves as a loading control.
(D) Cell-cycle arrest analysis of p53+/+ and p533KR/3KR MEFs either left untreated or exposed to 5 or 10 Gy of γ-irradiation. 23 hours after irradiation, MEFs were pulsed with 10 μM BrdU for 45 minutes, and then cells were fixed and processed for immunofluorescence analysis for BrdU. BrdU-positive cells representing cells in S phase of cell cycle during BrdU incorporation after 5 or 10 Gy of γ-irradiation are quantified. Values shown are the averages ±SD of three independent replicates.
(E) Immunoblot assays of p53, phopho-p53, Puma, p21, cleaved caspase 3 and β-actin protein in the lysates prepared from the thymus of p53+/+, p533KR/3KR and p53−/− mice 4 hours after 12.5 Gy of γ-irradiation.
(F) Western blot analysis of apoptosis in the thymus of p53+/+, p53K117R/K117R, p533KR/3KR and p53−/− mice upon DNA damage treatment. Mice were either untreated or exposed to 12.5 Gy of γ-irradiation; thymocytes were isolated at different time points after irradiation and analyzed for the level of cleaved caspase 3. β-actin was used as a loading control.
See alsow Figure S5 and Figure S6.
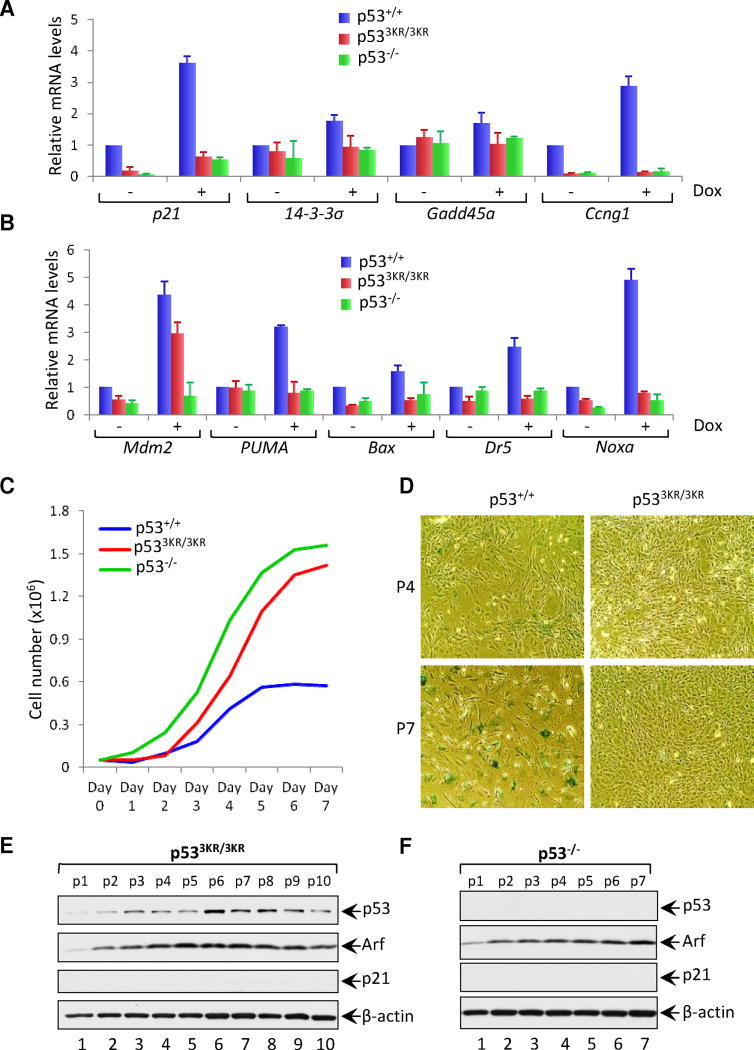
Finally, to confirm whether p533KR is also defective for induction of other p53 target genes involved in cell cycle arrest and apoptosis, mRNA levels in wildtype and mutant MEFs were compared by quantitative real-time RT-PCR. Significantly, none of the key p53 targets involved in cell-cycle arrest (i.e., p21, 14-3-σ, cyclin G1/ccng1, and GADD45) or apoptosis (Bax, PUMA, NOXA and DR5) were induced by doxorubicin in p533KR cells (Figures 5A and 5B). Indeed, the expression levels of these target genes were virtually indistinguishable in p533KR-mutant cells and p53-null cells. Taken together, these data indicate that the p533KR mutation abrogates both p53-mediated cell cycle arrest and apoptosis without affecting either genotoxin-induced p53 stabilization or transactivation of certain target genes such as Mdm2.
Figure 5. p533KR Loses Its Ability to Induce Cellular Senescence in p53 3KR/3KR MEFs.
(A and B) qRT-PCR analysis of indicated mRNAs in p53+/+, p533KR/3KR and p53−/− MEFs either untreated or treated with 0.2 μg/ml Dox for 8 hours. Results shown are the relative amount of specific mRNA normalized first to β-actin and then to the untreated wildtype sample values from three independent experiments with different MEF lines. Error bars represent ± SEM.
(C) Cell growth rate analysis of p53+/+, p533KR/3KR and p53−/− MEFs. 3 × 104 MEFs of different genotypes were seeded into 6-well plates at day 0 and counted daily.
(D) Images of SA-β-gal staining of p53+/+ and p533KR/3KR MEFs at different passages cultured according to the 3T3 protocol. MEFs at indicated passages were fixed and stained for β-galactosidase activity.
(E and F) Immunoblot analysis of p53, Arf and p21 in p533KR/3KR (E) and p53−/− (F) MEFs at indicated passages cultured according to the 3T3 protocol. β-actin serves as a loading control.
See also Figure S7.
The p533KR/3KR mutant fails to induce p53-dependent senescence
In addition to cell cycle arrest and apoptosis, numerous studies have identified senescence as an alternative mechanism of p53-mediated tumor suppression. In this regard, we found that doxorubicin-induced transactivation of several p53 target genes, including PAI-1 and PML, was also defective in p533KR/3KR cells (Figures S7A and S7B). Therefore, we examined whether p533KR/3KR MEFs are competent to undergo p53-dependent cellular senescence. As shown in Figure 5C, p533KR/3KR MEFs proliferated slightly slower than p53−/− MEFs, but dramatically faster than wildtype MEFs. As expected, wildtype MEFs reached senescence around passage 7; in contrast, both p53−/− and p533KR/3KR MEFs continued to proliferate and showed no evidence of senescence (Figure 5D). Notably, p53 and Arf transcription was gradually induced on passage of p533KR/3KR MEFs, similar to p53-null cells, but induction of p21 expression was not observed in p533KR/3KR MEFs (Figures 5E and 5F). In addition, we examined oncogene-induced senescence in these cells. As expected, very high levels of senescence were observed in p53-wt-MEF cells upon oncogene (H-ras) expression (Figure S7C and S7D); in contrast, we were unable to detect senescence of either p53-3KR or p53-null cells under these conditions. Taken together, these data demonstrate that p533KR/3KR cells fail to undergo p53-dependent senescence.
p533KR/3KR mice do not succumb to the early-onset spontaneous tumor formation commonly observed in p53-null mice
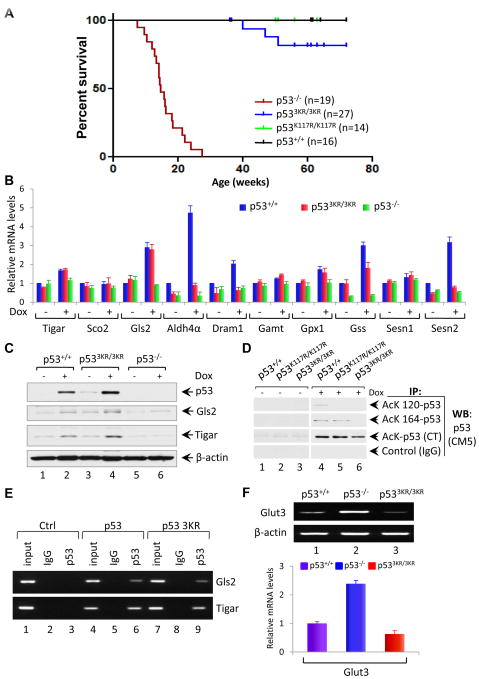
Although p533KR polypeptides retain the ability to bind DNA in a sequence-specific fashion and transactivate expression of the Mdm2 promoter, p533KR/3KR cells are defective for p53-dependent cell cycle arrest, apoptosis, and senescence. Therefore, to ascertain whether any of these canonical p53 functions are required for tumor suppression, we compared spontaneous tumor formation in cohorts of p53 wildtype (p53+/+), null (p53−/−) mutant (p53K117R/K117R and p533KR/3KR) mice. In accord with published data (Donehower et al., 1992; Jacks et al., 1994; Lozano, 2010), p53-null mice rapidly developed spontaneous tumors, primarily thymic lymphomas, and most died by 6 months of age. Although K117 acetylation is required for the p53-mediated apoptotic response in vivo, we observed no differences between p53K117R/K117R and wildtype mice with respect to either normal development or spontaneous tumor formation (Figure 6A). These results suggest that other functions of p53 (e.g., cell-cycle arrest and/or senescence) are sufficient to prevent early-onset spontaneous tumor formation. However, most p533KR/3KR animals remained healthy up to at least 16 months of age. Thus, despite their inability to mediate p53-dependent cell cycle arrest, apoptosis, or senescence, none of the p533KR/3KR mice succumbed to early-onset spontaneous tumors (Figure 6A). Indeed, only three of the twenty-seven p533KR/3KR mice developed tumors (Figure 6A), and sequence analysis uncovered de novo mutations of the p533KR gene in the tumor tissues of these three p533KR/3KR mice (Figure S7E), further validating that p53-3KR retains its function as a tumor suppressor. These data indicate that the combined loss of p53-mediated cell cycle arrest, senescence and apoptosis is not sufficient to abrogate the tumor suppression activity of p53.
Figure 6. p533KR Retains Its Tumor Suppressor Activity and the Ability to Activate Metabolic Targets.
(A) Kaplan-Meier survival curves of p53+/+, p53K117R/K117R, p533KR/3KR and p53−/− mice.
(B) qRT-PCR analysis of indicated mRNAs in p53+/+, p533KR/3KR and p53−/− MEFs either untreated or treated with 0.2 μg/ml Dox for 8 hours. Data are averages ± SEM of each mRNA quantities normalized first to β-actin and then to the untreated wildtype sample values from three independent MEF lines.
(C) Immunoblot assays of p53, Gls2 and Tigar protein in the whole cell lysates prepared from p53+/+, p533KR/3KR and p53−/− MEFs either untreated or treated with 0.2 μg/ml Dox for 8 hours. β-actin serves as a loading control.
(D) Analysis of acetylation of p53 at K117, K161 and K162 in p53K117R/K117R and p533KR/3KR MEF cells. p53+/+ and p533KR/3KR and p53K117R/K117R MEFs either untreated or treated with 0.2 μg/ml Dox, 1.0 μM TSA and 5 mM Nicotinamide for 6 hours. Cell lysates were immunoprecipitated with either anti-AcK120-p53, anti-AcK164-p53, anti- AcK CT-p53 antibodies or control IgG, and blotted with anti-p53 (CM5) antibody using ReliaBLOT systems from Bethyl Laboratories.
(E) ChIP assay for the binding of p53 or p53-3KR mutant to the consensus sites of p53 metabolic targets Gls2 and Tigar in H1299 cells transfected with plasmids DNA expressing mouse p53 wildtype (WT) or p53-3KR mutant.
(F) RT-PCR (upper panel) and qRT-PCR (lower panel) analysis of Glut3 expression in p53+/+, p533KR/3KR and p53−/− primary MEFs at passage 2. Data are represented as averages ± SEM from three independent experiments.
See also Figure S7
p533KR retains its ability to regulate the expression of metabolic p53 target genes
Cell cycle arrest, senescence, and apoptosis are widely viewed as the major mechanisms of p53-mediated tumor suppression; yet, our data suggest that these classical p53 responses are not essential for tumor suppression. Thus, other p53 functions, particularly those retained by p533KR, may be more relevant to suppression of early-onset tumorigenesis. To explore this possibility, we examined p533KR-mediated transcriptional activation in a panel of p53 target genes implicated in unconventional functions of p53. Notably, quantitative real-time RT-PCR revealed that the transcription of two known p53 target genes involved in metabolic regulation, GLS2 and TIGAR (Hu et al., 2010; Suzuki et al., 2010; Bensaad et al., 2006), was fully induced by doxorubicin in p533KR/3KR cells (Figure 6B). GLS2 encodes a mitochondrial glutaminase that modulates mitochondrial respiration and ATP generation by catalyzing the hydrolysis of glutamine to glutamate, while TIGAR encodes a fructose bisphosphatase that down-regulates glycolysis by reducing cellular levels of fructose-2, 6,-bisphosphate. Further analysis showed that the steady-state levels of GLS2 and TIGAR proteins were induced by stress in both wildtype and p533KR/3KR MEFs, but not in p53−/− cells (Figure 6C). To validate the p53 acetylation status in in these cells, p53+/+ and p533KR/3KR MEFs were first treated with 0.2 μg/ml Dox for 8 hours and were then processed for immunoprecipitation analysis. Cell lysates were immunoprecipitated with anti-AcK120-p53, anti-AcK164-p53 or anti-AcK CT-p53 antibodies, and blotted with anti-p53 (CM5) antibody. As expected, acetylation of p53 at each of these sites was readily induced by DNA damage in wild type MEFs (Lane 4, Figure 6D). In p533KR/3KR MEFs, however, damage- induced acetylation of the C-terminal lysines was retained, but no detectable acetylation was observed at either K117 or K161 (lane 6). Likewise, K117 acetylation was specifically abrogated in p53117R/117R MEFs, while acetylation of the other sites remained normal (Lane 5). Moreover, chromatin immunoprecipitation assays (CHIP) revealed that both wild type p53 and p53-3KR proteins bind equally well to the promoters of the GLS2 and TIGAR metabolic target genes. Thus, although p533KR fails to transactivate most p53 target genes, including those involved in p53-mediated cell cycle arrest, apoptosis, and senescence, it retains the ability to induce the metabolic target genes GLS2 and TIGAR in response to stress. Finally, increased glucose uptake is a common characteristic of tumor cells that preferentially metabolize glucose by aerobic glycolysis instead of oxidative phosphorylation, and several studies have shown that p53 expression leads to transcriptional downregulation of the GLUT3 glucose transporter (Kawauchi et al., 2008; Schwartzenberg-Bar-Yoseph et al., 2004). As expected, GLUT3 expression is dramatically increased in p53−/− cells relative to wildtype MEFs (Figure 6F). Significantly, the levels of GLUT3 transcript remain low in p533KR/3KR cells, suggesting that p533KR can inhibit GLUT3 expression in a manner similar to wildtype p53 (Figure 6F). Together, these data demonstrate that p533KR retains the capacity to regulate the expression of multiple metabolic p53 target genes.
The inhibitory effects of p533KR in regulating glucose uptake, glycolysis, reactive oxygen species (ROS) and colony formation
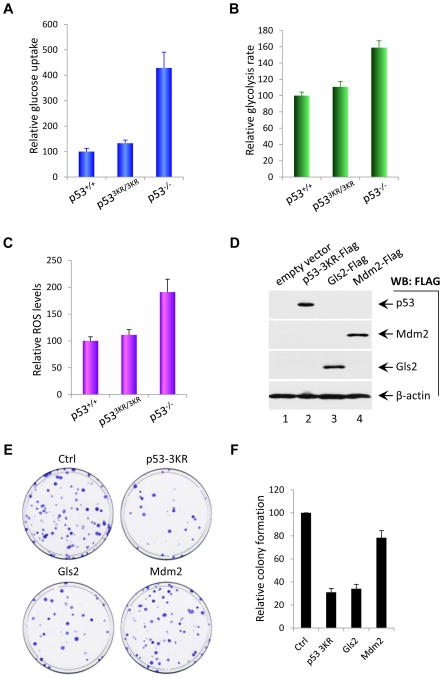
Cancer cells consume large quantities of glucose for glycolysis. It has been reported that inactivation of p53 lead to high glucose uptake (Zhang et al., 2011). Since p53-3KR is able to inhibit the expression of GLUT-3, p533KR/3KR cells are likely to retain the ability to suppress glucose uptake. To confirm this prediction, we compared uptake of 2-[3H]-deoxyglucose in p53+/+, p533KR/3KR and p53−/− MEFs. As shown in Figure 7A, relative glucose uptake was approximately 3-fold higher in p53−/− MEFs than p53+/+ MEFs, validating that p53 loss significantly promotes glucose uptake (Zhang et al., 2011). In contrast, we failed to detect a significant increase in p533KR/3KR MEF cells, suggesting that p533KR retains the ability to suppress glucose uptake normally. Moreover, we compared the glycolysis rate in p53+/+, p533KR/3KR and p53−/− MEFs by monitoring the conversion of 5-[3H] glucose to 3H2O, as described previously (Ashcroft et al., 1972). As shown in Figure 7B, the glycolysis rate of p53−/− MEF cells was about 60% higher than that of wildtype MEFs, indicating that p53 loss indeed increases the rate of glycolysis in vivo. In contrast, the glycolysis rate of p533KR/3KR cells was very similar to that observed in wildtype MEFs. Thus, p533KR retains the ability to suppress both glucose uptake and glycolysis.
Figure 7. p533KR Inhibits Glucose Uptake, Glycolysis, Reactive Oxygen Species Level and Colony Formation.
(A) Glucose uptake measurement of p53+/+, p533KR/3KR and p53−/− MEF cells determined by the uptake of 2-[3H]-deoxyglucose. Results shown are the averages ± SD of three different experiments.
(B) Glycolysis rate analysis of p53+/+, p533KR/3KR and p53−/− MEF cells determined by monitoring the conversion of 5-[3H] glucose to 3H2O as described in the Experimental Procedures. Values represent averages ± SD of three different experiments.
(C) Measurement of the reactive oxygen species (ROS) levels of p53+/+, p533KR/3KR and p53−/− MEF cells was performed by using mouse ROS ELISA kit as described in the Experimental Procedures. Data was reported as averages ± SD of three different experiments.
(D) Western blot analysis of p53-3KR, Mdm2 and Gls2 proteins in H1299 cells transfected with FLAG-tagged expression plasmids as indicated using anti-FLAG antibody. β-actin was used as a loading control.
(E) Colony formation assay of H1299 cells transfected with p53-3KR, Mdm2 and Gls2. H1299 cells were transfected with either empty vector, or FLAG-p53-3KR, FLAG-Mdm2, or FLAG-Gls2 expression plasmids for 48 hrs, and then split and subject to colony formation assay visualized by crystal violet staining.
(F) Quantification of colonies formed in H1299 cells transfected with empty vector, FLAG-p53-3KR, FLAG-Mdm2, and FLAG-Gls2 expression plasmids after 12 days culture in the presence of G418. Graphs are presented as percentage of colonies of empty vector transfected cells and values are shown as average percentage ± SD of three different experiments.
Reactive oxygen species (ROS) are produced as a normal byproduct of cellular metabolism and function as signaling molecules involved in tumorigenesis. Previous studies indicate that both GLS2 and TIGAR regulate the levels of reactive oxygen species (ROS) in cancer cells (Bensaad et al., 2006; Hu et al., 2010; Suzuki et al., 2010). Thus, we compared the levels of ROS in p53+/+, p533KR/3KR and p53−/− MEFs. As shown in Figure 7C, the ROS levels of p53−/− MEFs were about 85% higher than those of wildtype cells, suggesting that p53 loss significantly increases ROS levels in vivo. Conversely, the ROS levels of p533KR/3KR MEF cells were indistinguishable from those of wildtype cells. Finally, we tested whether expression of p53-3KR is able to suppress colony formation of cancer cells in a manner analogous to GSL2 expression, as reported (Hu et al., 2010; Suzuki et al., 2010). To this end, p53-null H1299 cells were transfected for 48 h with either an empty expression vector, Flag-GLS2 expression vector alone, or Flag-p53-3KR expression vector and Flag-Mdm2 expression vector (Figure 7D). As shown in Figure 7E and 7F, p53-3KR expression significantly reduced colony formation. Notably, in contrast to GLS2 expression, Mdm2 expression in these p53-null cells had no obvious effect, suggesting that Mdm2 induction by p53-3KR may not contribute to its tumor suppression activity (Figure 7E). Together, these data suggest that p53-3KR retains its tumor suppressor activity, at least in part, through its ability to regulate energy metabolism and ROS levels in vivo.
Discussion
Since p53 mutations are the most common genetic lesions associated with human cancer, a major objective of molecular oncology is to elucidate the mechanisms by which p53 suppresses tumor formation. Our studies of mice expressing acetylation-defective p53 mutants revealed that acetylation differentially regulates p53 function in cell cycle arrest, apoptosis, and senescence. This circumstance afforded us the opportunity to test whether these p53 functions are required for p53-mediated tumor suppression. Surprisingly, we found that the combined loss of p53-dependent cell cycle arrest, apoptosis, and senescence is not sufficient to abrogate the tumor suppression activity of p53. In particular, our results show that (i) loss of K117 acetylation on mouse p53 (corresponding to K120 of human p53) specifically abrogates transactivation of proapoptotic p53 gene targets; (ii) p53K117R is defective for p53-mediated apoptosis, but retains the cell cycle arrest and senescence functions of p53; (iii) unlike p53-null mice, p53K117R/K117R animals do not develop early-onset tumors, suggesting that apoptosis is largely dispensable for p53-mediated tumor suppression, at least in this biological setting; (iv) loss of acetylation at three sites (p533KR: K117, K161, and K162) in the DNA-binding domain of mouse p53 abrogates p53-mediated cell cycle arrest, apoptosis, and senescence, (v) p533KR mice do not develop early-onset tumors, indicating that p53-dependent cell cycle arrest, apoptosis, and senescence are not absolutely required for p53 tumor suppression; and (vi) p533KR retains the ability to modulate energy metabolism and reduce reactive oxygen species (ROS) by regulating metabolic p53 target genes in vivo. Thus, by demonstrating that the canonical functions of p53 in cell cycle arrest, apoptosis, and senescence are not essential for tumor suppression, these results challenge the prevailing view of how p53 lesions promote human cancer.
Reassessing the mechanisms of p53-mediated tumor suppression
Through its ability to induce cell cycle arrest, apoptosis, or senescence in response to various types of stress, p53 is thought to suppress tumor formation by preventing inappropriate expansion of cells with malignant potential. Although non-transcriptional functions of p53 may also contribute to tumor suppression, recent studies of mice bearing defined mutations of the p53 transactivation domains indicate that its role as a transcription factor is essential for p53-mediated tumor suppression (Brady et al., 2011). Indeed, many prominent p53 transcriptional target genes contribute to specific p53-dependent functions in cell cycle arrest (e.g., p21, 14-3-3σ), apoptosis (e.g. PUMA, NOXA, DR5), and senescence (e.g. p21, PAI-1, PML), and a major objective of current research is to determine how p53 differentially transactivates its target genes to elicit the most appropriate cellular response (Vosuden and Prives, 2009; Kruse and Gu, 2009). Although our current understanding is incomplete, it appears that multiple parameters, including tissue specificity and the nature of the inducing stress, coordinately determine whether p53 activation leads to cell cycle arrest, apoptosis, or senescence.
The ability of p53 to induce cell cycle arrest is mediated in large part by transcriptional activation of p21Cip1, which encodes an inhibitor of cyclin-dependent kinases (El-Deiry et al., 1993; Bunz et al., 1998). The induction of p21 expression is sensitive to even low levels of p53 protein, suggesting that a temporary G1 or G2 block, as induced by mild damage or stress, allows cells to survive until damage has been resolved or stress removed. In addition, p21 is also critical for p53-dependent senescence—an irreversible cell cycle arrest that can be elicited by DNA damage, oncogene activation, or telomere dysfunction. However, unlike p53-null mice, p21-null animals are not prone to early-onset tumorigenesis, although p21 deficiency extends longevity and rescues stem cell function in mice with dysfunctional telomeres (Deng et al., 1995; Choudhury et al., 2007). This observation implies that the apoptotic functions of p53 may be more relevant to tumor suppression, or may contribute to tumor suppression in a compensatory fashion when p53-mediated cell cycle arrest and/or senescence are compromised. Nevertheless, early-onset tumor formation is not observed in mice expressing an apoptosis-defective mutant (p53R172P) that retains the cell-cycle arrest and cellular senescence functions of p53 (Liu et al., 2004; Cosme-Blanco et al., 2007; Van Nguyen et al., 2007). Likewise, mice lacking PUMA, a p53-target gene required for p53-dependent apoptosis in a broad range of cell types (Yu and Zhang, 2008), are still resistant to early-onset tumorigenesis (Michalak et al., 2008). Interestingly, a p53 mutant (p5325,26) deficient for p53-mediated cell cycle arrest and apoptosis, but not senescence, was recently shown to retain the ability to inhibit KrasG12D- induced lung carcinogenesis (Brady et al., 2011). Thus, despite the fact that the canonical functions of p53 in cell cycle arrest, apoptosis, and senescence are widely accepted as major mechanisms of p53-mediated tumor suppression, it has never been formally tested whether they are rate-limiting steps in vivo.
Our data show that acetylation-defective p533KR polypeptides are stabilized and phosphorylated normally in response to DNA damage, and that they retain the ability to transactivate at least some p53 target genes, including Mdm2, GLS2, and TIGAR. Nevertheless, p533KR fails to induce p53-mediated cell cycle arrest, apoptosis, or senescence, and it does not transactivate key p53 target genes involved in these processes, confirming that acetylation is critical for prominent aspects of p53 function. Moreover, the unique phenotype of the p533KR mutant allowed us to test for the first time whether tumor suppression can occur when p53-mediated cell cycle arrest, apoptosis, and senescence are abrogated simultaneously. Surprisingly, p533KR/3KR mice do not develop the early-onset thymic lymphomas characteristic of p53-null animals. This result indicates that other unconventional functions of p53 are at least equally important for tumor suppression. Since p53 lesions are associated with a broad spectrum of human cancers, it will be interesting to see whether the unconventional p53 functions that inhibit early-onset lymphomagenesis also contribute to suppression of other tumor types.
Numerous studies demonstrate that cell cycle arrest, apoptosis and senescence clearly contribute to p53-mediated tumor suppression. Nonetheless, is less clear under which biological settings these aspects of p53 function are required for tumor suppression. For example, PUMA is a critical target for p53-mediated apoptosis, but PUMA knockout mice fail to develop early onset spontaneous tumors. Yet, loss of PUMA accelerates Eμ-Myc-mediated lymphomagenesis (Garrison et al., 2008; Michalak et al., 2009). Clearly, our studies demonstrate that p53-mediated cell cycle arrest, apoptosis and senescence are not essential to suppress early onset lymphomagenesis in mice. Nevertheless, since p53 lesions are observed in a broad spectrum of human cancers, it will be interesting to ascertain whether these aspects of p53 function are critical for tumor suppression in other physiological settings.
A role for p53-mediated metabolic regulation in tumor suppression?
Although p53-mediated cell cycle arrest, apoptosis, and senescence are presumed to be the primary mechanisms by which p53 inhibits oncogenesis, the phenotype of p533KR/3KR mice indicates that other, as yet undetermined, functions of p53 are also important for tumor suppression. In principle, the relevant aspects of p53 function should be retained by the p533KR mutant polypeptide. As such, it is intriguing that p533KR, which fails to transactivate a number of genes involved in p53-mediated cell cycle arrest, apoptosis, and senescence, still maintains the capacity to regulate p53 target genes implicated in metabolic control, including GLS2, TIGAR, and GLUT3. In resting cells under normal aerobic conditions, the pyruvate generated by glycolysis can be fed into the mitochondrial tricarboxylic acid (TCA) cycle for efficient ATP generation via oxidative phosphorylation. However, proliferating cells undergo a metabolic reprogramming to “aerobic glycolysis” in which glycolytic flux is increased, oxidative phosphorylation is suppressed, pyruvate is converted primarily to lactate, and intermediates of the TCA cycle are diverted into anabolic pathways to facilitate cell growth (Vander Heiden et al., 2009; Cairns et al., 2011). A similar metabolic switch to aerobic glycolysis, termed the Warburg effect, is also observed in tumor cells, and recent studies suggest that p53 normally inhibits this process by limiting glycolytic flux through multiple mechanisms (Gottlieb and Vousden, 2010; Feng and Levine, 2010; Warburg; 1956). For example, p53 suppresses the expression of glucose transporters such as GLUT3 (Kawauchi et al., 2008; Schwartzenberg-Bar-Yoseph et al., 2004), although the precise mechanism of this repression needs further elucidation. Interestingly, GLUT3 and other types of glucose transporters are overexpressed in various human tumors (Meller et al., 2007; Macheda et al., 2005; Medina et al., 2002). p53 also modulates production of glutamine, an alternative to glucose as a fuel for bioenergetic pathways, by transactivating the gene for GLS2, a mitochondrial glutaminase that converts glutamine to glutamate (Hu et al., 2010; Suzuki et al., 2010). As such, GLS2 promotes energy metabolism by providing a source of α-ketoglutarate for the TCA cycle and antioxidant defense by providing glutamate for glutathione synthesis. Notably, GLS2 is downregulated in human cancer and GLS2 overexpression reduces the in vitro proliferation and colony forming potential of tumor cells (Hu et al., 2010; Suzuki et al., 2010). The gene encoding TIGAR was also identified as a key p53 target implicated in energy metabolism and ATP generation (Bensaad et al., 2006). By lowering levels of fructose-2, 6,-bisphosphate, TIGAR normally functions to reduce glycolytic flux and direct glucose to the pentose phosphate pathway. Thus, p53 inactivation promotes metabolic changes similar to the Warburg effect by increasing both glycolytic flux and lactate production (Gottlieb and Vousden, 2010; Feng and Levine, 2010).
Although cell cycle arrest, senescence and apoptosis are abrogated in p533KR/3KR cells, p533KR retains its ability to suppress glucose uptake and glycolysis. Moreover, consistent with the fact that both GLS2 and TIGAR are critically involved in regulating ROS in vivo, p533KR retains its capacity to reduce ROS levels and expression of p533KR leads to suppression of colony formation ability of cancer cells. In light of these results, p533KR may block tumor formation, at least in part, by inhibiting the metabolic shift to aerobic glycolysis and reducing ROS levels in vivo. As such, the ability of p53 to regulate the metabolic state and antioxidant defense may serve as independent mechanisms of tumor suppression that are sufficient to suppress early-onset spontaneous tumorigenesis when the p53-mediated pathways of cell-cycle arrest, apoptosis, and senescence are simultaneously inoperative. Clearly, the molecular mechanisms by which p53 regulates its metabolic target genes need further elucidation. Based on our studies, the requirements for activating p53-mediated metabolic regulation are apparently different from those that modulate cell cycle arrest, apoptosis and senescence. It will be interesting to know whether other known acetylation sites, such as C-terminus lysine residues and K320, or other types of p53 modification, such as phosphorylation or methylation, modulate the expression of these metabolic targets of p53 in vivo.
Experimental Procedures
Generation and Characterization of Mice
To generate knockin mice, W4/129S6 mouse embryonic stem (ES) cells (Taconic, Hudson, NY, USA) were electroporated with targeting vectors containing 5′ homology region with exons 1-4, a neomycin resistance gene cassette flanked by two LoxP sites (LNL) and a 3′ homology region with exons 5-11 with the desired point mutations. ES cells were selected by G418 and screened for homologous recombination by Southern blotting with Bglll-digested genomic DNA using standard protocols. Southern blot probes were generated from PCR amplification in the regions outside the targeting vector. The correctly targeted ES clones containing the targeted allele with desired mutation(s) were injected into C57BL/6 blastocysts, which were then implanted into pseudopregnant females to generate chimeras. Germline transmission was confirmed by breeding chimeras with C57BL/6 mice. Germline-transmitted mice were bred with Rosa26Cre mice to remove the LNL cassette and generate mice with only the targeted p53K117R or p533KR mutations. To further validate the mutations created in p53K117R/K117R or p533KR/3Kr mice, we sequenced p53 mRNA from MEFs derived from these mice and only the desired point mutations and no additional mutations were found. The offspring were PCR genotyped using a primer set (Forward: 5′-CTTCCTGCAGTCTGGGACAGC C-3′, Reverse: 5′-GCAGCTGGGCCTACAGCACACG-3′). All mice work was performed in compliance with the IACUC (Institutional Animal Care and Use Committee) of Columbia University.
Protein Purification and Mass Spectrometry
To purify the acetylated mouse p53 protein, H1299 cells were cotransfected with pCin4-p53-Flag and CMV-Tip60 and cultured for 20 hr, then the transfected H1299 cells were treated with 1 μM TSA and 5 mM nicotinamide for 6 hrs before harvest. Cells were collected and lysed in the Flag-lysis buffer (50 mM Tris-HCl [pH 7.9], 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% sarkosyl, 10% glycerol, and fresh proteinase inhibitor cocktail [Sigma]) plus 2 μM TSA and 10 mM Nicotinamide, and the cell extracts were immunoprecipitated with the anti-Flag monoclonal antibody-conjugated M2 agarose beads (Sigma). The Flag peptide-eluted material was resolved by 8% SDS-PAGE. The p53 bands were excised from the gel and subjected to tryptic digestion and mass spectrometry.
Colony Formation Assay
H1299 cells were transfected with either empty vector or FLAG-tagged-p53,-GLS2 and - Mdm2 expression plasmids for 48 hrs. Cells were then split and seeded into 10-cm dishes at the density of 1,000 cells per dish and cultured under the selection of G418 (600 μg/ml) for 12 days. Cells were stained with crystal violet solution, and visible colonies were counted.
Measurement of Glucose Uptake and Glycolysis Rate
Glucose uptake of p53+/+, p533KR/3KR and p53−/− MEF cells were measured by determining the uptake of 2-[3H]-deoxyglucose (American Radiolabeled Chemicals) by MEF cells as previously described (Zhang C, et al 2011). Briefly, MEF cells were seeded in triplicate at 2.5 × 105 cells per well in six-well plates. After incubation at 37°C overnight, the cells were washed twice with Krebs buffer [116 mM NaCl, 4.6 mM KCl, 1.16 mM KH2PO4, 25 mM NaHCO3, 2.5 mM CaCl2.2H2O, 1.16 mM MgSO4.7H2O, 0.2% bovine serum albumin (pH 7.4)]. Then, 1 ml Krebs buffer was added to each well and the cells were incubated at 37°C for 30 min, and then another 1 ml Krebs buffer containing 2 μCi/ml [3H]2-deoxy-D-glucose was added to each well. Cells treated with 20 μM glucose transport inhibitor cytochalasin B (Sigma) were used as a negative control. Cells were incubated at 37°C for 20 min and then washed four times with ice-cold Krebs buffer, and lyzed with 0.5 ml 1% SDS. 2-[3H]-deoxyglucose was quantified by scintillation counting. Glycolysis rate of p53+/+, p533KR/3KR and p53-/- MEF cells was determined by monitoring the conversion of 5-[3H] glucose to 3H2O as described previously (Ashcroft et al., 1972). To measure glycolysis rate, 2.5 × 105 MEFs were seeded in each well of six-well plate and cultured overnight. MEF cells were washed once with phosphate-buffered saline (PBS) and once with Krebs buffer, and 1 ml of Krebs buffer was added to cells. After the cells were incubated for 30 min at 37°C, 0.5 ml of Krebs buffer containing 10 mM glucose and 10μCi of 5-[3H]-glucose were added. Following incubation for 1 hr at 37°C, 500 μl of 0.2 N HCl was added to the cells to stop the reaction, then scrape the cells and 500 μl cell suspension aliquots in triplicate were transferred to uncapped 1.5 ml Eppendorf tubes which then were put into a scintillation vial containing 1.0 ml of H2O in such a way that the water in the vial and the contents of the 1.5 ml Eppendorf tube were not allowed to mix. The vials were sealed, and diffusion was allowed to occur for a minimum of 24 hrs at room temperature. The amounts of diffused 3H2O in the vial, representing the glycolysis rate, were determined by scintillation counting.
Measurement of ROS
5 × 105 p53+/+ and p533KR/3KR and p53−/− MEFs were seeded in 6 cm-dishes, following incubation at 37 °C overnight, the cells were collected and lysed for ROS assay using Mouse Reactive Oxygen Species ELISA kit (E03R0070) from Life Science Advanced Technologies, Inc.. Briefly, 100 μl of samples or standards with known concentration of ROS were added to the triplicate microtiter plate wells precoated with anti-ROS antibody, and then 50 μl ROS-HRP conjugate was added to each well, mix and incubate microtiter plate at 37°C for 1hr, then the wells were washed with 400 μl 1 × washing solution for five times and incubated for 15 minutes with addition of 50 μl substrates for HRP enzyme. The enzyme-substrate reaction was terminated by adding 50 μl of stop solution to each well and the optical density (O.D.) at a wavelength of 450 nm was measured immediately using μQuant Scanning Microplate Spectrophotometer (BioTek Instruments, Inc.). The concentration of ROS in each sample is then determined by comparing the O.D. of each sample to the standard curve.
Supplementary Material
Highlights.
K120 acetylation of p53 is essential for p53-mediated apoptosis
Loss of acetylation at multiple sites abrogates p53-dependent cell cycle arrest, senescence, and apoptosis.
Cell cycle arrest, apoptosis and senescence are not absolutely required for p53′s tumor suppression.
The unconventional activities of p53 are critical for suppression of spontaneous tumorigenesis.
Acknowledgments
We thank Dr. Victor Lin from HICCC Mouse Facility for excellent services in developing p53 knockin mice and Dr. M. Szabolcs for mouse pathology analysis. We thank Dr. Z.H. Feng from UMDNJ for providing the reagents. This study was supported by grants from NIH/NCI and the Leukemia and Lymphoma Society. W.G. is also supported by an Ellison Medical Foundation Senior Scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashcroft SJ, Weerasinghe LC, Bassett JM, Randle PJ. The pentose cycle and insulin release in mouse pancreatic islets. Biochem J. 1972;126(3):525–532. doi: 10.1042/bj1260525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Berger SL. Keeping p53 in check: a high-stakes balancing act. Cell. 2010;142:17–19. doi: 10.1016/j.cell.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Choudhury AR, Ju Z, Djojosubroto MW, Schienke A, Lechel A, Schaetzlein S, Jiang H, Stepczynska A, Wang C, Buer J, et al. Cdkn1a deletion improves stem cell function and lifespan of mice with dysfunctional telomeres without accelerating cancer formation. Nat Genet. 2007;39:99–105. doi: 10.1038/ng1937. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cosme-Blanco W, Shen MF, Lazar AJ, Pathak S, Lozano G, Multani AS, Chang S. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007;8:497–503. doi: 10.1038/sj.embor.7400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, Yue W, Yu J, Zhang L, Onciu M, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Vousden KH. p53 regulation of metabolic pathways. Cold Spring Harb Perspect Biol. 2010;2:a001040. doi: 10.1101/cshperspect.a001040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kawauchi K, Araki K, Tobiume K, Tanaka N. p53 regulates glucose metabolism through an IKK-NF-kappaB pathway and inhibits cell transformation. Nat Cell Biol. 2008;10:611–618. doi: 10.1038/ncb1724. [DOI] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kortlever RM, Higgins PJ, Bernard R. Plasinogen activator inhibitor-1 is a critical downstream traget of p53 in the induction of replicative senescence. Nat Cell Biol. 2006;8:877–884. doi: 10.1038/ncb1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci U S A. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wu L, Corsa CA, Kunkel S, Dou Y. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36:290–301. doi: 10.1016/j.molcel.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano G. Mouse models of p53 functions. Cold Spring Harb Perspect Biol. 2010;2:a001115. doi: 10.1101/cshperspect.a001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer. Biol Res. 2002;35:9–26. doi: 10.4067/s0716-97602002000100004. [DOI] [PubMed] [Google Scholar]
- Meller J, Sahlmann CO, Scheel AK. 18F-FDG PET and PET/CT in fever of unknown origin. J Nucl Med. 2007;48:35–45. [PubMed] [Google Scholar]
- Mellert HS, McMahon SB. hMOF, a KAT(8) with many lives. Mol Cell. 2009;36:174–175. doi: 10.1016/j.molcel.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Michalak EM, Villunger A, Adams JM, Strasser A. In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 2008;15:1019–1029. doi: 10.1038/cdd.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, Strasser A, Adams JM, Scott CL. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 2009;16:684–696. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeliga M, Sidoryk M, Matyja E, Kowalczyk P, Albrecht J. Lack of expression of the liver-type glutaminase (LGA) mRNA in human malignant gliomas. Neurosci Lett. 2005;374(3):171–173. doi: 10.1016/j.neulet.2004.10.051. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nguyen T, Puebla-Osorio N, Pang H, Dujka ME, Zhu C. DNA damage-induced cellular senescence is sufficient to suppress tumorigenesis: a mouse model. J Exp Med. 2007;204:1453–1461. doi: 10.1084/jem.20062453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(1):S71–83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.