Abstract
The benzimidazole d-ribonucleosides TCRB and BDCRB are potent and selective inhibitors of human cytomegalovirus (HCMV) replication. Two HCMV strains resistant to these compounds were selected and had resistance mutations in genes UL89 and UL56. Proteins encoded by these two genes are the two subunits of the HCMV “terminase” and are necessary for cleavage and packaging of viral genomic DNA, a process inhibited by TCRB and BDCRB. We now report that both strains also have a previously unidentified mutation in UL104, the HCMV portal protein. This mutation, which results in L21F substitution, was introduced into the genome of wild-type HCMV by utilizing a recently cloned genome of HCMV as a bacterial artificial chromosome. The virus with this mutation alone was not resistant to BDCRB, suggesting that this site is not involved in binding benzimidazole nucleosides. As in previous proposals for mutations in UL104 of murine cytomegalovirus and HCMV strains resistant to BAY 38-4766, we hypothesize that this mutation could compensate for conformational changes in mutant UL89 and UL56 proteins, since the HCMV terminase is likely to interact with the portal protein during cleavage and packaging of genomic DNA.
HCMV is a cause of significant morbidity and mortality in immunocompromised patients such as organ transplant recipients (12, 36) and AIDS patients (20). This virus has been linked with development of atherosclerosis and restenosis following angioplasty (21, 39). Additionally HCMV is a leading cause of birth defects and infections in children (4).
Drugs currently approved for treatment of HCMV in the United States include ganciclovir (7), its prodrug valgancilcovir (8), cidofovir (13), foscarnet (6), and fomivirsen (25). The first four compounds inhibit HCMV DNA polymerase, and fomivirsen is a phosphorothioate oligonucleotide complementary to HCMV IE2 mRNA. Drawbacks of these drugs include poor bioavailability, toxicity, and emergence of resistant viral strains (1, 7, 9, 11, 13). Therefore, there is a need for better drugs with a different mode of action, good bioavailability, and a safer pharmacological profile.
The synthesis and activity of benzimidazole d-ribonucleosides TCRB and BDCRB have been previously reported by Townsend et al. (Fig. 1) (30). These compounds are potent inhibitors of HCMV in cell culture, and they have a novel mechanism of action. The step in the viral replication cycle that they inhibit is the cleavage and packaging of genomic DNA into capsids (17, 32). Two HCMV strains resistant to TCRB and BDCRB were selected in our laboratory by passage in the presence of TCRB (17). Both of these strains, termed D10 and C4, had a mutation in gene UL89; isolate C4 also had a mutation in UL56. The proteins encoded by these genes form the two subunits of the HCMV putative terminase (27). The large subunit encoded by UL56 is involved in DNA binding and capsid association (2, 27), whereas the small subunit encoded by UL89 is required for cleavage of genomic DNA (27). The ATPase activity of the large subunit UL56 is partially inhibited by BDCRB (28), and high concentrations of BDCRB partially inhibit the UL89-associated nuclease activity (27). The ORFs of four other genes thought to be essential for viral DNA cleavage and packaging (UL51, UL52, UL77, and UL104) also were sequenced in D10 and C4 HCMV strains, and no mutations were found (17).
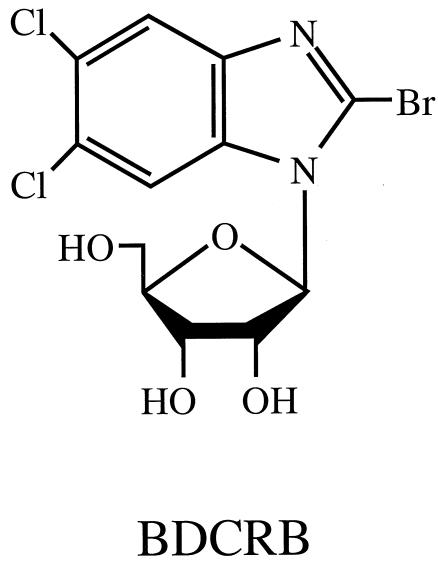
FIG. 1.
Structure of the benzimidazole ribonucleoside BDCRB.
Recently we selected another HCMV strain, termed G2, for resistance to the benzimidazole l-ribonucleoside maribavir. G2 also is resistant to TCRB and BDCRB because it was derived from C4. It was found that G2 has an L21F mutation in ORF UL104 (15). Moreover, we found that we had previously failed to identify this UL104 mutation in C4 from which G2 was derived. Because C4 contains this mutation and is resistant to TCRB and BDCRB, it could be involved in resistance to these benzimidazole d-ribonucleosides.
The product of the UL104 homologue in HSV-1, UL6 (23), is a portal protein through which viral DNA passes as it enters the capsid. This protein appears to serve as a docking site for the cleavage and packing complex, since it interacts with the HSV-1 putative terminase subunits UL15 and UL28 (35, 38). In addition, recent evidence that pUL104 colocalizes in the nucleus with pUL56 indicates that pUL104 is the HCMV portal protein (A. Dittmer and E. Bogner, Ninth Int. Cytomegalovirus Workshop and First Int. Betaherpesvirus Workshop, abstr. F08, 2003), thereby providing more rationale for the hypothesis that a mutation in UL104 could be involved in resistance to TCRB and BDCRB.
In addition, pUL104 may be involved in the activities of other novel compounds with activities against HCMV. The nonnucleoside sulfonamide inhibitor, BAY 38-4766, was found to target DNA maturation via the UL89 and UL56 gene products (5). HCMV and MCMV strains resistant to this compound had mutations in UL56, UL89, and UL104, but mutations in UL104 alone were not sufficient to confer resistance to this compound (5). In contrast, the homologue of HCMV UL104 in HSV-1 (UL6) was found to be involved in the mechanism of action of a novel class of thiourea compounds that inhibit HSV-1 DNA cleavage and encapsidation (33). Analogs of these thiourea compounds also target DNA maturation in VZV through ORF54, the HCMV UL104 homologue (34). HSV-1 and VZV strains resistant to these compounds are resistant due to mutations in UL6 and ORF54, respectively.
Consequently, we investigated the role of the UL104 L21F mutation in resistance of HCMV isolates to TCRB and BDCRB. This mutation was introduced into the genome of wild-type HCMV, and the resulting isolate was tested for sensitivity to BDCRB and other benzimidazole nucleosides.
(Portions of this work were reported at the XVth International Round Table, “Nucleosides, Nucleotides, Nucleic Acids,” Leuven, Belgium, September 2002.)
MATERIALS AND METHODS
Abbreviations.
The following abbreviations have been used: BAY 38-4766, 3-hydroxy-2,2-dimethyl-N-[4-({[5-(dimethylamino)-1-naphthyl]sulfonyl}amino)-phenyl]propanamide, BDCRB, 1-(β-d-ribofuranosyl)-2-bromo-5,6-dichloroben-zimidazole; FBS, fetal bovine serum; HCMV, human cytomegalovirus; HFF, human foreskin fibroblasts; HSV-1, herpes simplex virus type 1; IC50 and IC90, 50 and 90% inhibitory concentrations, respectively; maribavir, 1-(β-l-ribofuranosyl)-2-isopropylamino-5,6-dichloro-benzimidazole or 1263W94; MCMV, murine cytomegalovirus; MEM(E), minimal essential media with Earle's salts; MOI, multiplicity of infection; ORF, open reading frame; TCRB, 1-(β-d-ribofuranosyl)-2,5,6-trichlorobenzimidazole; UMJD 1311, 1-(α-l-5-deoxy-lyxofuranosyl)-2,5,6-trichlorobenzimidazole; VZV, varicella-zoster virus.
Chemicals.
BDCRB (30) and UMJD 1311 (22) were synthesized in the laboratory of L. B. Townsend as previously described. BAY 38-4766 was synthesized by a method similar to that reported (1a). Maribavir was synthesized at GlaxoSmithKline (16) and was provided through the courtesy of K. K. Biron. Ganciclovir was purchased from Hoffmann La Roche (Palo Alto, Calif.).
Cell culture procedures.
HFF were derived in our laboratory. They were grown in MEM(E) and 10% FBS at 37°C in a humidified atmosphere of 3% CO2-97% air. They were regularly passaged at 1:2 dilutions by using conventional procedures with 0.05% trypsin plus 0.02% EDTA in HEPES buffered saline (29, 31).
Viral strains and virological procedures.
HCMV strain Towne, isolate P0, was kindly provided by M. F. Stinski, University of Iowa. HCMV strains C4, D10, and r56 were derived in our laboratory from the parent Towne strain and are resistant to both TCRB and BDCRB (17). Two other TCRB- and BDCRB-resistant HCMV strains, 1038rA and 1038rB, were isolated at GlaxoSmithKline and kindly provided to us by K. K. Biron (32). AD169-RV HCMV was obtained by harvesting viral progeny from HFF cells transfected with AD169-BAC and plasmids expressing Cre recombinase (pBRep-Cre) and HCMV pp71 (pCGN71). Access to AD169-BAC was kindly provided by U. H. Koszinowski, Ludwig-Maximilians-Universität München, Munich, Germany. Plasmid pBRep-Cre was constructed by W. Brune (14), University of Wuerzburg, Wuerzburg, Germany. Plasmid pCGN71 was constructed in the laboratory of T. Shenk, Princeton University, Princeton, N.J. Stocks of HCMV were prepared by infecting HFF cells at an MOI of 0.01 PFU per cell, and stock viral titers were determined by using monolayer cultures of HFF cells as described previously (26, 31).
DNA sequencing.
Primers for PCR amplification and sequencing were designed by using the PRIME program in the Genetics Computer Group package (Wisconsin Package Version 10.3; Accelrys Inc., San Diego, Calif.) and were based on the published sequence of strain AD169 of HCMV. After PCR amplification, products were separated on a 0.8% agarose gel, extracted, and purified by using the QIAquick Gel Extraction kit (Qiagen, Valencia, Calif.). Sequencing PCR was done with the BigDye Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.) in a manner similar to that previously reported (17), except that sequencing reaction products were separated and detected with an ABI Prism 310 Genetic Analyzer. Sequences were aligned and edited by using Sequencher software (GeneCodes, Ann Arbor, Mich).
HCMV antiviral assays.
For plaque reduction assays, HFF were planted at 85,000 cells per well in 24-well cluster dishes. The next day, they were infected with HCMV at 100 PFU per well in MEM(E) plus 5% FBS. One to 2 h postinfection, media containing selected drug dilutions and final FBS concentrations of 5 and 0.5% methylcellulose were added. All drug dilutions were tested at least in duplicate by using five to six different drug concentrations. After incubation at 37°C for 9 to 11 days, cell monolayers were stained with crystal violet and plaques were enumerated under light microscopy. The number of plaques observed in the presence of each drug concentration was compared to the number observed in the absence of drug in order to determine the effects of the drug.
For yield reduction assays, HFF cells were plated at 10,000 cells per well in 96-well cluster dishes, incubated overnight, and infected with HCMV at an MOI of 0.5. After virus adsorption, the media were replaced with fresh media containing test compounds in eight 1:3 dilutions starting from a 100 μM drug concentration. Plates were incubated for 7 days and subjected to one cycle of freezing and thawing, and titers were determined by transferring 100-μl aliquots from each of the wells to a fresh 96-well monolayer culture of HFF cells followed by serial dilution across the plate. Cultures were incubated for 7 days, cells were stained, and the numbers of plaques were determined (26, 31).
Dose-response relationships were used to quantify drug effects. For plaque reduction assays, the percent inhibition of plaque number was plotted against log10 drug concentrations. For yield reduction assays, the log10 of the percent inhibition of viral titer was plotted against the log10 drug concentrations. IC50s and IC90s were interpolated from the linear portions of the regression lines.
Growth characteristics.
HFF cells were plated at 100,000 cells per well in 24-well cell culture plates. The next day they were infected with either wild-type AD169-RV or UL104L21F rec HCMV at an MOI of 0.01. Over a period of 10 days, the cultures were removed from the 37°C incubator and frozen at −80°C. All time points were done in duplicate. After all the cultures were collected, they were thawed, and titers were determined in duplicate by 12 serial threefold dilutions across 96-well plates (26). Seven days later, they were stained with crystal violet and titers were calculated.
Recombinant HCMV construction.
The genome of HCMV strain AD169 cloned by Gabrielle Han as a bacterial artificial chromosome (AD169-BAC) (3, 14) in U. H. Koszinowski's laboratory was used in recombinant HCMV construction. It was electroporated into Escherichia coli DY380 cells (19) (D. L. Court, Frederick Cancer Research and Development Center, Frederick, Md.) by W. Brune and kindly provided to us. The zeocin resistance gene driven by the bacterial EM7 promoter was amplified by PCR from pZeo (10) by using primers designed to bind to UL104 (104-FOR and 104-REV), which at each end had 50 bp of sequence homology to the target in AD169-BAC (104-FOR, 5′-CAGCAACCGCAGGAAGCTCATCGTCTGCCCCGTGGGGAAAATGTCGATGATGTTGACAATTAATCATCGGCAT-3′; 104-REV, 5′-AGAGGAGGCGGAGGAGTGAACGGTCGTC GTTGCCGCGGCGGTAGTTG-CGGGAATTCAGTCCTGCTCCTCGGCCA-3′). These PCR products were electroporated into E. coli DY380 cells containing the AD169-BAC, and mutants were selected by growing bacteria on Luria-Bertani agar plates supplemented with the antibiotic zeocin (25 μg/ml). The preparation of DY380 electroporation-competent cells and electroporation conditions were as described previously by Yu et al. (37). The genome coordinates of the sequences that were deleted and replaced with the zeocin resistance gene are 151936 to 152136. To confirm deletion in UL104, genomic DNA was digested with EcoRI restriction endonucleases and compared to the wild-type digest.
A DNA fragment containing gene UL104 and the first 834 nucleotides of UL105 (nucleotides 151339 to 152760) was PCR amplified from AD169 genomic DNA by using primers with either EcoRI- or XhoI-cut sites (underlined), as follows: 104EcoRI, 5′-GCGCGAATTCAGCGCACCTTCTTG-AAGAC-3′; 104XhoI, 5′-GCGCCTCGAGTAAAAAAACACCACCACCTG-3′. This PCR fragment was ligated into EcoRI- and XhoI-cut sites of the pBluescript SK(−) vector from Stratagene, resulting in pBSK(104-105). By use of the QuickChangeTM site-directed mutagenesis kit (Stratagene), the L21F mutation was introduced. The resulting plasmid pBSK(104-105)L21F was sequenced to verify the mutation. The oligonucleotides used for mutagenesis were 5′-CGTCGACAAGGTGAAGTCCCTCTCGCG-3′ and 5′-CGCGAGAGGGACTTCACCTTGTCGACG-3′.
In order to rescue the deletion virus, the UL104Δ-BAC (5 μg) was electroporated into HFF cells together with pBSK(104-105)L21F (5 μg) plus plasmids pBRep-Cre and pCGN71 (1 μg each). Electroporation was performed by using a Bio-Rad Gene Pulser II set at 260 V and a capacitance extender at 975 μF. Media were changed the next day, and the virus was harvested approximately 28 days later, 3 days after 100% cytopathic effect was observed.
Nucleotide sequence accession numbers.
Nucleotide and amino acid sequences were obtained from GenBank. Accession numbers are as follows: HCMV AD169, X17430; HCMV strain Towne ORF UL104, AF047524.
RESULTS
UL104 mutation.
Two HCMV strains, of which the more resistant strain (C4) has mutations in UL56 and UL89, have been described by Krusky et al. as being resistant to TCRB and BDCRB (17), whereas the less resistant virus from which C4 was isolated (D10) has a mutation only in UL89 (32). In the present study we again sequenced UL104 in both of these strains and identified a G-to-C change at position 63, resulting in a putative L21F mutation in this ORF. Sequencing UL104 from the wild-type Towne strain used to isolate D10 and wild-type AD169 virus used in our laboratory revealed that this mutation was not present in either strain. Because C4 was derived from D10, the mutation would be expected to occur in both strains.
We also sequenced this portion of UL104 from three other HCMV strains resistant to TCRB and BDCRB. Strains 1038rA and 1038rB had been isolated in the laboratory of K. K. Biron and are resistant to TCRB and BDCRB due to mutations in UL89 (32). Resistance to a third strain, r56, is from a mutation in UL56 inserted into wild-type Towne by recombination (17). Sequencing revealed that the L21F mutation was not present in any of these strains.
Construction of mutant virus.
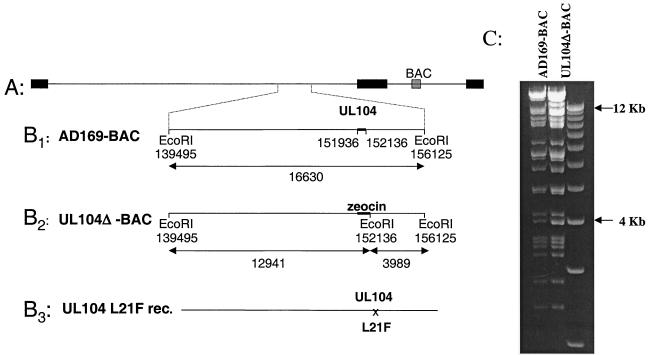
In order to determine if the L21F mutation in UL104 of HCMV alone was sufficient to cause resistance to TCRB and BDCRB, a virus with just this mutation was constructed. First, a deletion in UL104 of HCMV cloned as a bacterial artificial chromosome (AD169-BAC) (Fig. 2A) was introduced by replacing 100 bp on each side of leucine 21 (genome coordinates 151936 to 152136) (Fig. 2B1) with a zeocin resistance gene driven by the bacterial EM7 promoter (Fig. 2B2). To confirm this replacement, a novel EcoRI-cut site was introduced within the zeocin cassette. This novel restriction site at nucleotide position 152136 resulted in the loss of the 16.6-kb fragment and the appearance of two new fragments of 12.9 and 4 kb (Fig. 2B3). The new 12.9-kb fragment can be clearly seen on the agarose gel (Fig. 2C), confirming that the zeocin cassette had replaced the 200-bp deletion in UL104. It is more difficult to see the new 4-kb fragment, since there is one already present in the wild-type AD169-BAC.
FIG. 2.
Construction of HCMV with the L21F mutation in UL104. (A) Genome of HCMV cloned as a bacterial artificial chromosome (AD169-BAC); (B1) expanded region surrounding the UL104 ORF; (B2) AD169-BAC in which a portion of UL104 was replaced with the zeocin cassette; (B3) AD169-BAC in which the zeocin cassette is replaced with the UL104 fragment containing the L21F mutation; (C) EcoRI digest of AD169-BAC and UL104ΔBAC.
Since the UL104 gene overlaps with ∼170 bp of UL105 (HCMV helicase), this gene was also disrupted by the 200-bp deletion in UL104. One would not expect HCMV to replicate without UL104 and UL105 genes, since the homologue of UL104 in HSV-1 (UL6) is essential for cleavage and packaging of viral DNA (18), and UL105 is required for HCMV DNA replication (24). In order to rescue this virus, a fragment containing the 200-bp deletion with the L21F mutation plus an additional ∼500 bp of sequence on each side was electroporated into HFF cells together with the UL104Δ-BAC DNA. Two weeks later, plaques confirming that the homologous recombination had occurred between the fragment containing the L21F mutation and UL104Δ-BAC were observed. A portion of UL104 from this virus (UL104 L21F rec) was sequenced in order to confirm the presence of the L21F mutation.
Phenotypic characterization.
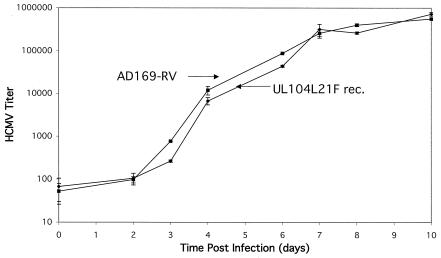
To determine if this mutation would change the growth characteristics of UL104 L21F rec HCMV, HFF cells were infected at an MOI of 0.01 and the growth of this virus was monitored over a period of 10 days (Fig. 3). This study revealed that UL104 L21F rec was not growth deficient compared to wild-type AD169-RV HCMV.
FIG. 3.
Growth study comparing wild-type HCMV AD169-RV and HCMV with L21F mutation in UL104 (UL104L21F rec). HFF cells were infected at an MOI of 0.01 PFU/cell, incubated at 37°C, and harvested at the times indicated over the course of 10 days. After all samples were collected, viral titers were determined as described in Materials and Methods.
In both plaque (Table 1) and yield (Table 2) reduction assays, UL104 L21F rec HCMV showed the same sensitivity to BDCRB as the wild-type AD169-RV virus. The sensitivity of the mutant virus to control compounds that act by different mechanisms (ganciclovir and the benzimidazoles maribavir and 1311) and a similar mechanism (BAY 38-4766) also did not differ from the sensitivity of the wild-type virus. We conclude that the L21F mutation in UL104 was not sufficient for resistance of HCMV to BDCRB.
TABLE 1.
Activities of BDCRB and control compounds against wild-type and mutant strains of HCMV in a plaque reduction assay
| Compound | Mean IC50 ± SD (μM)a
|
|
|---|---|---|
| AD169-RV (wild-type) | UL104 L21F rec (mutant) | |
| BDCRB | 0.4 ± 0.3 | 0.4 ± 0.1 |
| TCRB | 2.2 ± 0.5 | 1.7 ± 0.1 |
| BAY 38-4766 | 0.5 ± 0.2 | 0.4 ± 0.04 |
| Maribavir | 0.4 ± 0.2 | 0.3 ± 0.2 |
| UMJD 1311 | 0.6 ± 0.3 | 0.6 ± 0.3 |
| Ganciclovir | 5.2 ± 0.6 | 6.2 ± 1 |
Calculated from either duplicate data from one experiment or by averaging data from three experiments done with BDCRB by using at least five drug concentrations each.
TABLE 2.
Activities of BDCRB and ganciclovir against wild-type and mutant strains of HCMV in a yield reduction assay
| Compound | Mean IC90 ± SD (μM)a
|
|
|---|---|---|
| AD169-RV (wild-type) | UL104 L21F rec (mutant) | |
| BDCRB | 0.4 ± 0.04 | 0.4 ± 0.04 |
| Ganciclovir | 4.5 ± 0.3 | 2.6 ± 0.3 |
Calculated from a yield reduction assay performed in quadruplicate by using seven drug concentrations and either wild-type HCMV strain AD169-RV or the HCMV strain with the L21F mutation in UL104 (UL104L21F rec).
DISCUSSION
The majority of antiviral drugs on the market for treatment of HCMV infections target viral DNA polymerase. Recently, however, two different classes of compounds that target the DNA cleavage and packaging step in the viral replication cycle have been described (5, 17, 32). They include benzimidazole ribonucleosides TCRB and BDCRB, and a nonnucleoside inhibitor, BAY 38-4766.Mutations conferring resistance to both classes of compounds mapped to UL56 and UL89 ORFs. MCMV and HCMV strains resistant to BAY 38-4766 also had mutations in UL104. Mutations in this ORF alone, however, were not sufficient to confer resistance (5).
It is becoming clear that UL104 and its homologue in HSV-1 (UL6) encode the portal protein through which genomic DNA must pass to enter the capsid (23, 35, 38; Dittmer and Bogner, Abstr. Ninth ICW and First IBW, 2003). Since the HCMV terminase is likely to interact with UL104 during cleavage and packaging of genomic DNA, Buerger et al. speculated that mutations in UL104 may compensate for the conformational changes in mutant UL89 and UL56 proteins even though these mutations alone were not sufficient to produce resistance to the drugs (5). In contrast, mutations in UL6 of HSV-1 and ORF54 of VZV (homologues of UL104) were sufficient to confer resistance on their own to thiourea compounds that inhibit cleavage and packaging of genomic DNA in these viruses (33, 34).
In the present study an L21F mutation in the UL104 ORF of an HCMV strain resistant to TCRB and BDCRB was identified. This mutation was a result of a G-to-C change at nucleotide 63 (TTG→TTC) resulting in the putative amino acid change L21F. At this position UL105 overlaps with UL104, resulting in a change in this ORF as well. There is no amino acid change in this ORF, however, because UL105 is read on the opposite strand and the C-to-G change at nucleotide 111 gives GTG, which like GTC, encodes valine.
A virus with just the L21F mutation in UL104 was constructed and tested for resistance. It was found that this mutation alone did not confer resistance of HCMV to BDCRB, TCRB, or to the related benzimidazole nucleosides maribavir and UMJD 1311, which act by different mechanisms. In addition, the virus with the UL104 mutation replicated in fibroblasts at the same rate as the virus from which it was derived. In an earlier study it was observed that virus strain r56—resistant to BDCRB because of a mutation in UL56 only—replicated at the same rate as the wild-type virus from which it was derived (17). Because r56 is a recombinant virus made from wild-type HCMV with no mutation in UL104, the UL104 mutation could not be needed to compensate for the mutation in UL56. However, we do not have a virus with a mutation in only UL89, so we cannot eliminate the possibility that the UL104 mutation compensates for the UL89 mutation that confers resistance to BDCRB and TCRB. Alternatively, it is possible that this is not a compensatory mutation for a mutation conferring resistance to benzimidazole nucleosides but is simply an unrelated, background mutation.
Nonetheless, mutations in UL104 have been found in virus isolates resistant to benzimidazole nucleosides and to the sulfonamides (5, 17, 32). The fact that neither we nor Buerger et al. (5, 17, 32) found an effect does not mean that one does not exist, especially since mutations in HSV-1 and VZV homologues did produce resistance to thioureas (33, 34).
Acknowledgments
This study was supported by National Institute for Allergy and Infectious Diseases grants U19-AI-31718 and P01-AI-46390 and by research funds from the University of Michigan. Use of GCG programs was supported by grant M01RR00042 to the University of Michigan General Clinical Research Center. G.K. gratefully acknowledges fellowship support from the University of Michigan College of Pharmacy.
We also thank Karen K. Biron (from GlaxoSmithKline) for helpful discussions.
REFERENCES
- 1.Amin, H. I., E. Ai, H. R. McDonald, and R. N. Johnson. 2000. Retinal toxic effects associated with intravitreal fomivirsen. Arch. Ophthalmol. 118:426-427. [PubMed] [Google Scholar]
- 1a.Bender, W., et al. December 2002. U.S. patent 6,498,183.
- 2.Bogner, E., K. Radsak, and M. F. Stinski. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britt, W. J., R. F. Pass, S. Stagno, and C. A. Alford. 1991. Pediatric cytomegalovirus infection. Transplant. Proc. 23:115-117. [PubMed] [Google Scholar]
- 5.Buerger, I., J. Reefschlaeger, W. Bender, P. Eckenberg, A. Popp, O. Weber, S. Graeper, H. D. Klenk, H. Ruebsamen-Waigmann, and S. Hallenberger. 2001. A novel nonnucleoside inhibitor specifically targets cytomegalovirus DNA maturation via the UL89 and UL56 gene products. J. Virol. 75:9077-9086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chrisp, P., and S. P. Clissold. 1991. Foscarnet: a review of its antiviral activity, pharmacokinetic properties, and therapeutic use in immunocompromised patients with cytomegalovirus retinitis. Drugs 41:104-129. [DOI] [PubMed] [Google Scholar]
- 7.Crumpacker, C. S. 1996. Ganciclovir. Drug Ther. 335:721-729. [DOI] [PubMed] [Google Scholar]
- 8.Curran, M., and S. Noble. 2001. Valganciclovir. Drugs 61:1145-1152. [DOI] [PubMed] [Google Scholar]
- 9.Deray, G., F. Martinez, C. Katlama, B. Levaltier, H. Beaufils, M. Danis, M. Rozenheim, A. Baumelou, E. Dohin, M. Gentilini, and C. Jacobs. 1989. Foscarnet nephrotoxicity—mechanism, incidence and prevention. Am. J. Nephrol. 9:316-321. [DOI] [PubMed] [Google Scholar]
- 10.Drocourt, D., T. Calmels, J. P. Reynes, M. Baron, and G. Tiraby. 1990. Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 18:4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erice, A. 1999. Resistance of human cytomegalovirus to antiviral drugs. Clin. Microbiol. Rev. 12:286-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grattan, M. T., C. E. Moreno-Cabral, V. A. Starnes, E. B. Stinson, and N. E. Shumway. 1989. Cytomegalovirus infection is associated with cardiac allograph rejection and atherosclerosis. JAMA 261:3561-3566. [PubMed] [Google Scholar]
- 13.Hitchcock, M. J. M., H. S. Jaffe, J. C. Martin, and R. J. Stagg. 1996. Cidofovir, a new agent with potent anti-herpesvirus activity. Antivir. Chem. Chemother. 7:115-127. [Google Scholar]
- 14.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komazin, G., R. G. Ptak, B. T. Emmer, L. B. Townsend, and J. C. Drach. 2003. Resistance of human cytomegalovirus to the benzimidazole l-ribonucleoside maribavir maps to UL27. J. Virol. 77:11499-11506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koszalka, W. G., S. D. Chamberlain, R. J. Harvey, L. W. Frick, S. S. Good, M. L. Davis, A. Smith, K. K. Biron, J. C. Drach, and L. B. Townsend. 1996. Benzimidazoles for the treatment of human cytomegalovirus. Antivir. Res. 30:A43. [Google Scholar]
- 17.Krosky, P. M., M. R. Underwood, S. R. Turk, K. W.-H. Feng, R. K. Jain, R. G. Ptak, A. C. Westerman, K. K. Biron, L. B. Townsend, and J. C. Drach. 1998. Resistance of human cytomegalovirus to benzimidazole nucleoside analogs maps to two open reading frames: UL89 and UL56. J. Virol. 72:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 19.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 20.McKenzie, R., M. W. D. Travis, S. A. Dolan, S. Pittaluga, I. M. Feuerstein, J. Shelhamer, R. Yarchoan, and H. Masur. 1991. The cause of death in patients with human immunodeficiency virus infection: a clinical and pathological study with emphasis on the role of pulmonary disease. Medicine 70:326-343. [DOI] [PubMed] [Google Scholar]
- 21.Melnick, J. L., E. Adam, and M. E. DeBakey. 1998. The link between CMV and atherosclerosis. Infect. Med. 15:479-486. [Google Scholar]
- 22.Migawa, M. T., J. L. Girardet, J. A. Walker II, G. W. Koszalka, S. D. Chamberlain, J. C. Drach, and L. B. Townsend. 1998. Design, synthesis, and antiviral activity of α-nucleosides: d- and l-isomers of lyxofuranosyl- and (5-deoxylyxofuranosyl) benzimidazoles. J. Med. Chem. 41:1242-1251. [DOI] [PubMed] [Google Scholar]
- 23.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry, C. M., and J. A. Balfour. 1999. Fomivirsen. Drugs 57:375-380. [DOI] [PubMed] [Google Scholar]
- 26.Prichard, M. N., S. R. Turk, L. A. Coleman, S. L. Englehardt, C. J. Shipman, and J. C. Drach. 1990. A microtiter virus yield reduction assay for the evaluation of antiviral compounds against human cytomegalovirus and herpes simplex virus. J. Virol. Methods 28:101-106. [DOI] [PubMed] [Google Scholar]
- 27.Scheffczik, H., C. G. Savva, A. Holzenburg, L. Kolesnikova, and E. Bogner. 2002. The terminase subunits pUL56 and pUL89 of human cytomegalovirus are DNA-metabolizing proteins with toroidal structure. Nucleic Acids Res. 30:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz, B., S. Rechter, J. C. Drach, L. B. Townsend, and E. Bogner. 2003. Identification of the ATP-binding site in the terminase subunit pUL56 of human cytomegalovirus. Nucleic Acids Res. 31:1426-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shipman, C. J. 1969. Evaluation of 4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid (HEPES) as a tissue culture buffer. Proc. Soc. Exp. Biol. 130:305-310. [DOI] [PubMed] [Google Scholar]
- 30.Townsend, L. B., R. V. Devivar, S. R. Turk, M. R. Nassiri, and J. C. Drach. 1995. Design, synthesis, and antiviral activity of certain 2,5,6-trihalo-1-(β-d-ribofuranosyl)benzimidazoles. J. Med. Chem. 38:4098-4105. [DOI] [PubMed] [Google Scholar]
- 31.Turk, S. R., C. Shipman, R. Nassiri, G. Genzlinger, S. H. Krawczyk, L. B. Townsend, and J. C. Drach. 1987. Pyrrolo[2,3-d]pyrimidine nucleosides as inhibitors of human cytomegalovirus. Antimicrob. Agents Chemother. 31:544-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Underwood, M. R., R. J. Harvey, S. C. Stanat, M. L. Hemphill, T. Miller, J. C. Drach, L. B. Townsend, and K. K. Biron. 1998. Inhibition of human cytomegalovirus DNA maturation by a benzimidazole ribonucleoside is mediated through the UL89 gene product. J. Virol. 72:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Zeijl, M., J. Fairhurst, T. R. Jones, S. K. Vernon, J. Morin, J. LaRocque, B. Feld, B. O'Hara, J. D. Bloom, and S. V. Johann. 2000. Novel class of thiourea compounds that inhibit herpes simplex virus type 1 DNA cleavage and encapsidation: resistance maps to the UL6 gene. J. Virol. 74:9054-9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visalli, R. J., J. Fairhurst, S. Srinivas, W. Hu, B. Feld, M. DiGrandi, K. Curran, A. Ross, J. D. Bloom, M. van Zeijl, T. R. Jones, J. O'Connell, and J. I. Cohen. 2003. Identification of small molecule compounds that selectively inhibit varicella-zoster virus replication. J. Virol. 77:2349-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wingard, J. R., S. Piantadosi, W. H. Burns, M. L. Zahurak, G. W. Santos, and R. Saral. 1990. Cytomegalovirus infections in bone marrow transplant recipients given intensive cytoreductive therapy. Rev. Infect. Dis. 12(Suppl. 7):S793-S804. [DOI] [PubMed] [Google Scholar]
- 37.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, D., and S. K. Weller. 1998. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J. Virol. 72:7428-7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou, Y. F., M. B. Leon, M. A. Waclawiw, J. J. Popma, Z. X. Yu, T. Finkel, and S. E. Epstein. 1994. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N. Engl. J. Med. 335:624-630. [DOI] [PubMed] [Google Scholar]