Abstract
The determinants of CD8+ cytotoxic T-lymphocyte (CTL) antiviral activity against human immunodeficiency virus type 1 (HIV-1) remain poorly defined. Although recent technological advances have markedly enhanced the ability to detect HIV-1-specific T cells, commonly used assays do not reveal their direct interaction with virus. We investigated two determinants of CTL antiviral efficiency by manipulating HIV-1 and measuring the effects on CTL suppression of viral replication in acutely infected cells. Translocation of a Gag epitope into the early protein Nef markedly increased the activity of CTL recognizing that epitope, in comparison to HIV-1 expressing the epitope normally in the late protein Gag. Because this epitope translocation resulted not only in earlier expression but also in loss of major histocompatibility complex class I downregulation by Nef, the activities of CTL against a panel of viral constructs differing in kinetics of epitope expression and class I downmodulation were compared. The results indicated that both the timing of epitope expression and the reduction of class I have profound effects on the ability of CTL to suppress HIV-1 replication in acutely infected cells. The epitope targeting of CTL and viral control of class I therefore likely play important roles in the ability of CTL to exert pressure on HIV-1.
Human immunodeficiency virus type 1 (HIV-1)-specific CD8+ cytotoxic T lymphocytes (CTL) are considered mediators of protective immunity and therefore a key focus of vaccine development efforts for HIV-1. The role of CTL in viral immunopathogenesis has prompted development of new methodologies to facilitate accurate measurement of epitope-specific immune responses. The technology for detecting CTL has advanced tremendously (5), and defining antiviral CTL responses can now be accomplished with unprecedented precision and ease. Chromium release assays for virus-specific killing by bulk peripheral blood mononuclear cells and limiting dilution precursor frequency assays have been supplanted by more highly specific and quantitative methods such as Elispot and peptide-major histocompatibility complex (MHC) tetramer binding.
One common characteristic of both past and recent assays, however, is the dependence on nonphysiologic sources of antigen, such as small synthetic peptides and recombinant vaccinia viruses. Common methods to measure HIV-1-specific CTL responses therefore detect the presence of CTL but do not evaluate their efficiency against HIV-1-infected cells. The use of synthetic and recombinant antigens to trigger CTL entirely bypasses the process of class I antigen processing and presentation. It is therefore likely that epitope presentation is variable and influenced by multiple factors and that different epitopes vary in presentation (31).
The interaction of HIV-1-specific CTL with HIV-1 is therefore an area that is still poorly understood. The antiviral properties of CTL and the determinants of antiviral efficiency remain unclear. Importantly, the role of epitope specificity is not known. Studies suggest that the specificity of CTL may be an important factor in the recognition of infected cells (34) and suppression of HIV-1 replication (35), despite equivalent activity against exogenously peptide-loaded cells. Another potentially key factor is the ability of viral Nef to downregulate the major histocompatibility complex class I molecule (MHC-I) and reduce the ability of CTL to act upon infected cells (8, 28, 30, 36).
Given the rapidity of the viral life cycle after infection of a target cell (15, 32), the window of susceptibility to cytolysis is probably small. The kinetics of protein and epitope expression are therefore likely to be a key determinant of the timing of infected cell susceptibility to killing by CTL relative to virion production. It is known that the Tat, Rev, and Nef proteins are expressed earliest after cellular infection (25). It is unclear, however, whether CTL recognizing epitopes in these early proteins differ functionally from those recognizing epitopes in late proteins (such as Gag). In this study, we examined the influence of epitope expression kinetics on the antiviral efficiency of CTL by manipulating the timing of epitope expression and evaluated the relative contributions of protein kinetics and Nef-mediated downregulation of class I.
MATERIALS AND METHODS
Viral constructs and stocks. (i) NL4-3 half-genome plasmids.
Plasmids p83-2 (containing HIV-1 NL4-3 sequences up to a unique EcoRI site) and p83-10 (containing NL4-3 sequences beyond the EcoRI site) were obtained from the National Institites of Health AIDS Research and Reference Reagent Program. Variants of these plasmids were produced by PCR mutagenesis as per the method of Saiki et al. (26). The p83-2.1 plasmid was derived from p83-2 to alter a common CTL epitope from SLYNTIAVL in NL4-3 to the clade B consensus sequence SLYNTVATL (nucleotide changes A to G at position 1033, G to A at position 1039, and T to C at position 1040).
The p83-2.1-SL9x plasmid contained nucleotide change C to A at position 1042, resulting in an amino acid change of L to I at position 85) in p83-2.1. The p83-10-NefSL9 plasmid contained an insertion of the nucleotides GTTCATTATATAATACAGTAGCAACCCTCTATTG in place of nucleotide 9382 of NL4-3, resulting in an insertion of the SL9 epitope into Nef. The p83-2.1/heat-stable antigen plasmid (containing murine CD24 inserted in the vpr reading frame of p83-2.1) was constructed by swapping a PflMI-EcoRI fragment (nucleotides 5297 to 5743) from NL-r-HSAS (16) into p83-2.1 (1). All plasmids were verified by sequencing.
Production of infectious virions.
As per a modification of the method of Gibbs et al. (12), the above p83-2.1 and p83-10 derivatives were linearized with EcoRI (10 μg each) and coelectroporated (270 V, 800 μF) into T1 or H9 cells with a Cell-Porator (Gibco BRL, Rockville, Md.). The p24 in the supernatants of the electroporated cells was monitored by quantitative p24 enzyme-linked immunosorbent assay (Dupont, Boston, Mass.) for 10 to 21 days, and virus stocks were harvested at peak. Coelectroporation of different plasmids allowed different combinations of Gag and Nef variants, with or without the heat-stable antigen reporter (1). Virus titer determination (TCID50/ml) was carried out as described previously (17).
Real-time quantitative reverse transcription-PCR.
Total RNA from T1 cells infected at excess multiplicity with HIV-1 NL4-3 was isolated at serial time points for reverse transcription and PCR amplification for viral nef, long terminal repeat/gag, and cellular β2-microblobulin RNA sequences with the Qiagen One Step reverse transcription-PCR kit (Qiagen, Chatsworth, Calif.) with the manufacturer's protocol. To quantitate the number of nef, long terminal repeat/gag, or β2-microglobulin transcripts, in vitro-transcribed standards were generated (Promega, Madison, Wis.). Serial dilutions of each in vitro transcript were amplified in parallel. The in vitro transcripts were detectible to below 10 RNA copies with PCR primers as previously described (7).
CTL clones.
CTL clones recognizing the HLA A2-restricted Gag p17 epitope SLYNTVATL were obtained at limiting dilution and maintained as previously described (35).
HIV-1 suppression assays.
Inhibition assays were performed as previously described (27), with modifications. The HLA A2-expressing cell line T1 was acutely infected with the indicated HIV-1 variant at a multiplicity of 0.1 TCID50 per cell (multiplicity of infection of 0.1) and washed twice. These cells then were cocultured with CTL at a ratio of 5.0 × 104 target cells with 0.63 × 104 CTL in 96-well U-bottomed plates, in a final volume of 200 μl per well. Then 100 μl of supernatant medium was removed and replaced with fresh medium at 2- to 4-day intervals. Viral replication was assessed by quantitative p24 antigen enzyme-linked immunosorbent assay on the removed supernatant fluids. Each condition was performed in triplicate.
Measurement of HLA A2 expression in acutely infected cells.
Reporter viruses expressing murine CD24 were used to analyze the effects of HIV-1 infection on MHC-I expression as previously described (1). Fluorescein isothiocyanate-conjugated rat anti-mouse CD24 (heat-stable antigen) monoclonal antibody clone M1/69 was obtained from BD PharMingen (San Diego, Calif.). Biotin-conjugated anti-HLA-A2 immunoglobulin M monoclonal antibody (BIH0648) was obtained from One Lambda, Inc. (Canoga Park, Calif.). Isotype control antibodies and the streptavidin-phycoerythrin conjugate were purchased from BD PharMingen.
T1 cells in the log phase of growth were acutely infected with the indicated viruses at multiplicity of infection of 1 and harvested after 4 days for costaining for heat-stable antigen and HLA A2. Isotype control antibody staining of infected and uninfected cells was used to establish the negative quadrant and appropriate compensation between FL1 and FL2 channels. Flow cytometric analysis was performed on a FacScan with CellQuest software (Becton Dickinson) running on a G4 Power Macintosh (Apple Computer).
RESULTS
Nef is expressed earlier than Gag or RT in acutely infected cells.
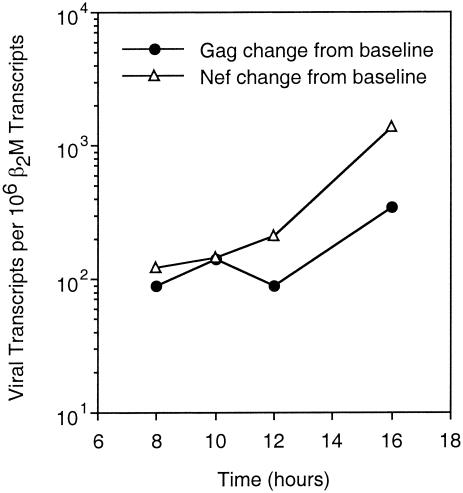
Nef is expressed from an RNA transcript that is not Rev-dependent for nuclear export, in contrast to Gag and RT, and therefore likely expressed earlier (9, 25). To measure the kinetics of Nef and Gag/RT expression, real-time quantitative reverse transcription-PCR was performed at serial time points after acute HIV-1 infection of T1 cells (Fig. 1). As expected, the rise in Gag/RT transcripts lagged behind that of Nef transcripts by several hours. These findings confirmed that Nef transcripts appear in the cytoplasm earlier than Gag/RT transcripts in acutely infected cells and support the concept that Nef is an early protein, while Gag and RT are late proteins in the viral life cycle. This raises the question of whether temporal differences in protein expression lead to functional consequences in the interaction of CTL with acutely infected cells.
FIG. 1.
Kinetics of Nef, Gag, and RT expression in acutely infected cells. T1 cells were acutely infected with NL4-3 and real-time quantitative reverse transcription-PCR was performed to measure the production of Gag-Pol and Nef transcripts over time.
A common Gag epitope can be transposed into Nef.
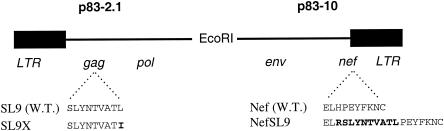
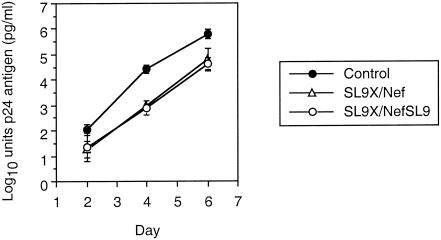
To assess the impact of epitope expression kinetics on the antiviral function of CTL, we manipulated the expression of a common Gag epitope by translocation into Nef (Fig. 2). In the p83-2.1 plasmid containing the Gag-Pol portion of the HIV-1 NL4-3-based genome, the HLA A2-restricted epitope SLYNTVATL (SL9) was functionally knocked out by altering the C-terminal anchor residue from leucine to isoleucine, SLYNTVATI (SL9x), resulting in alteration of the HLA A2-binding motif. In the p83-10 plasmid containing the Env-Nef portion of NL4-3, the SL9 epitope was inserted into the C terminus of Nef (NefSL9). Cotransfection of these plasmids in different combinations allowed production of HIV-1 virions containing Gag with SL9 and normal Nef (wild-type, SL9/Nef), Gag with SL9x and Nef (SL9x/Nef), and Gag with SL9x and NefSL9 (SL9x/NefSL9). These genetic manipulations did not appear to affect viral replication or spread in our tissue culture system (Fig. 3), indicating the feasibility of producing replication-competent HIV-1 containing a single copy of the wild-type Gag epitope transposed into Nef. Interestingly, the SLYNTVATP variant was nonviable (data not shown), consistent with severe fitness constraints from single amino acid changes in Gag (11).
FIG. 2.
Construction of HIV-1 with a transposed SL9 epitope in Nef and knockout of SL9 in Gag. The half genomes of modified HIV-1 NL4-3 contained in plasmids p83-2.1 and p83-10 are shown. Wild-type versions of each plasmid contain the SL9 epitope in Gag and unmodified Nef (last 10 amino acids shown), respectively. The SL9x mutation was introduced by altering the codon for the HLA A2 anchor residue from leucine to isoleucine in p83-2.1. The NefSL9 mutation was created by inserting sequences coding for 10 amino acids including the SL9 epitope into the 3′ end of nef.
FIG. 3.
Growth curves for HIV-1 containing Gag and Nef mutations. Virions produced by cotransfecting these plasmid constructs were evaluated for their growth after acute infection of T1 cells. The growth curves reflect the mean of two independent experiments (error bars represent one standard deviation in log10 units). Viral growth, as reflected by the slope of virion concentration, was similar for the three viruses.
Transposition of SL9 into Nef markedly enhances the antiviral activity of SL9-specific CTL.
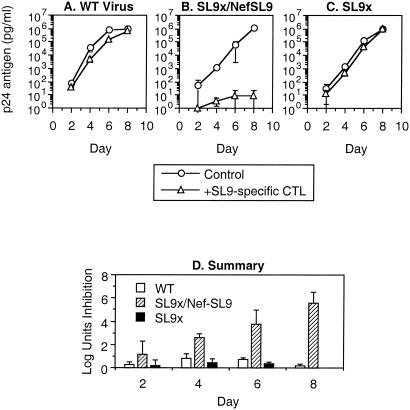
To test the impact of epitope translocation and kinetic alteration on the antiviral function of SL9-specific CTL, the susceptibilities of SL9/Nef, SL9x/Nef, and SL9x/NefSL9 to inhibition by SL9-specific CTL were assessed in a coculture system measuring the effect of CTL on viral replication and spread (Fig. 4). This system was chosen because standard CTL assays such as chromium release and Elispot only measure CTL triggering at a single point in time, while this assay reflects the net effect of CTL over exposure during the entire life cycle of HIV-1. At a low effector-to-target cell ratio (1:8, chosen to allow inefficient baseline suppression), the CTL were mildly inhibitory against the wild-type virus, decreasing replication by less than 1 log10 unit. Against the SL9x/Nef virus, minimal inhibition was observed, indicating that the mutation in the SL9 within Gag markedly reduced the recognition of the epitope by the CTL. The SL9x/NefSL9 virus, however, was highly sensitive to the CTL, with replication suppressed by up to 5 log10 units. These results strongly suggested that earlier expression of the SL9 epitope within Nef boosts the antiviral efficiency of the CTL compared to the usually later expression of SL9 in Gag.
FIG. 4.
Enhancement of antiviral activity of SL9-specific CTL by expression of epitope in the nef reading frame. HIV-1 containing combinations of unmodified or translocated SL9 was tested for its inhibition by SL9-specific CTL. Target cells acutely infected with the indicated viruses were cocultured with an SL9-specific CTL clone at an effector-to-target cell ratio of 1:8 (a condition in which suppression of the wild-type virus was inefficient). (A to C) Viral replication in the presence or absence of CTL is shown as p24 antigen production over time. (D) CTL-mediated suppression of replication in log10 units at each time point is plotted. All data are given as the mean of triplicates, and error bars represent one standard deviation. This experiment is representative of three independent experiments.
The Nef containing the Gag SL9 epitope loses its ability to downregulate the class I molecule.
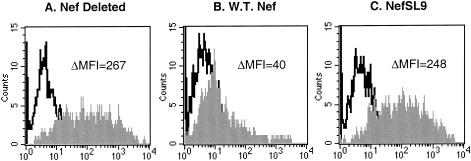
Among the many functions ascribed to Nef, downregulation of the major histocompatibility complex class I molecule (MHC-I) has been shown to interfere with the activity of CTL against HIV-1. Although the C terminus of Nef contains no sequences reported to affect MHC-I downregulation, CD4 downregulation, or cell signaling (14, 18, 22), we checked whether the transposition of SL9 into Nef might interfere. The effect of the SL9 epitope translocation on MHC-I was evaluated with reporter HIV-1 constructs (Fig. 5), to investigate the possibility that loss of this function contributed to increased CTL efficiency. Unexpectedly, this change completely abolished the ability of Nef to downregulate MHC-I. Cells infected with HIV-1 with truncated Nef expressed high levels of HLA A2 (Fig. 5A), whereas those infected with virus containing unmodified Nef were markedly reduced for expression (Fig. 5B). HIV-1 with NefSL9 downregulated MHC to the same extent as Nef-truncated virus (Fig. 5C). These results indicated that HIV-1 with the translocated SL9 epitope in Nef completely loses Nef-mediated MHC-I downregulatory activity, and therefore opened the possibility of comparatively examining the impacts of both kinetic and MHC-I regulation on the antiviral activity of CTL.
FIG. 5.
MHC-I downregulation in cells infected by virus containing the NefSL9 constuct. T1 cells were acutely infected with virus from which Nef was deleted (A), wild-type virus (B), or virus containing the NefSL9 mutation (C), each containing a reporter gene in the vpr reading frame. Flow cytometry was performed and infected cells were identified by gating on the reporter; histograms demonstrating the expression of the HLA on infected cells are shown. Background isotype antibody staining is shown in the open histograms, and specific HLA A2 staining is shown in the shaded histograms; the net fluorescence intensity (above isotype) is indicated. These data are representative of two independent experiments.
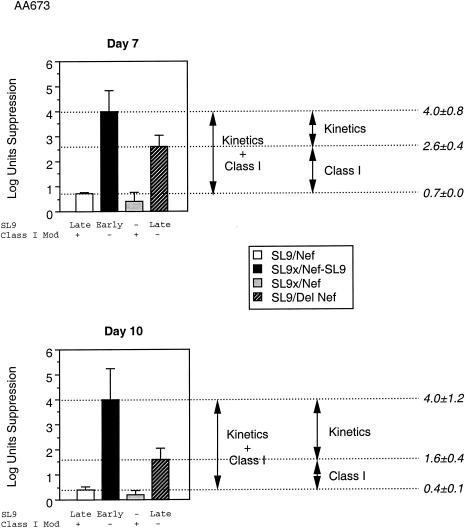
MHC-I downregulation and epitope expression kinetics both affect the antiviral activity of CTL.
To evaluate simultaneously the components of epitope kinetics and MHC-I regulation in the observed difference in antiviral activity of CTL, we tested a panel of HIV-1 mutants that allowed controlled comparisons of MHC-I and epitope kinetic effects (Fig. 6). These mutants differed in kinetics of SL9 expression and/or downregulation of MHC-I. Virus SL9/Nef (wild-type) expressed SL9 in the later Gag protein and downregulated MHC-I. Virus SL9x/NefSL9 expressed SL9 in the earlier Nef protein but did not downregulate MHC-I. Virus SL9/ΔNef expressed SL9 in the later Gag protein but did not downregulate MHC-I. Comparisons of: SL9/Nef versus SL9/ΔNef allowed evaluation of MHC-I downregulation effects, SL9/ΔNef versus SL9x/NefSL9 allowed evaluation of epitope kinetic effects, and SL9/Nef versus SL9x/NefSL9 allowed evaluation of both factors in combination. The results demonstrated that both factors play an important role in determining the antiviral efficiency of CTL. Consistent with a previous study (36), Nef exerted a 10- to 100-fold effect in this system. Epitope kinetics also appeared to exert a 10- to 100-fold change in the antiviral efficiency of CTL recognizing the SL9-epitope. Thus, the timing of epitope expression is a crucial determinant of CTL efficiency, similar to that of the MHC-I modulation by Nef.
FIG. 6.
Antiviral activity of SL9-specific CTL against virus containing the Nef-SL9 construct or Nef deletion. Suppression of replication of a panel of HIV-1 mutants was assessed as in Fig. 4 above. Two time points are plotted. The status of SL9 expression and MHC-I downregulatory function is indicated below the bar for each virus. These data are representative of two independent experiments.
DISCUSSION
This study demonstrates that specific properties of epitopes may have an extreme influence on the ability of CTL to control HIV-1 replication. Previous studies strongly suggested that expression levels of proteins containing CTL epitopes affects the ability of those CTL to recognize infected cells (31). Tsomides et al. found that chronically infected cells present an RT epitope an average of 17 copies per cell surface, compared to approximately 200 per cell surface for the SL9 Gag epitope. Both Tsomides et al. and Yang et al. further demonstrated that CTL recognizing these epitopes differ in their ability to kill infected cells, and Yang et al. (34) found that these CTL differ by orders of magnitude in their ability to inhibit viral replication (35). However, despite these marked antiviral differences between Gag- and RT-specific CTL, the clones studied were equivalent in their ability to kill exogenously peptide-loaded target cells (34). These findings therefore indicated that the antiviral efficiency of CTL depends on the quantity of protein expression within infected cells. The present data address the further impact of the kinetics of protein expression.
Those earlier reports on SL9-specific CTL by Tsomides et al. and Yang et al. found that the SL9 epitope is expressed in quantity on infected cells to allow 100% efficiency of infected cell lysis, therefore likely in excess for CTL recognition. Here we find, however, that the antiviral efficiency of SL9-specific CTL can be yet further increased by changing the kinetics of SL9 epitope expression to match those of Nef rather than Gag. Transposition of the epitope into an early protein therefore augments the antiviral function of CTL, suggesting that differences in epitope kinetics may have a profound influence on the ability of CTL to act upon acutely infected cells.
A similar approach was used by van Baalen et al. (3), who translocated an RT epitope into Nef. This study found that HIV-1 with the translocated epitope was more efficiently recognized than the wild-type virus, and concluded that the earlier kinetics of epitope expression resulted in enhanced antiviral efficiency of the RT-specific CTL. Potential caveats of this study, however, were the effects of enhanced expression of the RT epitope (in the light of the low levels of RT expression compared to Gag and Nef), and potential effects of epitope translocation on the MHC-I regulatory effects of Nef. The results of our current study address these caveats, confirming and extending the results.
Although we cannot entirely exclude that translocation of SL9 into Nef mediated this effect by quantitative enhancement of epitope expression, several facts argue against this. The prior findings that SL9 epitope presentation by acutely infected cells allows highly efficient recognition by CTL indicate that expression of SL9 is already in excess (31, 34). Furthermore, we cut SL9 expression by Gag in the SL9-translocated virus by altering an HLA A2-binding anchor amino acid, resulting in a single copy of wild-type SL9 in the SL9-translocated virus. Presumably, because Gag is the most plentifully expressed viral protein in infected cells (9), we did not increase the overall level of SL9 epitope expression. It is possible that we somehow enhanced the efficiency of epitope processing, although the SL9-Nef construct contained the same two amino acids immediately upstream of the epitope, and proteasome processing is dependent on the amino acids flanking the epitope (10, 19, 20, 24, 29). Finally, when we tested virus containing SL9 in both Gag and Nef, the antiviral activity of the SL9-specific CTL was not noticeably enhanced compared to virus containing SL9 in Nef alone. These points all suggest that increased SL9 expression is not the mechanism of improved antiviral activity against HIV-1 with the Nef-SL9 translocation.
We therefore propose that the earlier expression of SL9 after translocation into Nef contributes to increased antiviral efficiency of SL9-specific CTL. The replicative cycle of HIV-1 is rapid, with a generation time estimated to be only one to two days (15, 32). This probably poses a considerable kinetic challenge to CTL, which must kill an infected cell before the first virion is released to stop replication. Previously it has been shown that Gag-, RT-, and Env-specific CTL can kill acutely infected cells approximately 12 h before peak virion production (34), indicating a tight kinetic race. Nef is an early protein that is expressed several hours before the later protein Gag (25), and therefore Nef epitopes would be expected to appear hours earlier on the cell surface for CTL recognition. This could therefore provide Nef-specific CTL a significant kinetic edge that translates to enhanced viral inhibition.
Surprisingly, SL9 translocation into the tail of Nef destroyed its activity in downregulating MHC-I. This was unexpected because this region of Nef has not been described to play a role in any of its reported functions. We used this finding to our advantage to test the relative contributions of earlier epitope expression and MHC-I downregulation by Nef to the antiviral activity of CTL. It was clear that epitope kinetics play an important role, when comparing Nef-defective viruses expressing SL9 late or early. It was also clear that MHC-I downregulation plays a role as well, when comparing Nef-defective to Nef-competent viruses both expressing SL9 late, in agreement with prior findings. Within the variability of the assays (as reflected by the standard deviations), the contributions of earlier epitope expression and MHC-I downregulation appeared roughly equivalent. These controlled comparisons therefore suggested that epitope kinetics (later in Gag versus earlier in Nef) and MHC-I modulation (reduction by Nef versus no reduction without Nef) could have similar impacts on CTL efficiency.
Because viral suppression in the context of disrupted Nef is still increased by earlier epitope expression (virus with SL9 in Nef versus virus with Nef deletion), the influence of epitope kinetics occurs independently of Nef-mediated downregulation of class I by HIV-1. These data suggest that the effects of epitope kinetics and class I modulation are similar in magnitude when compared in isolation if it is assumed that these effects occur independently. It is possible, however, that this assumption leads to a significant underestimation of the effects of epitope kinetics in the physiologic situation, because earlier epitope expression in the face of normal Nef function may have the added benefit of allowing sufficient epitope presentation before Nef-mediated class I downmodulation. Thus, in the context of normal Nef function, it could be that CTL recognition of early (Nef, Tat, and Rev) versus late epitopes (Gag, Pol, and Env) not only allows the advantage of better viral clearance by earlier killing, but also more efficient cell clearance by allowing recognition before class I downmodulation. Whether this is the case will require further experimental investigation.
Our findings on epitope kinetics could relate to some interesting findings in vivo. In the SIV model, CTL recognizing the early proteins Tat and Nef appear to pressure SIV consistently to escape by mutation, whereas CTL directed against the late protein Gag do not (2, 21). This has been interpreted to suggest that the early protein CTL exert more selective pressure against the virus. Human data are far more limited and difficult to interpret, but CTL in early infection more commonly target Nef, as opposed to Gag in later infection (13), and studies of CTL escape mutation seem to have more consistently demonstrated rapid escape in early infection (4, 23) than late (6). These findings are consistent with our experimental data suggesting that early proteins may be easier targets for CTL (37).
These results indicate that CTL may differ dramatically in their ability to control HIV-1 replication depending on their epitope specificity, due to epitope kinetics. The timing of epitope expression is a key determinant of how well CTL can inhibit HIV-1 production by infected cells. Our data highlight the caveats of utilizing measurements of CTL reactivity against synthetic or recombinant antigens as “correlates of immunity” in immunopathogenesis and vaccine studies (33). It is likely that there are numerous epitope-dependent determinants of the efficacy of HIV-1-specific CTL, including the quantity and kinetics of epitope expression. These likely include factors such as viral protein expression, efficiency of epitope production (processing and transport) and class I binding, T-cell receptor avidity for the epitope/MHC-I complex, tolerance for epitope changes, and fitness costs for epitope mutation and escape. A deeper understanding of such issues is important for interpreting the immunopathogenic significance of CTL responses and possibly for the design and implementation of vaccines and immune-based therapies.
Acknowledgments
IL-2 was generously provided through the National Institutes of Health AIDS Research and Reference Reagent Repository. This study was funded by NIAID grants AI43203 and AI51970.
We thank C. Uittenbogaart, P. Krogstad, O. Martinez, and B. Jamieson for helpful critical reviews.
REFERENCES
- 1.Ali, A., B. D. Jamieson, and O. O. Yang. 2003. Half-genome human immunodeficiency virus type 1 constructs for rapid production of reporter viruses. J. Virol. Methods 110:137-142. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 3.Baalen, C. A., C. Guillon, M. van Baalen, E. J. Verschuren, P. H. Boers, A. D. Osterhaus, and R. A. Gruters. 2002. Impact of antigen expression kinetics on the effectiveness of HIV-specific cytotoxic T lymphocytes. Eur. J. Immunol. 32:2644-2652. [DOI] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Brander, C., and P. J. R. Goulder. 2000. The evolving field of HIV CTL epitope mapping: New approaches to the identification of novel epitopes, p. I1-19. In B. T. M. Korber, C. Brander, B. F. Haynes, R. Koup, C. Kuiken, J. P. Moore, B. D. Walker, and D. Watkins (ed.), HIV molecular immunology database, vol. 2000. Los Alamos National Laboratory: Theoretical Biology and Biophysics, Los Alamos, N.Mex.
- 6.Brander, C., K. E. Hartman, A. K. Trocha, N. G. Jones, R. P. Johnson, B. Korber, P. Wentworth, S. P. Buchbinder, S. Wolinsky, B. D. Walker, and S. A. Kalams. 1998. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J. Clin. Investig. 101:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brooks, D. G., S. G. Kitchen, C. M. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7:459-464. [DOI] [PubMed] [Google Scholar]
- 8.Collins, K. L., B. K. Chen, S. A. Kalams, B. D. Walker, and D. Baltimore. 1998. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature 391:397-401. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, B. R. 1995. Regulation of HIV gene expression. AIDS 9:S19-S32. [PubMed] [Google Scholar]
- 10.Eggers, M., B. Boes-Fabian, T. Ruppert, P. M. Kloetzel, and U. H. Koszinowski. 1995. The cleavage preference of the proteasome governs the yield of antigenic peptides. J. Exp. Med. 182:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., J. M. Orenstein, A. J. Buckler-White, and M. A. Martin. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J. Virol. 68:5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:343-350. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, M. M. Addo, S. He, J. S. Mukherjee, M. N. Phillips, M. Bunce, S. A. Kalams, R. P. Sekaly, B. D. Walker, and C. Brander. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg, M. E., A. J. Iafrate, and J. Skowronski. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 17:2777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 16.Jamieson, B. D., and J. A. Zack. 1998. In vivo pathogenesis of a human immunodeficiency virus type 1 reporter virus. J. Virol. 72:6520-6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, V. A., and B. D. Walker. 1990. HIV-infected cell fusion assay, p. 92-94. In B. D. Walker (ed.), Techniques in HIV research. Stockton Press, New York, N.Y.
- 18.Mangasarian, A., V. Piguet, J. K. Wang, Y. L. Chen, and D. Trono. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J. Virol. 73:1964-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miconnet, I., C. Servis, J. C. Cerottini, P. Romero, and F. Levy. 2000. Amino acid identity and/or position determines the proteasomal cleavage of the HLA-A*0201-restricted peptide tumor antigen MAGE-3271-279. J. Biol. Chem. 275:26892-26897. [DOI] [PubMed] [Google Scholar]
- 20.Mo, A. X., S. F. van Lelyveld, A. Craiu, and K. L. Rock. 2000. Sequences that flank subdominant and cryptic epitopes influence the proteolytic generation of MHC class I-presented peptides. J. Immunol. 164:4003-4010. [DOI] [PubMed] [Google Scholar]
- 21.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horten, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 22.Piguet, V., and D. Trono. 1999. A structure-function analysis of the nef protein of primate lentiviruses, p. 448-459. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), Human retroviruses and AIDS 1999. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 23.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rammensee, H. G., T. Friede, and S. Stevanoviic. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178-228. [DOI] [PubMed] [Google Scholar]
- 25.Ranki, A., A. Lagerstedt, V. Ovod, E. Aavik, and K. J. Krohn. 1994. Expression kinetics and subcellular localization of HIV-1 regulatory proteins Nef, Tat and Rev in acutely and chronically infected lymphoid cell lines. Arch. Virol. 139:365-378. [DOI] [PubMed] [Google Scholar]
- 26.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 27.Salter, R. D., D. N. Howell, and P. Cresswell. 1985. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21:235-246. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz, O., V. Marechal, S. Le Gall, F. Lemonnier, and J. M. Heard. 1996. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat. Med. 2:338-342. [DOI] [PubMed] [Google Scholar]
- 29.Toes, R. E., A. K. Nussbaum, S. Degermann, M. Schirle, N. P. Emmerich, M. Kraft, C. Laplace, A. Zwinderman, T. P. Dick, J. Muller, B. Schonfisch, C. Schmid, H. J. Fehling, S. Stevanovic, H. G. Rammensee, and H. Schild. 2001. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J. Exp. Med. 194:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomiyama, H., H. Akari, A. Adachi, and M. Takiguchi. 2002. Different effects of Nef-mediated HLA class I down-regulation on human immunodeficiency virus type 1-specific CD8+ T-cell cytolytic activity and cytokine production. J. Virol. 76:7535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsomides, T. J., A. Aldovini, R. P. Johnson, B. D. Walker, R. A. Young, and H. N. Eisen. 1994. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J. Exp. Med. 180:1283-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, et al. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 33.Yang, O. O. 2003. Will we be able to ‘spot’ an effective HIV-1 vaccine? Trends Immunol. 24:67-72. [DOI] [PubMed] [Google Scholar]
- 34.Yang, O. O., S. A. Kalams, M. Rosenzweig, A. Trocha, N. Jones, M. Koziel, B. D. Walker, and R. P. Johnson. 1996. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J. Virol. 70:5799-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, O. O., S. A. Kalams, A. Trocha, H. Cao, A. Luster, R. P. Johnson, and B. D. Walker. 1997. Suppression of human immunodeficiency virus type 1 replication by CD8+ cells: evidence for HLA class I-restricted triggering of cytolytic and noncytolytic mechanisms. J. Virol. 71:3120-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, O. O., P. T. Nguyen, S. A. Kalams, T. Dorfman, H. G. Gottlinger, S. Stewart, I. S. Chen, S. Threlkeld, and B. D. Walker. 2002. Nef-mediated resistance of human immunodeficiency virus type 1 to antiviral cytotoxic T lymphocytes. J. Virol. 76:1626-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, O. O., P. T. Sarkis, A. Trocha, S. A. Kalams, R. P. Johnson, and B. D. Walker. 2003. Impacts of avidity and specificity on the antiviral efficiency of HIV-1-specific CTL. J. Immunol. 171:3718-3724. [DOI] [PubMed] [Google Scholar]