Abstract
Retrovirus assembly and budding involve a highly dynamic and concerted interaction of viral and cellular proteins. Previous studies have shown that retroviral Gag proteins interact with actin filaments, but the significance of these interactions remains to be defined. Using equine infectious anemia virus (EIAV), we now demonstrate differential effects of cellular actin dynamics at distinct stages of retrovirus assembly and budding. First, virion production was reduced when EIAV-infected cells were treated with phallacidin, a cell-permeable reagent that stabilizes actin filaments by slowing down their depolymerization. Confocal microscopy confirmed that the inhibition of EIAV production correlated temporally over several days with the incorporation dynamics of phallacidin into the actin cytoskeleton. Although the overall structure of the actin cytoskeleton and expression of viral protein appeared to be unaffected, phallacidin treatment dramatically reduced the amount of full-length Gag protein associated with the actin cytoskeleton. These data suggest that an association of full-length Gag proteins with de novo actin filaments might contribute to Gag assembly and budding. On the other hand, virion production was enhanced when EIAV-infected cells were incubated briefly (2 h) with the actin-depolymerizing drugs cytochalasin D and latrunculin B. Interestingly, the enhanced virion production induced by cytochalasin D required a functional late (L) domain, either the EIAV YPDL L-domain or the proline-rich L domains derived from human immunodeficiency virus type 1 or Rous sarcoma virus, respectively. Thus, depolymerization of actin filaments may be a common function mediated by retrovirus L domains during late stages of viral budding. Taken together, these observations indicate that dynamic actin polymerization and depolymerization may be associated with different stages of viral production.
Equine infectious anemia virus (EIAV) is a member of the lentivirus subfamily of retroviruses that also includes human immunodeficiency virus type 1 (HIV-1). Like all retroviruses, EIAV produces three predominant polyprotein precursors in the infected cells, Gag, Gag-Pol, and Env, which form the structural and enzymatic proteins of the viral particle (18). A general model for retrovirus budding was proposed 25 years ago (5); however, viral and cellular components and processes that mediate virion assembly and budding remain poorly defined.
The Gag polyprotein of retroviruses is sufficient to direct retroviral assembly and budding processes. Several functional domains within the Gag polyprotein have been identified that perform different roles during virus assembly. For example, N-terminal myristylation of the MA protein is important for targeting the Gag polyprotein to the plasma membrane (M domain) (6, 23). The CA interaction (I) domain plays a role in the multimerization of Gag proteins (8, 17). NC motifs participate in genomic RNA incorporation, and various Gag late assembly (L) domains are critical for the release of budding virions from the plasma membrane (1, 19, 49, 59). The L-domain function of Gag (22, 49, 58) is particularly interesting since mutant retroviruses with a deficient L domain produce viral particles that remain tethered to the plasma membranes of infected cells, suggesting a critical function of the L domain for virion-cell separation (22). To date, three types of L domain have been identified from various retroviruses. Rous sarcoma virus (RSV) (58) and other enveloped viruses (11, 24, 27, 60) use a PPPY motif, whereas HIV-1, HIV-2, and SIV utilize a PTAP motif (22). In contrast to these proline-rich L domains, EIAV utilizes a YPDL motif (49). Despite these different sequences, both PPPY and PTAP L domains are able to replace the functions of the YPDL L domain in supporting EIAV replication (37), indicating that these L domains might access a common pathway through different portals.
Recent studies on retrovirus assembly and budding have revealed several cellular proteins involved in the budding process (20, 50, 56, 62). Previously, we reported that adaptor protein complex AP-2, a major component of the endocytic pathway, is recruited to the budding sites of EIAV-infected cells (50), suggesting that EIAV adapts cellular endocytic machinery to facilitate the budding process. Recently, Tsg101 has been demonstrated to interact with the HIV-1 PTAP L domain to mediate HIV-1 budding (13, 20, 56). Tsg101 is a subunit of the ESCRT-I complex (for endosomal sorting complex required for transport I), which functions in the multivesicular body (MVB) sorting pathway (30). Recruitment of early (AP-2) and late (ESCRT-I) endocytic machinery by various retroviral L domains indicates that these cellular functions might have been adapted for retrovirus budding, although their precise functions in mediating retrovirus release remain to be defined.
Limited evidence that the dynamic cellular actin network participates in retroviral replication has also been reported (7, 26). For example, actin and actin-binding proteins have been identified within highly purified HIV-1 virions (44-46) and other retroviruses (12), presumably as a result of specific viral-cellular protein interactions. Direct interactions between retroviral Gag and actin filaments have been shown in cells infected with murine leukemia virus (15, 16) or HIV-1 (26, 40, 52). Extensive depolymerization of cellular actin filaments by cytochalasin D (cytoD), an actin-disrupting drug, in murine mammary tumor virus or HIV-1-infected cells results in a 50% reduction in virion production (41, 55), suggesting that an intact actin cytoskeleton is essential for retrovirus assembly and budding (48). However, the precise role of a dynamic cellular actin network in the various steps of retroviral replication remains to be defined.
To gain more insight into the function of the actin cytoskeleton in retrovirus assembly and budding, we designed a series of experiments to examine the effects of various actin filament-modulating agents on EIAV virion production. These studies provide novel insights into the cell biology of retroviral budding and demonstrate for the first time distinct roles for actin at various stages of EIAV assembly.
MATERIALS AND METHODS
Reagents and cell lines.
All actin-modulating reagents were purchased from Sigma (St. Louis, Mo.) or Calbiochem (San Diego, Calif.). These compounds included the actin filament-depolymerizing drugs cytoD and latrunculin B (latB), the microtubule-depolymerizing drug nocodazole, and the actin filament-stabilizing drug phallacidin. Stock solutions were prepared in dimethyl sulfoxide and diluted >500-fold in culture medium for assays.
Equine dermal (ED) cells were obtained from the American Type Culture Collection (catalog no. CCL 57), Rockville, Md., and cultured as described previously (9). EIAV chronically infected ED cells were generated by transfecting ED cells with infectious molecular EIAVuk provirus (10), followed by several rounds of passage in the culture medium (9). Isolation and infection of primary equine blood monocyte-derived macrophage (MDM) cells were previously described (37, 51).
Phallacidin treatment on EIAV-infected ED cells.
EIAV-infected ED cells grown on six-well plates at 80 to 90% confluence were incubated with the culture medium (9) containing 25 μM phallacidin for 20 h at 37°C. The media were then collected and replaced with drug-free medium daily. Virion production was measured by determining the reverse transcriptase (RT) activity of viral particles released into the culture medium.
RT assay.
The extracellular RT activity in clarified media or associated with virions pelleted by ultracentrifugation from clarified culture medium was measured to determine the levels of virion production as described previously (39). Duplicate samples were tested in each experiment, and the Student t test method was used to evaluate statistical significance of the mean value.
Fluorescence microscopy.
To examine incorporation of phallacidin into actin filaments in live cells, EIAV-infected ED cells grown on eight-well Lab-Tek chambered coverglass slips at 50 to 60% confluence were incubated with the culture medium containing 25 μM BODIPY FL phallacidin (Molecular Probes, Eugene, Oreg.) at 37°C for 20 h. The extracellular fluorescent phallacidin was then removed by several washes, and the cells were observed daily for 5 days by using a Leica TCS NT confocal microscope to monitor drug incorporation into actin filaments.
To examine effects of actin-modulating drugs on actin filaments in EIAV-infected ED cells, drug-treated cells grown on a coverglass were fixed and permeabilized, and the cellular actin cytoskeleton was stained by BODIPY FL phallacidin according to the manufacturer's recommendations. The images of stained filamentous actin network were recorded by using a Leica TCS NT confocal microscope.
EIAV Gag protein expression assays.
To examine the general expression levels of viral proteins, both treated and untreated EIAV-infected cells grown on six-well plates were dissolved in 250 μl of lysis buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% deoxycholic acid, 1% Triton X-100, protease inhibitor cocktail, 0.1% sodium dodecyl sulfate [SDS]). The cellular lysates were then resolved by electrophoresis through a 4 to 15% gradient gel (Bio-Rad, Hercules, Calif.) and immunoblotted with a reference immune serum from a naturally infected horse (42). Horseradish peroxidase-conjugated goat anti-horse immunoglobulin G F(ab′)2 (Jackson ImmunoResearch, West Grove, Pa.) was used as the secondary antibody. The immunoblots were developed by incubation with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, Ill.). The amounts of viral proteins were digitally quantified by using a Kodak imaging station model 1000. In addition, viral particles pelleted from clarified supernatant were examined similarly by SDS-polyacrylamide gel electrophoresis (PAGE) to analyze protein composition and processing. Mouse monoclonal antiactin antibody purchased from Sigma was used to normalize cellular contents of actin proteins.
Fractionation of cells by Triton X-100 treatment.
To selectively isolate viral proteins associated with the actin cytoskeleton, EIAV-infected ED cells grown on six-well plates were incubated with 1% Triton X-100 in phosphate-buffered saline with 0.5 mM MgCl2 at room temperature for 30 min as previously described (25). The supernatant containing monomeric actin and other solubilized cellular components was collected and designated the soluble fraction. The residual portion containing the actin cytoskeleton and its associated proteins was solubilized with lysis buffer and spun at 20,800 × g for 2 min to remove nuclei. The postnuclear supernatant was designated the insoluble fraction. Viral proteins from both fractions were analyzed by SDS-PAGE and immunoblotting.
DNA mutagenesis.
To generate stably transduced producer ED cells, a neomycin-resistant gene from pRC/CMV (catalog no. V750SG; Invitrogen, Carlsbad, Calif.) was inserted into the proviral cmvEIAVuk plasmid to provide a selection marker. Briefly, the BamHI/EcoRI fragment of pRC/CMV containing the neomycin resistance gene with simian virus 40 promoter and polyadenylation signal was isolated and filled in with Klenow polymerase to a generate blunt-ended DNA fragment. This DNA fragment was then ligated to the blunt-ended cmvEIAVuk proviral DNA (9) that had been linearized by EcoRI digestion, filled in by Klenow polymerase, and dephosphorylated by calf intestinal alkaline phosphatase.
Two groups of proviruses were generated. The L-domain-positive group included the parental cmvEIAVuk provirus and cmvEIAVuk with either a PTAP or a PPPY L domain, all of which contain a functional L domain (37). The L-domain-negative group contained cmvEIAVuk proviruses expressing a mutated YPDL motif or truncated p9 proteins (Q3 and L22, designating the site of the engineered termination codon) with the YPDL motif deleted (9). All plasmid DNAs were isolated by using the Qiagen Midiprep kit (Qiagen, Valencia, Calif.), and the mutations were confirmed by DNA sequencing. Cos-1 cells are not susceptible to EIAV infection, so there is only a single round of viral production, and no reinfection.
Generation of stably transduced producers.
ED cells were transfected with cmvEIAVuk proviral plasmids containing the neomycin resistance gene by using GenePorter 2 (Gene Therapy Systems, San Diego, Calif.) according to the manufacturer's recommendations. The transfected cells were cultured without selection for 3 days prior to the addition of 600 μg of G418 (Geneticin [catalog no. 11811]; Gibco-BRL, Carlsbad, Calif.)/ml. The G418-selected ED cells stably producing EIAV were cultured continuously under antibiotic selection.
RESULTS
Incorporation of fluorescent phallacidin into filamentous actin of living cells.
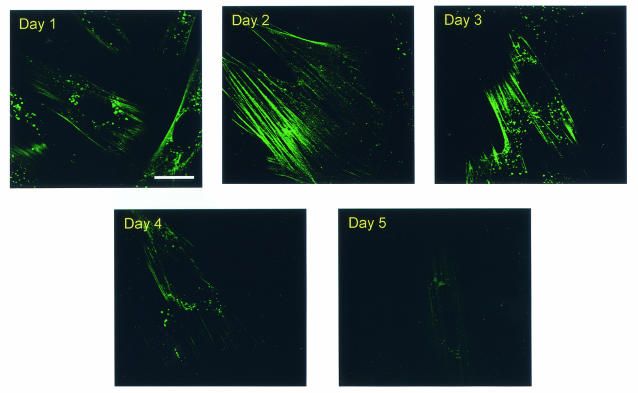
To evaluate the role of cellular actin dynamics in retroviral budding, we initially evaluated the effects of stabilizing the actin cytoskeleton on EIAV virion production. Phallacidin was chosen for this purpose because it is reported to bind F-actin with high affinity and to stabilize the F-actin cytoskeleton by preventing depolymerization (54). Moreover, phallacidin and its derivatives are able to enter live cells with the negligible cytotoxicity (3, 4, 31, 57). We first examined the kinetics of phallacidin integration into actin filaments in living EIAV-infected cells by using fluorescent BODIPY FL phallacidin. EIAVuk-infected ED cells were incubated for 20 h with 25 μM BODIPY FL phallacidin. The medium was then replaced with fresh culture medium to remove extracellular fluorescent phallacidin, and the fluorescent staining of cellular actin filaments in live cells was observed daily over a 5-day period by confocal microscopy (Fig. 1). These data indicated that staining of cellular actin filaments was clearly observed upon the removal of phallacidin (day 1 in Fig. 1). Interestingly, the intensity of filamentous staining appeared to increase at 2 and 3 days posttreatment and then decrease by days 4 and 5. Cellular filamentous actin staining by fluorescent phallacidin was relatively homogeneous in the cell cultures. These data confirmed that phallacidin is able to enter live ED cells and to incorporate into filamentous actin. Although the phallacidin-treated cells multiplied slightly slower than the untreated controls, cell division was not completely arrested during the experimental period. More importantly, these data demonstrated a distinct transient integration of phallacidin into cellular actin filaments in living EIAV-infected ED cells over a 5-day period.
FIG. 1.
Staining of cellular actin cytoskeleton in live EIAV-infected ED cells by fluorescent phallacidin. EIAVuk-infected ED cells grown on Lab-Tek chambers were incubated at 37°C for 20 h with 25 μM BODIPY FL phallacidin. The fluorescent drugs were then replaced with drug-free culture medium. Images of fluorescent actin filaments were captured daily over a 5-day period by using Leica confocal microscope and identical imaging conditions. Bar, 15 μm.
Since dynamic actin cytoskeleton is essential for normal cellular function, we next examined the effects of internalized phallacidin on cell viability using the standard trypan blue exclusion assay. Notably, the phallacidin-treated cells remained ca. 90% viable over the 10 days of the experiment period and displayed normal morphology under a microscope, confirming the previous report that phallacidin internalized by live cells produced no significant cell toxicity (4).
Effects of phallacidin treatment on EIAV production.
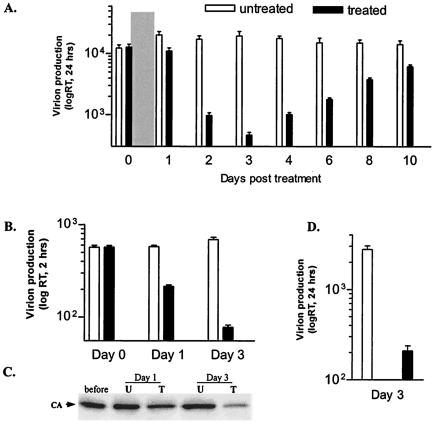
To examine the effects of stabilized actin cytoskeleton on EIAV replication, EIAVuk-infected ED cells were first incubated with 25 μM phallacidin for 20 h, at which time the culture medium was collected and replaced with phallacidin-free medium daily. Extracellular RT activity associated with EIAV virion in the culture medium was measured to quantify daily EIAV production. As summarized in Fig. 2A, virion production was reduced by 40% after 20 h of drug treatment (day 1) compared to an untreated control cell culture. Subsequently, daily virion production continued to decrease and reached its lowest level, ∼20-fold reduction, at day 3. Virion production then steadily increased over the next 4 days, ultimately returning to the level of untreated control (Fig. 2A). These data revealed that the kinetics of EIAV virion production in phallacidin-treated cells correlated temporally with the dynamics of phallacidin incorporation into and dissociation from cellular actin filaments (cf. Fig. 1).
FIG. 2.
Effects of phallacidin treatment on EIAV production. (A) Daily EIAV production from phallacidin-treated (▪) and -untreated (□) EIAVuk-infected ED cells. Parallel cell cultures grown on a six-well plate were incubated with 25 μM phallacidin at 37°C for 20 h (indicated by the gray bar), at which time drug-free culture medium was added and replaced daily. Virion production during each 24-h interval was assayed by determining the RT activity of virions released into the culture medium. (B) EIAV production at 2-h intervals. Parallel cells were grown and treated as for panel A except that the culture medium used for RT assay was collected at 2-h intervals. (C) Western blot assay showing EIAV production at the indicated time points. EIAV particles pelleted from equal amounts of culture medium (see panel B) at the indicated time points were resolved through a 4 to 15% SDS-PAGE gel and probed by the reference serum. The intensity of the virus-specific capsid protein was used to quantify virion production. (D) EIAV production from infected equine macrophages at the maximal inhibition time (day 3). The data represented the mean values of at least three independent experiments done in duplicate.
We also measured virion production over a shorter time period (2 h) to confirm the inhibitory effect of phallacidin on EIAV production (Fig. 2B and C). EIAV-infected cells were treated with phallacidin as described above except the culture medium was replaced and assayed at 2-h intervals. Virion production levels at three time points are shown in Fig. 2B. The parallel cell cultures produced similar amounts of virions before phallacidin treatment (Fig. 2B, day 0). After 20 h of incubation and removal of phallacidin (day 1), EIAV produced from phallacidin-treated cells over a 2-h period was reduced by 60%. At 3 days posttreatment, an ∼20-fold reduction of RT activity was observed compared to the untreated control (Fig. 2B). EIAV particles released into the medium were also collected by centrifugation and examined by Western blot with the EIAV immune serum (42). Figure 2C demonstrates the amount of the major viral protein in mature virions found in supernatant at the indicated time points. Compared to untreated control cells, there is a >10-fold reduction in virion production at day 3, which parallels the results of RT assays.
A potential concern with any drug treatment of cells is the specificity of the observed effect under the experimental conditions. To verify the specificity of phallacidin on EIAV budding, we repeated the phallacidin treatment on EIAV-infected equine macrophages, the natural targets for EIAV infection. Parallel cultures of equine MDMs were infected with EIAV by using a standard protocol (37) and treated as described in Fig. 2A. At the maximum inhibition time (day 3), an ∼15-fold reduction in virion production was detected (Fig. 2D). These data indicated that the suppression of EIAV production by phallacidin is not limited to the ED producer cells but is equally evident in the natural target cell for EIAV. These results demonstrate for the first time that stabilization of actin filaments by phallacidin markedly suppresses EIAV virion production, indicating a role for dynamic actin filaments in retroviral assembly and budding.
Effects of phallacidin treatment on viral protein expression and association with filamentous actin.
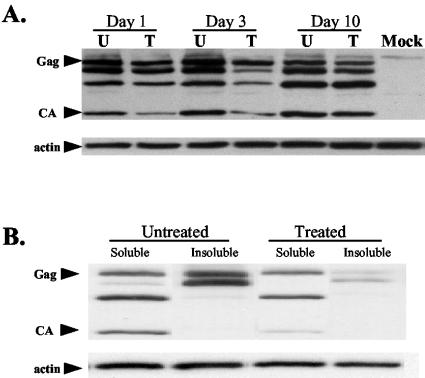
To study the mechanism by which the phallacidin-stabilized actin cytoskeleton inhibits virion production, we examined the expression level of viral proteins in phallacidin-treated and -untreated cells by Western blot (Fig. 3A). To identify EIAV-specific viral proteins, total cellular lysates at the indicated times were resolved through SDS-4 to 15% PAGE gels and immunoblotted with the EIAV immune serum (42). Cellular actin content, quantified with a monoclonal mouse anti-β-actin antibody, was used to normalize protein concentration in treated and untreated cells (Fig. 3A, lower panel). Specific EIAV proteins expressed in treated and untreated cells from three time points (days 1, 3, and 10) are shown in Fig. 3A. These data indicated a 15% reduction in EIAV protein expression at day 1, a 35% reduction at day 3, and no significant reduction at day 10 (Fig. 3A). Although the observed reduction in EIAV Gag protein expression in phallacidin-treated cells may contribute to the observed reduction in virion production, the 35% reduction in protein expression cannot explain the 10- to 20-fold reduction in virion production. In addition, it is important to note that full-length Gag polyproteins, processed intermediates, and mature capsid proteins were detected in all of the lysates, indicating that expression of EIAV proteins and protease processing were not evidently impaired by phallacidin treatment.
FIG. 3.
Effects of phallacidin on expression of EIAV proteins. (A) EIAV proteins in total cell lysates of treated (lanes T) and untreated (lanes U) cells at the indicated times. Parallel EIAV-infected ED cells were incubated with 25 μM phallacidin for 20 h and then cultured in the absence of drugs. Total cellular lysates of both treated and untreated cells at 1, 3, and 10 days posttreatment were analyzed by Western blotting to detect cell-associated EIAV proteins. Arrows indicate full-length Gag polyprotein and capsid proteins. The same blot was stripped and probed with an actin-specific antibody to normalize the amount of extract in each lane. Uninfected ED cells were used as the mock control. (B) Distribution of EIAV proteins in Triton X-100-soluble and -insoluble fractions at the maximum inhibition time (day 3). EIAV proteins associated with either fraction were detected as in panel A.
We next examined the effects of phallacidin treatment on the association of viral proteins with cellular actin filaments to evaluate potential roles of the actin cytoskeleton in Gag polyprotein assembly. For this purpose, cells were treated with Triton X-100, and soluble and insoluble fractions were collected (32, 35). The Triton X-100-solubilized fraction contained monomeric actin and cytosol proteins. The insoluble fraction contained the filamentous actin cytoskeleton and its associated proteins (see Materials and Methods). At the maximum inhibition time (day 3), EIAV-specific proteins associated with the soluble and insoluble fractions were analyzed by Western blotting, and representative data are presented in Fig. 3B. An approximately equal amount of actin was detected in the soluble and insoluble fractions from untreated cells, and phallacidin treatment appeared to have no significant effect on the distribution of actin proteins in both fractions. In untreated control cells, there were approximately equal amounts of Gag proteins in soluble and insoluble fractions. Phallacidin treatment did not significantly affect the amounts of Gag proteins present in the soluble fraction (Fig. 3B). In contrast, phallacidin treatment markedly reduced the amount of Gag proteins observed in the insoluble fraction. This result revealed that phallacidin treatment altered the ratio of insoluble Gag to soluble Gag such that <10% of total Gag remained associated with the insoluble fraction compared to 50% of Gag associated with the insoluble fraction in untreated control cells. Therefore, the 35% reduction of total Gag by phallacidin treatment was not evenly distributed to both soluble and insoluble fractions. Rather, the phallacidin treatment specifically reduced by ∼5-fold the amount of Gag associated with the insoluble fraction. Thus, the reduced virion production by phallacidin treatment predominantly correlated with the reduced amounts of Gag proteins associated with the actin cytoskeleton, suggesting that stabilized actin cytoskeleton impair viral assembly and budding process.
These assays also revealed a difference in the extent of proteolytic processing between actin-associated and free Gag polyproteins. Proteins associated with the actin cytoskeleton were mainly full-length Gag proteins; the processed Gag intermediates and mature viral proteins, such as capsid protein, were excluded from the insoluble fraction. These observations suggest that interaction between Gag polyproteins and actin filaments is lost upon cleavage of the polyprotein to mature virion proteins. The data also indicate that the Gag polyprotein-actin filament interaction is markedly reduced by stabilization of actin filaments, suggesting a role for dynamic actin in targeting Gag polyprotein to the budding sites on plasma membrane.
Effects of actin depolymerization on EIAV budding.
The preceding experiments indicated that specific interaction of Gag with actin filaments contributes to the EIAV assembly process. To evaluate further the involvement of actin cytoskeleton in EIAV budding, we also examined the effect of actin depolymerization on EIAV budding. Two cell-permeable specific actin-depolymerizing drugs, cytoD and latB, were used for this assay. Nocodazole, a cell-permeable drug that specifically disrupts microtubules but does not affect actin filaments (61), was also examined, since microtubules comprise another major component of the cell cytoskeleton that can be associated with microbial pathogen transport (21, 47).
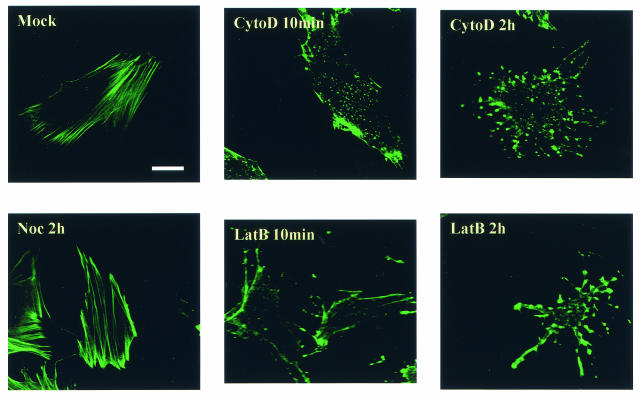
To confirm the specificities of these drugs, the cellular actin cytoskeleton was examined by fluorescence microscopy of EIAV-infected ED cells after various times of treatment with the respective drugs. Treated cells were fixed and permeabilized, and cellular filamentous actin was localized by BODOPY FL phallacidin staining and observed by confocal fluorescence microscopy (Fig. 4). In untreated ED cells (Fig. 4, Mock), well-organized actin filaments were clearly evident. In contrast, both cytoD and latB disrupted the majority of cellular filamentous actin after only a 10-min exposure to the drugs. After the cells were treated for 2 h, virtually all of the filamentous actin was depolymerized, as indicated by the punctuated BODIPY FL phallacidin staining. In contrast, the filamentous actin network was not affected by nocodazole treatment (Fig. 4, Noc). These data clearly demonstrate the rapid and specific effect of cytoD and latruculin B on actin filament disruption under the experimental conditions.
FIG. 4.
Actin cytoskeleton after treatment of EIAV-infected ED cells with various drugs. Cells grown on coverslips at 50% confluence were incubated with the indicated drugs for 10 min or 2 h and then fixed, permeabilized, and stained with BODIPY FL phallacidin to visualize cellular filamentous actin. The images were captured by a Leica confocal microscope. Bar, 15 μm. Mock, untreated cells; Noc, nocodazole.
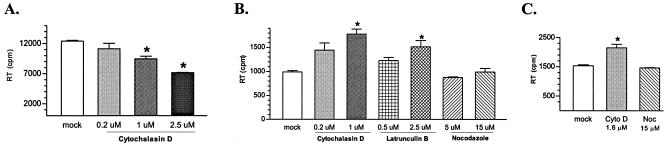
It has previously been reported that treatment of HIV-1-infected cells with 5 μM cytoD for 24 h reduced viral budding by ca. 50% (55). To test whether EIAV budding was also inhibited by long-term cytoD treatment, parallel cultures of ED cells chronically infected with EIAV were incubated with increasing concentrations of cytoD for 28 h, and viral production during this period was monitored by extracellular RT activity of the culture medium. EIAV production was reduced by cytoD incubation in a dose-dependent manner with an average of 50% reduction at 2.5 μM cytoD (Fig. 5A). This result is similar to that observed with HIV-1 and supports the hypothesis that actin polymerization participates in retroviral assembly and budding.
FIG. 5.
Effects of actin depolymerization on EIAV production. (A) Effects of long-term (28 h) actin depolymerization on EIAV production. Infected ED cells were incubated with increasing concentrations of cytoD for 28 h at 37°C. EIAV production was measured by RT activity in the culture medium. (B) Effects of short-term (2.5-h) actin depolymerization on EIAV budding. The infected ED cells were incubated with the different drugs at the indicated concentrations for 2.5 h at 37°C, and the level of EIAV production was determined by measuring the RT activity in the culture medium. (C) Effects of short-term actin depolymerization on EIAV production from infected MDM. Infected macrophages were treated with the indicated reagents for 2.5 h at 37°C, and EIAV production was evaluated. Each experiment was repeated independently at least three times in duplicate. The data represent the mean value with the standard deviation of the duplicates. The asterisks indicate that the difference between the mean value is statistically significant (P < 0.05) compared to the untreated control.
In light of the rapid kinetics of actin depolymerization by cytoD, we evaluated the effects of short-term cytoD treatment on EIAV budding. Parallel EIAV-infected ED cells were incubated with the increasing concentrations of cytoD for 2.5 h, and extracellular RT activity in the culture medium was quantified as a measure of virion production. Surprisingly, incubation with cytoD for 2.5 h stimulated virion production as indicated by increased RT activity (Fig. 5B). For example, virion production was increased by ca. 80% in cells treated with 1.0 μM cytoD compared to untreated cells. Increased virion production was also observed with viruses pelleted from cytoD-treated cell culture medium, as quantified by Western blotting (data not shown). These data suggest that brief depolymerization of actin filaments facilitates the release of budding particles.
To determine whether the increased viral budding induced by the brief cytoD treatment was a common effect of actin filament-disrupting drugs, we also tested latB, another reagent that specifically depolymerizes actin filaments (Fig. 5B). Again, a 50% increase in virion production was detected from cells treated with latB for 2.5 h, and the enhancement was dose dependent, confirming that short-term depolymerization of actin filaments facilitates viral budding. In contrast, nocodazole demonstrated no detectable effects on EIAV budding at standard treatment concentrations (5 to 15 μM) for 2.5 h; drug-treated and untreated cells produced similar levels of extracellular virus, as measured by RT activity (Fig. 5B).
Several studies have demonstrated that retroviral phenotypes related to replication and assembly can be highly cell dependent (14). To assess the relevance of the drug effects observed in ED cells, we repeated the cytoD treatment on primary equine MDM, the natural in vivo targets for EIAV infection (Fig. 5C). Virion production from cytoD-treated macrophages was increased by 50% compared to untreated MDM cultures. Nocodazole treatment caused no detectable changes in EIAV budding. These data clearly demonstrate the specific enhancement of EIAV budding by depolymerization of filamentous actin induced by cytoD or latB, further supporting a specific role for the actin network in EIAV budding.
Involvement of L-domain function in actin depolymerization-induced enhancement of virion production.
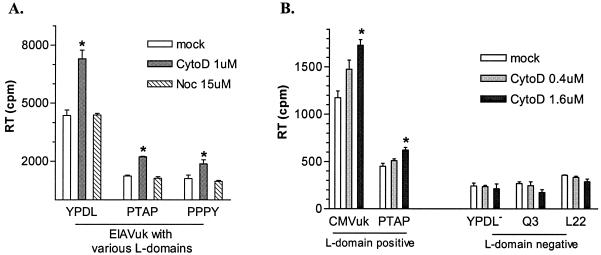
Based on the preceding results, we speculated that actin depolymerization might be essential during the late stages of virion budding and release and that the observed enhancement of virion production might be correlated with drug-induced actin depolymerization at the site of viral budding. In light of the fact that L domains of different retroviruses are responsible for the late stage of virion release, we hypothesized that depolymerization of actin might be part of the L-domain function to facilitate fission and release budding viral particles from plasma membranes. To test this model, we first examined three EIAV proviruses carrying variant L domains, i.e., the wild-type EIAV and two chimeric EIAV constructs with the parental YPDL L domain replaced by either a PTAP (HIV-1) or PPPY (RSV) L domain (37). Previous studies from our lab have demonstrated that these proviruses are replication competent in ED cells (37). ED cells chronically infected with these proviruses were used for the assay. Despite varied absolute amounts of virion production among the L-domain variants (37), short-term cytoD treatment increased virion production from all of the producer cells an average of ∼2-fold compared to the respective untreated control producer cells (Fig. 6A). Importantly, nocodazole had no apparent effect on the levels of EIAV budding mediated by any of the L-domain motifs. These data confirmed the specificity of cytoD-induced enhancement of viral budding and indicated that the effect was not limited to a YPDL L domain contained in EIAV but was equally evident with proline-rich L domains of HIV-1 and other retroviruses.
FIG. 6.
Involvement of L-domain function in enhanced EIAV budding induced cytoD treatment. (A) Equal amounts of ED cells chronically infected with EIAVuk proviruses containing specific L domains (EIAV YPDL, HIV-1 PTAP, or RSV PPPY) (37) were incubated with the indicated agents at 37°C for 2.5 h. Virion production was examined by measuring the RT activity in culture medium after treatment. (B) Equal amounts of L-domain-positive and -negative producer cells were incubated with cytoD at the indicated concentrations for 2.5 h at 37°C, and viral particles in the culture medium were then collected by centrifugation. The pelleted virions were resuspended in 1× phosphate-buffered saline. The RT activity associated with the resuspended virions was tested as a measure of EIAV production. The data represent the mean value with standard deviation of duplicate experiments. The astrisks indicate a statistically significant difference (P < 0.05) compared to the untreated control.
We next tested whether a functional L domain was required for the observed enhancement in EIAV budding by cytoD treatment. L-domain-positive and -negative EIAV producer ED cells were generated for this assay. To compensate for the intrinsically low levels of EIAV budding in L-domain-deficient proviruses (9, 49), we used a cmvEIAVuk proviral construct in which viral gene expression is driven by the cytomegalovirus (CMV) promoter at levels substantially higher than those achieved by the EIAVuk proviruses under long terminal repeat control (9). The L-domain-positive group included the parental cmvEIAVuk provirus and cmvEIAVuk, with a PTAP L-domain substitution for the YPDL motif (37). The L-domain-negative group contained cmvEIAVuk proviruses lacking a functional L-domain with either mutated YPDL or truncated p9 proteins Q3 and L22 (where stop codons were introduced to preterminate synthesis of p9 protein before the YPDL L domain) (9). The data summarized in Fig. 6B demonstrate increased viral budding in a dose-dependent manner from both members of the L-domain-positive group. A statistically significant (P < 0.05) increase of 25% in viral budding was observed with 0.4 μM cytoD, and a 60% increase was observed with 1.0 μM cytoD. The budding efficiency of the L-domain-negative producers was lower than that of the L-domain-positive producers due to the lack of the L-domain function to facilitate budding. Nevertheless, viral budding from the L-domain mutants was reproducibly detectable (9), and cytoD treatment on the L-domain-negative producers had no significant effect on EIAV budding in all three L-domain-negative producer cells (Fig. 6B). These data indicate that the cytoD-induced enhancement of EIAV budding requires a functional L domain, suggesting that depolymerization of actin filaments during the fission of viral particles may be a function of the L domain and can be enhanced by brief cytoD treatment.
DISCUSSION
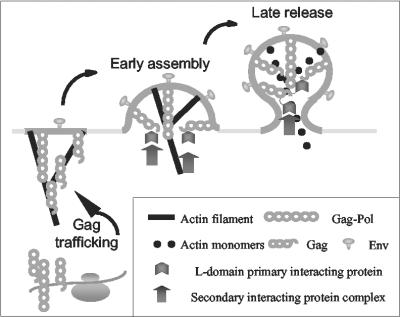
A general model for retroviral assembly was proposed nearly 25 years ago (5), but the precise mechanisms by which the efficient trafficking and assembly of viral proteins are achieved have remained largely undefined. Recent advances in studies of retroviral budding suggest that cellular factors associated with the endocytic pathway are specifically recruited by the L domains of various retroviruses to facilitate the budding process (20, 33, 50, 56). These studies suggest a highly concerted and complex interaction of viral proteins with components of the cellular trafficking machinery components to specifically adapt and direct these cell functions to viral purposes. The primary cellular factors associated with retroviral budding include various endocytic proteins and components of the MVB sorting pathway, although the exact functions of these cell proteins in retroviral budding are still under investigation. In addition, previous demonstrations of specific binding between retroviral Gag proteins and cellular actin filaments (40, 43, 52) has led to the hypothesis that the latter may contribute to retroviral budding process as well. In the present EIAV studies, we now demonstrate for the first time that a dynamic actin cytoskeleton is necessary for virion production, perhaps with dual functions of Gag polyprotein trafficking and virion release, the latter via a viral L-domain-dependent interaction. Therefore, we propose that targeting of EIAV Gag polyproteins from ribosomes to the plasma membrane requires polymerizing actin comets and that depolymerization of actin filaments associated with budding virions is critical to the release of viral particles via an L-domain-dependent interaction (Fig. 7).
FIG. 7.
Model for the role of dynamic actin cytoskeleton in retroviral budding. In this model, EIAV Gag and Gag-Pol polyproteins are trafficked via dynamic actin comets to the budding site on the plasma membrane. Gag polyproteins specifically recruits cellular components of the endocytic machinery via L-domain interactions during assembly and budding process. The recruited cellular factors facilitate late stages of virion budding, including depolymerization of actin filaments to release the virions from the plasma membrane.
Several lines of evidence support the hypothesis that depolymerization of actin filaments is involved in late stages of EIAV budding. We demonstrate here that 2.5 h of treatment of EIAV-producing ED and MDM cells with two different actin-depolymerizing drugs, cytoD and latB, reproducibly increased virion production by ca. 50%. The specificity of this phenomenon was supported by two important additional observations. First, the microtubule disrupting drug nocodazole had no effect on virion production from EIAV-infected ED cells, revealing a clear difference in the role of the distinct actin and microtubule cytoskeleton networks in EIAV budding. Second, the budding enhancement by the depolymerizing drugs was shown to be dependent on the presence of a functional L domain; the budding levels from EIAV constructs lacking an L domain were not increased by short-term cytoD (Fig. 6B). Interestingly, similar levels of enhancement of virion budding were observed with YPDL, PTAP, and PPPY L domains in the context of the EIAV p9 protein. We interpret these results to suggest that L-domain interactions with other cellular factors (e.g., endocytic or MVB sorting proteins) lead to a depolymerization of actin filaments at the site of viral budding to release assembled virions from the plasma membrane. Recently, it has been demonstrated that dynamic actin filaments are associated with endocytic processes (2, 34, 36, 38). Based on this model, it should be possible to examine the actin structures associated with retroviral budding sites for colocalization with viral proteins and other candidate cellular factors.
The EIAV studies described here are consistent with earlier reports that long-term (i.e., >20-h) disruption of the actin network by cytoD treatment decreases retroviral particle production by ca. 50% in HIV-1- or mouse mammary tumor virus-producing cells (41, 55), indicating an association of actin filament integrity with retroviral assembly. These data suggest that a dynamic actin network facilitates trafficking of Gag polyproteins via actin polymerization from ribosomes to budding sites on the plasma membrane. Association of Gag polyproteins with the actin filaments could target the Gag polyproteins specifically to lipid rafts, where actin cytoskeleton exists at a highly dynamic state (28, 53). This model of Gag polyprotein trafficking appears to be consistent with our findings that EIAV production is markedly inhibited by phallacidin, which stabilizes actin filaments in viable cells. The fact that long-term treatments with actin-destabilizing and -stabilizing drugs both decrease EIAV particle production indicates a critical role for a properly regulated and dynamic actin filament network in retroviral particle production.
In addition to the actin cytoskeleton, cellular components involved in early endocytosis (50) and the late MVB sorting pathway (20) have been identified to specifically interact with retroviral Gag proteins in facilitating the late budding process. MVB formation in late endosomes resembles retroviral budding topologically but occurs at sites other than the plasma membrane used for retroviral budding. Conversely, endocytosis occurs at the plasma membrane but proceeds in a topology opposite to retroviral budding. Although more studies are required to elucidate how various retroviruses adapt one or both of these cellular trafficking networks to achieve virion production from Gag polyproteins, it is relevant to note that dynamic actin cytoskeleton function is essential to both endocytosis and MVB processing by providing the necessary mechanical force for movement (29, 34, 36). The current EIAV studies provide new fundamental information on the specificity and potential role of the actin network in retroviral replication. Further elucidation of the determinants of viral Gag and actin interactions can provide a higher-resolution portrait of retroviral assembly mechanisms and potentially elucidate new functional targets for antiviral drug development.
Acknowledgments
This research was supported by grant number R01 CA49296 from the National Cancer Institute of the National Institutes of Health. C.C. was supported as a postdoctoral trainee in the Molecular Microbial Persistence and Pathogenesis Training Program funded by NIH grant T32 AI49820 from the National Institute of Allergy and Infectious Diseases.
We acknowledge Sean Alber and Laura Sysko from the Center for Biological Imaging and the DNA Sequencing Core of the University of Pittsburgh for excellent technical assistance.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apodaca, G. 2001. Endocytic traffic in polarized epithelial cells: role of the actin and microtubule cytoskeleton. Traffic 2:149-159. [DOI] [PubMed] [Google Scholar]
- 3.Barak, L. S., R. R. Yocum, E. A. Nothnagel, and W. W. Webb. 1980. Fluorescence staining of the actin cytoskeleton in living cells with 7-nitrobenz-2-oxa-1,3-diazole-phallacidin. Proc. Natl. Acad. Sci. USA 77:980-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barak, L. S., R. R. Yocum, and W. W. Webb. 1981. In vivo staining of cytoskeletal actin by autointernalization of nontoxic concentrations of nitrobenzoxadiazole-phallacidin. J. Cell Biol. 89:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolognesi, D. P., R. C. Montelaro, H. Frank, and W. Schafer. 1978. Assembly of type C oncornaviruses: a model. Science 199:183-186. [DOI] [PubMed] [Google Scholar]
- 6.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinskaya, A., B. Brichacek, A. Mann, and M. Stevenson. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, R. F., C. Leroux, S. J. Cook, S. L. Berger, D. L. Lichtenstein, N. N. Ghabrial, R. C. Montelaro, and C. J. Issel. 1998. Development and characterization of an in vivo pathogenic molecular clone of equine infectious anemia virus. J. Virol. 72:1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damsky, C. H., J. B. Sheffield, G. P. Tuszynski, and L. Warren. 1977. Is there a role for actin in virus budding? J. Cell Biol. 75:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edbauer, C. A., and R. B. Naso. 1983. Cytoskeleton-associated Pr65gag and retrovirus assembly. Virology 130:415-426. [DOI] [PubMed] [Google Scholar]
- 16.Edbauer, C. A., and R. B. Naso. 1984. Cytoskeleton-associated Pr65gag and assembly of retrovirus temperature-sensitive mutants in chronically infected cells. Virology 134:389-397. [DOI] [PubMed] [Google Scholar]
- 17.Franke, E. K., H. E. Yuan, K. L. Bossolt, S. P. Goff, and J. Luban. 1994. Specificity and sequence requirements for interactions between various retroviral Gag proteins. J. Virol. 68:5300-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 19.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, J. M., I. G. Goldberg, and T. L. Benjamin. 2003. Cell penetration and trafficking of polyomavirus. J. Virol. 77:2615-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heuser, J. E., and M. W. Kirschner. 1980. Filament organization revealed in platinum replicas of freeze-dried cytoskeletons. J. Cell Biol. 86:212-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ibarrondo, F. J., R. Choi, Y. Z. Geng, J. Canon, O. Rey, G. C. Baldwin, and P. Krogstad. 2001. HIV type 1 Gag and nucleocapsid proteins: cytoskeletal localization and effects on cell motility. AIDS Res. Hum. Retrovir. 17:1489-1500. [DOI] [PubMed] [Google Scholar]
- 27.Ikeda, M., A. Ikeda, L. C. Longan, and R. Longnecker. 2000. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 268:178-191. [DOI] [PubMed] [Google Scholar]
- 28.Janmey, P. A., W. Xian, and L. A. Flanagan. 1999. Controlling cytoskeleton structure by phosphoinositide-protein interactions: phosphoinositide binding protein domains and effects of lipid packing. Chem. Phys. Lipids 101:93-107. [DOI] [PubMed] [Google Scholar]
- 29.Jeng, R. L., and M. D. Welch. 2001. Cytoskeleton: actin and endocytosis—no longer the weakest link. Curr. Biol. 11:R691-R694. [DOI] [PubMed] [Google Scholar]
- 30.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 31.Katzmann, D. J., C. J. Stefan, M. Babst, and S. D. Emr. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenworthy, A. 2002. Peering inside lipid rafts and caveolae. Trends Biochem. Sci. 27:435-437. [DOI] [PubMed] [Google Scholar]
- 33.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. A. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of rous sarcoma virus and are required for Gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanzetti, L., P. P. Di Fiore, and G. Scita. 2001. Pathways linking endocytosis and actin cytoskeleton in mammalian cells. Exp. Cell Res. 271:45-56. [DOI] [PubMed] [Google Scholar]
- 35.Laux, T., K. Fukami, M. Thelen, T. Golub, D. Frey, and P. Caroni. 2000. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 149:1455-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, E., and P. De Camilli. 2002. Dynamin at actin tails. Proc. Natl. Acad. Sci. USA 99:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the Gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lichtenstein, D. L., K. E. Rushlow, R. F. Cook, M. L. Raabe, C. J. Swardson, G. J. Kociba, C. J. Issel, and R. C. Montelaro. 1995. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J. Virol. 69:2881-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, B., R. Dai, C. J. Tian, L. Dawson, R. Gorelick, and X. F. Yu. 1999. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J. Virol. 73:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maldarelli, F., N. W. King, Jr., and M. J. Yagi. 1987. Effects of cytoskeletal disrupting agents on mouse mammary tumor virus replication. Virus Res. 7:281-295. [DOI] [PubMed] [Google Scholar]
- 42.Montelaro, R. C., B. Parekh, A. Orrego, and C. J. Issel. 1984. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J. Biol. Chem. 259:10539-10544. [PubMed] [Google Scholar]
- 43.Mortara, R. A., and G. L. Koch. 1989. An association between actin and nucleocapsid polypeptides in isolated murine retroviral particles. J. Submicrosc. Cytol. Pathol. 21:295-306. [PubMed] [Google Scholar]
- 44.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359-374. [DOI] [PubMed] [Google Scholar]
- 45.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 46.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ploubidou, A., and M. Way. 2001. Viral transport and the cytoskeleton. Curr. Opin. Cell Biol. 13:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raabe, M. R., C. J. Issel, and R. C. Montelaro. 1998. Equine monocyte-derived macrophage cultures and their applications for infectivity and neutralization studies of equine infectious anemia virus. J. Virol. Methods 71:87-104. [DOI] [PubMed] [Google Scholar]
- 52.Rey, O., J. Canon, and P. Krogstad. 1996. HIV-1 Gag protein associates with F-actin present in microfilaments. Virology 220:530-534. [DOI] [PubMed] [Google Scholar]
- 53.Rozelle, A. L., L. M. Machesky, M. Yamamoto, M. H. Driessens, R. H. Insall, M. G. Roth, K. Luby-Phelps, G. Marriott, A. Hall, and H. L. Yin. 2000. Phosphatidylinositol 4, 5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10:311-320. [DOI] [PubMed] [Google Scholar]
- 54.Sampath, P., and T. D. Pollard. 1991. Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry 30:1973-1980. [DOI] [PubMed] [Google Scholar]
- 55.Sasaki, H., M. Nakamura, T. Ohno, Y. Matsuda, Y. Yuda, and Y. Nonomura. 1995. Myosin-actin interaction plays an important role in human immunodeficiency virus type 1 release from host cells. Proc. Natl. Acad. Sci. USA 92:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wieland, T. 1983. The toxic peptides from Amanita mushrooms. Int. J. Pept. Protein Res. 22:257-276. [DOI] [PubMed] [Google Scholar]
- 58.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zegers, M. M., K. J. Zaal, S. C. van IJzendoorn, K. Klappe, and D. Hoekstra. 1998. Actin filaments and microtubules are involved in different membrane traffic pathways that transport sphingolipids to the apical surface of polarized HepG2 cells. Mol. Biol. Cell 9:1939-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]