Abstract
The genome of Junonia coenia densovirus (JcDNV) shares with members of the genus Densovirus the property of possessing structural (VP) and nonstructural (NS) genes in opposite orientations. The three NS genes located in the 5′ half on one strand encode three NS proteins assumed to be involved in viral DNA replication: NS-1 and NS-2, which are common to all DNVs, and a 28-kDa polypeptide, NS-3, with a unique sequence. Whereas the essential role played by JcDNV NS-1 in viral DNA replication has been clearly established (C. Ding, M. Urabe, M. Bergoin, and R. M. Kotin, J. Virol. 76:338-345, 2002), nothing is known of the biological function(s) of NS-3. To investigate this function, we designed constructs derived from pBRJ, a plasmid encompassing an infectious sequence of JcDNV DNA (M. Jourdan, F. X. Jousset, M. Gervais, S. Skory, M. Bergoin, and B. Dumas, Virology 179:403-409, 1990), with partial or complete deletion of NS-3 sequence or with the ATG initiation codon mutated by site-directed mutagenesis. Transfection of these constructs to sensitive Ld 652 cells or Spodoptera littoralis larvae prevented the accomplishment of a productive cycle. We clearly established that the blocking of the replicative cycle in the absence of NS-3 expression occurred at the level of viral DNA replication. Replication of viral DNA could be restored by cotransfecting Ld 652 cells with a plasmid expressing JcDNV-NS-3 protein in trans. Time course analysis showed that NS-3 is produced early (6 h posttransfection) in the replicative cycle, and its production parallels that of replicative-form viral DNA. Finally, we present evidence that NS-1 and NS-2 proteins are synthesized at apparently the same levels whether or not NS-3 is expressed.
Densoviruses (DNVs) are autonomously replicating parvoviruses known to be highly pathogenic for two phyla of invertebrates: insects and crustacea (3, 13). Like their vertebrate counterparts, DNVs are small icosahedral nonenveloped viruses with a single-stranded linear DNA genome 4 to 6 kb in length (3, 4, 26). Based on the size, organization of coding sequences, and structure of extremities of their genome, DNVs are distributed in three genera within the subfamily Densovirinae (5). Members of the genus Densovirus, with the Junonia coenia DNV (JcDNV) as the prototype, have a 6-kb genome, a long (>500-nucleotide [nt]) inverted terminal repeat (ITR), and an ambisense organization. The four structural proteins VP1, VP2, VP3, and VP4 are nested in a single large open reading frame (ORF), ORF1, located in the 5′ half of one strand, whereas in the 5′ half of the complementary strand ORF2, ORF3, and ORF4 encode the three nonstructural (NS) proteins NS-1, NS-2, and NS-3, respectively (11). VP and NS genes are transcribed from the P9 and P93 promoters, respectively. The four overlapping capsid polypeptides are synthesized from an unspliced 2.5-kb mRNA by a “leaky-scanning” mechanism whereby ribosomes initiate translation at the first, second, third, or fourth/fifth in-frame AUG codon. Thus, the four VPs share the same C-terminal sequence and differ only in their N-terminal region. The frequency of translation initiation events at each AUG codon is very likely responsible for the different levels of expression of the four polypeptides, as reflected by the presence of VP1, VP2, VP3, and VP4 in an approximate 1:9:9:41 ratio in the virion (27). The three NS proteins are synthesized from two mRNAs, an unspliced 2.4-kb mRNA coding for NS-3 and a spliced 1.7-kb mRNA coding for NS-1 and NS-2 (unpublished data). Among other functions, these proteins are likely to be involved in viral DNA replication. Sequence analyses revealed that NS-1 proteins of DNVs share homologies with NS-1 proteins of vertebrate parvoviruses, including NTP-binding and helicase activity domains (1, 3, 11, 26). Recent in vitro studies of JcDNV NS-1 biochemical properties have confirmed its involvement in viral DNA replication by showing that it binds to specific DNA sequences within the A-A′ region of the terminal palindrome and, like its vertebrate counterparts, has multiple functions (including a strand- and site-specific nicking activity), and has an ATP-dependent DNA helicase activity (10). In contrast, no clear function could be assigned so far to NS-2 and NS-3 proteins of DNVs. These proteins share no homology with NS proteins of vertebrate parvoviruses or with protein sequences from data banks. In the present study we attempted to elucidate the function of NS-3 in the life cycle of JcDNV by generating deletion mutants and analyzing their ability to undergo a replicative cycle. Our results demonstrate that NS-3 is absolutely required for viral DNA replication and the production of virus particles, both in Ld 652 cells and in Spodoptera littoralis larvae.
MATERIALS AND METHODS
NS-3 deletion and mutagenesis.
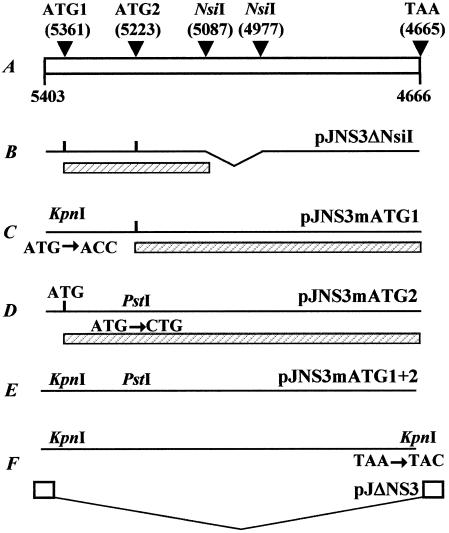
All constructs derived from pBRJ, a plasmid encompassing an infectious sequence of JcDNV DNA (19). Partial deletion of NS-3 (pJNS3ΔNsiI construct) was obtained by digestion of pBRJ with NsiI and religation with T4 DNA ligase (1 U of DNA ligase in 20 μl of DNA mixture for 30 min at room temperature). pBRJ contains two NsiI restriction sites, both located in the central coding region of NS-3, and this deletion eliminates a 112-bp fragment of this sequence (see Fig. 3). For complete deletion of NS-3 coding sequence (pJΔNS3 construct), the ATG initiation codon and the TAA stop codon were mutated by in vitro site-directed mutagenesis, by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. The primer pair 5′-GAAACCTGGCAATAGAGGTACCGATTATATTCTGGAACCAC-3′ and 5′-GTGGTTCCAGAATATAATCGGTACCTCTATTGCCAGGTTTC-3′ (ATG1 mutation) and the primer pair 5′-GTTGGTGTCTCCATTGTTCATCGGTACCTAATGTCTACTATTAGG-3′ and 5′-CCTAATAGTAGACATTAGGTACCGATGAACAATGGAGACACCAAC-3′ (TAA mutation) were designed so to create two KpnI restriction sites (indicated in boldface). pBRJ was used as a template, and cycling conditions were 95°C for 50s, 60°C for 50s, and 68°C for 25 min for 18 rounds. After treatment with DpnI, the PCR products were used to transform XL-10 Gold Ultra-Competent cells. The colonies were screened for mutated plasmid and pJΔNS3 was obtained by digestion with KpnI and religation with T4 DNA ligase. Mutations of ATG1 (pJNS3mATG1), ATG2 (pJNS3mATG2), and ATG1+2 (pJNS3mATG1+pJNS3mATG2) were generated according to the same protocol with the same primer pair mentioned above for ATG1 mutation and the primer pair 5′-CTAGATCATGCTGCAGATTAAGTATAGGTTTAGGCAAAATTGAC-3′ and 5′-GTCAATTTTGCCTAAACCTATACTTAATCTGCAGCATGATCTAG-3′ for ATG2 mutation. These last primers contained the PstI restriction site (indicated in boldface) for easy identification. All constructs were confirmed by DNA sequencing.
FIG. 3.
Schematic representation of JcDNV ORF4 (A) showing the positions of the two first ATG codons and the NsiI restriction sites used to delete the NsiI restriction fragment (B), to mutagenize the first (C), the second (D), and both ATG codons (E), and the TAA stop codon for complete deletion of the NS-3 coding sequence (F). Putative NS-3 polypeptides generated by constructs pJNS3ΔNsiI, pJNS3mATG1, and pJNS3mATG2 are indicated by hatched boxes (▨).
JcNS-3 and MlNS-3 expression system.
The JcDNV NS-3 coding sequence (nt 5361 to 4667 of the JcDNV sequence, see Fig. 2A) was amplified by PCR with the primer pair 5′-GGTGTCTCGCGGCCGCATCTGTAATTAATGTCTAC-3′ and 5′-GAATAGGATCCGTATGTCTATTGCCAGG-3′ containing NotI and BamHI restriction sites (indicated in boldface) near their 5′ extremities, respectively. The purified PCR product was double digested by NotI and BamHI and inserted at the BamHI-NotI restriction sites of pA3ΔSmATG, a plasmid derived from pBmAcat (18) containing a shortened sequence of the Bombyx mori cytoplasmic actin 3 promoter, the ATG translation initiation codon of actin 3 mutated to AGG, and a downstream multiple cloning site (F. X. Jousset, unpublished results). In this construct (pA3JcNS3), NS-3 is inserted with its own ATG and expressed under the control of B. mori A3 promoter. The Mythimna loreyi DNV (MlDNV) NS-3 coding sequence was amplified by PCR with pMl28, a plasmid encompassing the MlDNV sequence (G. Fédière, unpublished results) as a template and the primer pair 5′-CCAGAAACTCTGGATCCTATGTCTATTGC-3′ and 5′-TCTCCATCGCGGCCGCGTAATTAATGTCTGC-3′. The PCR product was digested with BamHI and NotI and inserted into pA3ΔSmATG as described above to generate pA3MlNS3.
FIG. 2.
(A) Nucleotide and amino acid sequence of NS-3 protein showing (i) the two first ATG codons and the TAA stop codon (boldface italic characters) mutated to generate pJNS3mATG1, pJNS3mATG2, pJNS3mATG1+2, and pJΔNS3 constructs, respectively; (ii) the two NsiI restriction sites used to generate pJNS3ΔNsiI construct; (iii) the oligopeptidic sequence used to prepare a specific anti-NS3 antiserum (boldface characters); (iv) putative zinc-finger motifs; (v) N-glycosylation sites (NXT/NXS); and (vi) phosphorylation site (RRYS) (underlined). (B) Alignment of NS-3 sequences of JcDNV (the present study), MlDNV(Fédière, unpublished), GmDNV (GenBank accession number NC_004286), and DsDNV (GenBank accession number NC_001899) by using the Multalin program (8).
NS-1, NS-2, and NS-3 cloning and production of antisera.
The NS-1, NS-2 and NS-3 coding sequences (nt 4656 to 3019, nt 4649 to 3822, and nt 5361 to 4667 of the JcDNV genome, respectively) were amplified by PCR with the following primer pairs: 5′-CGAGGATCCATGAACAATGGAGACACCAAC-3′ and 5′-GAGAAGCTTACGGTAATGGAGCTACTCTTGG-3′ for NS-1, 5′-TTACAGAGGATCCATGGAGACACCAAC-3′ and 5′-GACACATAATAAGCTTTTAATAACGCTTTTGTC-3′ for NS-2, and 5′-CAGAATATAGGATCCATGTCTATTGCCAG-3′ and 5′GGTGTCTCCAAGCTTCATCTGTAATTAATGTCTAC-3′ for NS-3. The PCR products digested with BamHI and HindIII were inserted at the BamHI and HindIII sites of pMal-C2 X vector (New England Biolabs, Inc., Beverly, Mass.). In these constructs, NS-1, NS-2, and NS-3 sequences are in frame with the malE gene encoding the maltose-binding protein (MBP). MBP-NS1, MBP-NS2, and MBP-NS3 fusion proteins were produced in Escherichia coli TB1 cells and purified by amylose affinity chromatography according to the manufacturer's instructions (New England Biolabs). Antisera were prepared against these purified proteins by injecting rabbits with 1 mg of the recombinant protein emulsified in Freund incomplete adjuvant. Two booster injections prepared in Freund incomplete adjuvant were given at 2-week intervals. Another antiserum was prepared against NS-3 by using the synthetic oligopeptide NH2-CIYENSPNKKRRYSSSS-COOH (amino acids [aa] 198 to 213) coupled to keyhole limpet hemocyanin (EPYTOP, Nimes, France).
Labeling and immunoprecipitation of NS-3.
At different intervals posttransfection, pBRJ-transfected Ld 652 cells (2 × 106 cells/25-cm2 flask) were washed three times with methionine-cysteine-free TC100 medium, and each flask was refed with 2.5 ml of methionine-cysteine-free TC 100 medium plus 20 μCi of Tran35S-label (ICN)/ml, followed by incubation at 28°C for 3 h. Cells were then harvested, and NS-3 was immunoprecipitated by using an antiserum prepared against NS-3 oligopeptide (50 μl/ml) and a protein A immunoprecipitation kit (Roche Biochemicals) according to the manufacturer's instructions. Immunoprecipitated NS-3 protein labeled with [35S]methionine-[35S]cysteine was separated by electrophoresis on a 12% polyacrylamide gel, and the gel was dried and autoradiographed at room temperature for 16 h.
SDS-PAGE and immunoblotting.
Ld 652 cells transfected with pBRJ or pJΔNS3 constructs were harvested, sedimented by centrifugation, and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer (125 mM Tris [pH 6.8], 2% SDS, 5% β-mercaptoethanol, 50% glycerol, 0.01% bromophenol blue). After denaturation at 100°C for 5 min, proteins were separated by SDS-PAGE (on a 12% acrylamide gel). MBP-NS3 fusion protein digested or not by protease Factor Xa (New England Biolabs) was also subjected to SDS-PAGE.
After separation, proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) by semidry blotting. Membranes were blocked by 30 min of incubation with 5% lowfat milk powder and 0.05% Tween 20 in TS buffer (0.05 M Tris, 0.2 M NaCl) at room temperature, incubated with a rabbit anti-NS-1 or NS-2 or NS-3 immune serum (diluted 1/250) for 40 min at room temperature, washed three times in phosphate-buffered saline containing 0.5% Tween 20, and further incubated with goat anti-rabbit antibody (diluted 1/500) conjugated with peroxidase (Diagnostics Pasteur) as a second antibody. Blots were stained by the 3-amino-9-ethylcarbazol chromogenic reaction to peroxidase (Sigma).
Transfection and infection of S. littoralis larvae and virus assays.
Transfection and infection of third-instar S. littoralis larvae were performed according to previously reported protocols (19). Briefly, larvae were transfected with pBRJ and pBRJ-derived constructs by injecting a mixture of plasmid DNA (ca. 1 μg per larva) and DEAE-dextran (2 mg/ml). For virus assays, Ld 652 cells transfected with different constructs were harvested 7 days posttransfection and sonicated, and the virions were pelleted by centrifugation (164,000 × g, 35 min) and resuspended in 100 μl of phosphate-buffered saline. Third-instar larvae were infected by inoculation of 13 μl of a diluted (1:10) viral suspension. The larvae were fed an artificial medium and maintained at 25°C and 70% relative humidity. Dead larvae were collected daily and kept at −20°C, and at 15 days posttransfection insects still living were frozen. Each larva or nymph was homogenized individually, and the presence of virus particles was assessed by enzyme-linked immunosorbent assay (ELISA) with an anti-JcDNV capsid protein antiserum.
Cell transfections.
Lymantria dispar Ld 652 cells (16) were maintained at 27°C in TC100 medium supplemented with 10% heat-inactivated fetal calf serum. Cells were transfected with plasmid DNA (10 μg per 25-cm2 tissue culture flask) by using DOTAP liposomal transfection reagent (Roche Biochemicals) according to a previously described protocol (20). For cotransfection experiments, 1 μg of pJΔNS3 DNA was mixed with 10 μg of pA3JcNS3 or pA3MlNS3 DNA, and transfections were completed as described here.
Analysis of intracellular JcDNV DNA.
At different times posttransfection, cells (8 × 105 to 2 × 106 cells/sample) were harvested and peleted by centrifugation (3,400 rpm for 5 min). The cell pellets were treated according to Hirt's protocol (17). Cells were resuspended in 0.4 ml of lysis solution (10 mM Tris-HCl [pH 7.4], 0.1 M EDTA, 0.6% SDS), and 0.25 ml of 5 M NaCl was added. After these components were mixed, a 10% volume of proteinase K (2 mg/ml) was added, followed by incubation for 10 min at 37°C, followed by further incubation for 60 min at 50°C and overnight at 4°C. The mixture was centrifuged at 400,000 × g for 5 min to pellet the cell debris and the cellular DNA. The low-molecular-weight DNA was phenol extracted from the supernatant and precipitated by a 0.7 volume of isopropanol. The DNA pellet was rinsed with 70% ethanol and finally resuspended in 30 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Then, 15 μl of extracted DNA was treated with restriction enzyme DpnI (Promega) to digest the plasmid DNA and subjected to electrophoresis through a 1% agarose gel in TEP buffer (90 mM Tris-phosphate, 20 mM EDTA; pH 8). The DNA was transferred to a positively charged nylon membrane (Roche Biochemicals) by the method of Southern (25). After prehybridization at 68°C for 2 h, the membrane was hybridized with digoxigenin-labeled JcDNV DNA probe by random priming (15). The blots were developed according to the supplier protocol (Roche Biochemicals).
RESULTS
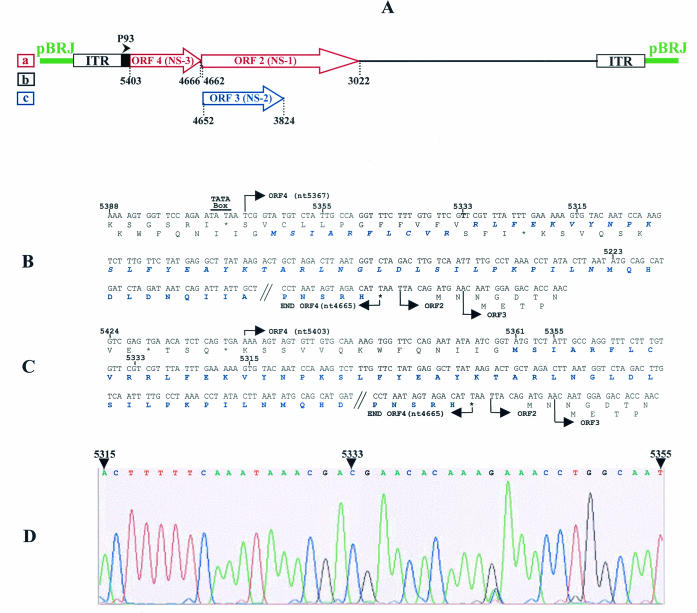
Correction of the JcDNV sequence.
The JcDNV genome shares with the bona fide members of the genus Densovirus a unique organization of the NS coding sequences in three ORFs located in the 5′ half of the strand complementary to the strand coding for structural polypeptides. The leftmost, ORF4, codes for NS-3, and two overlapping ORFs, ORF2 and ORF3, encode NS-1 and NS-2, respectively (Fig. 1A). ORF4 and ORF2 are in the same frame and are separated by only a TAA stop codon. Consistent with this organization, NS polypeptides are translated from two mRNAs, a 2.4-kb unspliced mRNA for NS-3 expression and a spliced 1.7-kb mRNA expressing NS-1 and NS-2 (unpublished data).
FIG. 1.
(A) Organization of the JcDNV sequence cloned in pBRJ showing the strand encoding the NS polypeptides NS-1, NS-2, and NS-3 under control of the P93 (map unit 93 of the viral sequence) promoter. The figures refer to the beginnings and ends of ORF2, ORF3, and ORF4 relative to the viral sequence (11). a, b, and c refer to the frames. (B and C) Sequence of the JcDNV ORF4 as previously published (11) (B) and after rectification (C). (D) Detail of the chromatography profile of NS-3 nucleotidic sequence in the region where the error was detected.
In the JcDNV sequence published by Dumas et al. (11), ORF4 starts at nt 5367 but the first in-frame ATG codon is only at nt 5223, i.e., 48 codons downstream (Fig. 1B). When we compared this sequence to that of Galleria mellonella DNV (GmDNV) ORF4 (GenBank accession number NC_004286), we noticed that, unlike JcDNV NS-3, GmDNV NS-3 coding sequence started very close to the beginning of ORF4 and that its NH2-terminal sequence showed strong homology with the untranslated 48 codons of the 5′-terminal sequence of JcDNV ORF4. This prompted us to verify the JcDNV sequence in this region. We found that the published sequence contained an “extra” A (T on the complementary strand) at position 5333 that is not present in the sequence (Fig. 1D) and which artificially introduced a frameshift in the N-terminal sequence. After we rectified this error, ORF4 was found to span from nt 5403 to nt 4665, and the coding sequence of NS-3 consisted of 232 aa residues starting at the ATG initiation codon at position 5361 and ending at the TAA stop codon at position 4665 (Fig. 1C). This sequence presents some interesting features including two zinc-finger motifs [C:(X)2:C:X3:C (aa 144 to 151) and C:(X)2:C:H:X:C (aa 184 to 190)], two putative N glycosylation sites (NXT/NXS, aa 162 to 164 and 194 to 196), and a putative phosphorylation site RRYS (aa 207 to 210) mediated by a cyclic AMP-dependent protein kinase (Fig. 2A). Comparisons of this sequence with those of the databases showed strong homologies with NS-3 of MlDNV (89% identity), GmDNV (85% identity) and, to a lesser extent, of Diatraea saccharalis DNV (67% identity) (Fig. 2B). Curiously, the DsDNV NS-3 sequence is truncated of the 34 C-terminal amino acid sequence common to NS-3 of JcDNV, MlDNV, and GmDNV. No further significant homologies could be detected with NS polypeptides of Densovirinae and Parvovirinae or with any protein of the data banks.
ORF4 is essential for virus infectivity.
To get an insight into the function of NS-3, we generated a series of mutants in the ORF4 coding sequence. All of the constructs derived from pBRJ (Fig. 1A), a plasmid encompassing an infectious sequence of the JcDNV genome (19). Plasmid pJNS3ΔNsiI (Fig. 3B) contains an NS-3 sequence with a deletion of a 112-bp NsiI internal fragment. This deletion interrupts the NS-3 coding sequence immediately after aa 94 by introducing a TGA stop codon at position 4967. The first or (and) the second ATG codons of ORF4 were mutated by site-directed mutagenesis to generate pJNS3mATG1, pJNS3mATG2, and pJNS3mATG1+2 constructions (Fig. 3C, D, and E, respectively). Complete deletion of the coding sequence of NS-3 (pJΔNS3 construction, Fig. 3F) was achieved by site-directed mutagenesis of NS-3 ATG1 initiation and TAA stop codons. Details on these constructions, including the primers used to generate them, are given in Materials and Methods. All of the constructs were sequenced in the appropriate regions to verify the presence of the expected mutations or deletions.
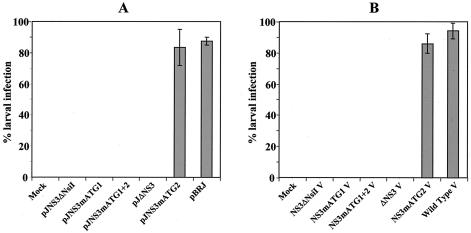
These constructs were transfected to groups of 12 third-instar S. littoralis larvae (1 μg of plasmid DNA/larva) according to a previously described protocol (19). At 2 weeks posttransfection, the presence of virus particles in individual insect was assessed by ELISA with a specific antiserum prepared against purified virions. As shown in Fig. 4A, partial or complete deletion of the NS-3 coding sequence (pJNS3ΔNsiI and pJΔNS3 constructs) or punctual mutations of ATG1 or ATG1+2 (pJNS3mATG1 and pJNS3mATG1+2 mutants) completely abolished the capacity of the constructs to generate a productive infection in transfected larvae. In contrast, averages of 84 and 86% of larvae transfected with pJNS3mATG2 and pBRJ DNA, respectively, were found to be positive by ELISA. In a parallel experiment, the five constructs were transfected to sensitive Ld 652 cells. Five days later, the cells were sonicated, and virions were pelleted and used to infect either Ld 652 cells or groups of 12 third-instar S. littoralis larvae. Virus production in cells was examined 3 days postinfection by using an immunofluorescence test (20). Immunofluorescence-positive cells were detected only in cell cultures infected with supernatant from pBRJ- and pJNS3mATG2-transfected cells (data no shown). Similarly, larval infection was monitored by collecting dead larvae or pupae for 15 days, and virus production was assessed by ELISA. The results (Fig. 4B) show that 86 and 94% of the larvae were found to be positive after inoculation with supernatants from pJNS3mATG2- or pBRJ-transfected cells, respectively. In contrast, not a single larva was detected as positive after injection with supernatants from pJNS3ΔNsiI-, pJΔNS3-, pJNS3mATG1-, and pJNS3mATG1+2-transfected cells. Taken together, these results clearly demonstrated that full-length NS-3 is absolutely required for productive infection both in cell culture and in animals. Mutation of the first ATG codon is enough to completely abolish the production of virus particles, whereas mutation of the second ATG alone does not affect the capacity of the virus to enter on a productive cycle. Thus, if we assume that in the absence of the first AUG codon the second AUG could act as a translational initiation codon, the function of the resulting polypeptide with a deletion of the first 46-aa sequence (Fig. 3C) is completely abolished. Similarly, expression of the 94 N-terminal amino acids of NS-3, as it very likely occurs in pJNS3ΔNsiI-transfected cells (Fig. 3B), is not sufficient to promote virus production.
FIG. 4.
(A) Infectivity tests of pJNS3ΔNsiI, pJΔNS3, pJNS3mATG1, pJNS3mATG1+ATG2, pJNS3mATG2, and pBRJ constructs transfected to fourth-instar S. littoralis larvae. Mock, mock-transfected larvae. Purified plasmid DNA (10 μg/larva) was injected with DEAE-dextran transfection reagent. At 15 days posttransfection, virus infection was detected in larval-pupal homogenates by an ELISA. Error bars indicate the standard errors of the means of at least three experiments. (B) Infectivity tests of virions produced in Ld 652-transfected cells. Five days posttransfection with the different constructs, cells were homogenized, and cell extracts were inoculated to fourth-instar S. littoralis larvae. Fifteen days later, virus infection was detected in larval-pupal homogenates by ELISA. Error bars indicate the standard errors of the means of at least three experiments.
NS-3 plays an essential role in viral DNA replication.
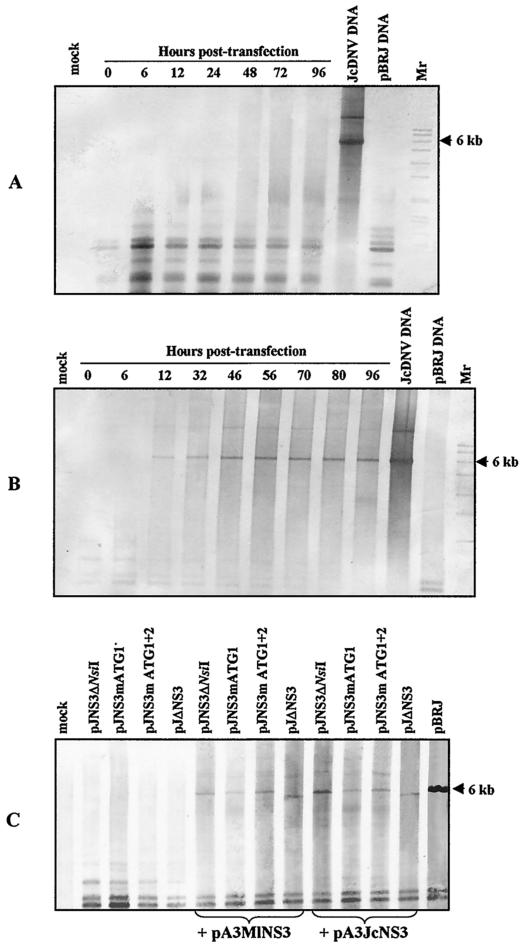
To further investigate the role of NS-3 and, in particular, to precisely determine at which step of the viral cycle its function is essential, we analyzed the effect of NS-3 deletion on DNA replication. After transfection of Ld 652 cells with pJΔNS3, the low-molecular-weight cell DNA (Hirt's supernatant) assumed to contain the replicative form (RF) of JcDNV genome was extracted at different intervals. After digestion by DpnI enzyme to fragment plasmid DNA and electrophoresis through a 1% agarose gel, the DNA was transferred to a nylon membrane and hybridized with a JcDNV DNA probe. No viral DNA replication could be detected by Southern blot analysis in cells transfected with pJΔNS3 during the 96-h course of the experiment (Fig. 5A). In contrast, viral DNA synthesis was clearly detected in pBRJ-transfected cells as soon as 12 h posttransfection (Fig. 5B).
FIG. 5.
(A and B) Time course (in hours) analysis of JcDNV RF DNA synthesis in pJΔNS3-transfected (A) and pBRJ-transfected (B) Ld 652 cells. Cells were harvested at different intervals posttransfection, low-molecular-weight DNA was extracted, digested with DpnI, and analyzed by Southern blotting with a viral DNA probe. The arrows indicate the 6-kbp RF of the viral DNA. Mock-transfected (Mock) cells served as a negative control; purified viral DNA and pBRJ DNA served as positive controls. Mr, molecular weight markers. (C) Southern blot analysis of JcDNV RF DNA extracted from Ld 652 cells cotransfected with NS-3 mutant constructs and either pA3JcNS3 or pA3MlNS3 DNA or cells transfected with the NS-3 mutant constructs only. Mock, mock-transfected cells. Approximately 2 × 106 cells were transfected with 1 μg of mutant plasmid DNA plus 10 μg of pA3JcNS3 or pA3MlNS3 or 1 μg of mutant plasmid DNA. At 3 days posttransfection, low-molecular-weight cell DNA was extracted, digested with DpnI, and analyzed by Southern blotting with a viral DNA probe. The arrow indicates the 6-kbp RF of the viral DNA. The less size of the viral DNA produced in pJΔNS3-transfected cells is clearly visible.
To confirm the role of NS-3 in viral replication, we cotransfected Ld 652 cells with each of the deletion constructs and a construct designated pA3JcNS3 expressing the JcDNV NS-3 coding sequence under the control of the B. mori actin 3 promoter known to be functional in this cell line (9). At 72 h posttransfection, we looked for viral RF DNA in the low-molecular-weight cell DNA as described above. The results presented in Fig. 5C clearly indicated that synthesis of viral DNA is fully recovered, regardless of the mutant plasmid used, by coexpression of NS-3 polypeptide in trans. Replication of viral DNA was also rescued by coexpression of the heterologous MlDNV NS-3 polypeptide (pA3MlNS3, Fig. 5C). As expected, no RF DNA was detected in cells transfected only with the mutant palsmids (Fig. 5C).
Immunodetection of NS-3 expression and impact on NS-1 and NS-2 synthesis.
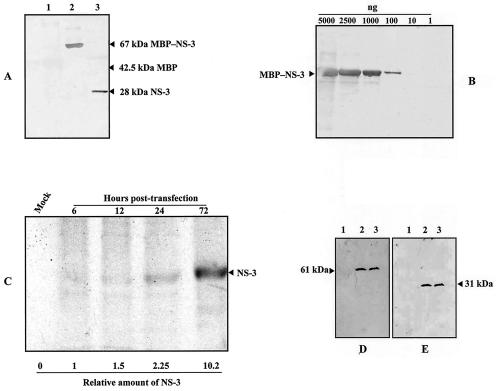
The results presented above suggested that NS-3 function occurred early in the viral life cycle. To confirm this point, attempts were made to detect NS-3 by immunological techniques. First, we looked for NS-3 expression in pBRJ-transfected Ld 652 cells by indirect immunofluorescence and Western blot analysis, with two anti-NS-3 immune sera prepared by injecting two rabbits with MBP-NS-3 fused protein produced in E. coli. None of these attempts was successful, probably because of the weakness of the antisera. A second attempt was made to obtain an antiserum by injecting a rabbit with the synthetic 16-mer oligopeptide (NH2-IYENSPNKKRRYSSSS-COOH [aa 198 to 213], Fig. 2A) located in the NS-3 C-terminal sequence. The specificity of this antiserum was assessed by Western blot with the chimeric MBP-NS-3 purified protein. As shown in Fig. 6A, the antiserum specifically recognized the NS-3 fraction after digestion of the fused MBP-NS-3 protein with Factor Xa, and approximate titration showed that up to 100 ng of MBP-NS-3 protein could be detected (Fig. 6B). Ultimately, we succeeded in detecting NS-3 in an immunoprecipitation assay by using this antiserum and 35S-labeled NS-3. The protein could be detected as early as 6 h posttransfection, and its expression increased with time during the 3 days of the experiment (Fig. 6C). Finally, we analyzed the impact of NS-3 deletion on the synthesis of the two other NS proteins, NS-1 and NS-2. Ld 652 cells transfected with pBRJ and pJNS3ΔNsiI were harvested 3 days posttransfection and analyzed by Western blotting with anti-NS-1 and anti-NS-2 antisera, respectively. As shown in Fig. 6D and E, no significant difference was observed in the level of expression of both proteins, suggesting that, at least at this time, NS-3 is not involved in the regulation of the two other NS polypeptides.
FIG. 6.
Immunodetection of NS proteins. (A) Detection by Western blot of purified MBP-NS-3 fusion protein (lane 2) or NS-3 protein alone after digestion with factor Xa (lane 3) by using a 1/250 dilution of the anti-NS-3 polyclonal antiserum prepared against a 17-mer synthetic oligopeptide as described in Materials and Methods. Lane 1, MBP control protein. (B) Dilutions of MBP-NS-3 fusion protein used to evaluate the strength of the antiserum. (C) Immunodetection of NS-3 in pBRJ-transfected Ld 652 cells. At various times (in hours) posttransfection, cells were labeled for 3 h with [35S]methionine-[35S]cysteine, and then NS-3 was immunoprecipitated by using 50 μl of anti-NS-3 antiserum and protein A, as described in Materials and Methods. The relative amounts of NS-3 were estimated by using the UTHSCSA ImageTool program. (D and E) Comparative analysis by Western blot of NS-1 and NS-2 expression in mock-transfected (lanes 1), pBRJ-transfected (lanes 2), and pJNS3ΔNsiI-transfected Ld 652 cells (lane 3). Three days posttransfection, cells were harvested, pelleted, and treated for analysis by SDS-PAGE (12% gel). Proteins were transferred onto nitrocellulose membranes, and NS-1 and NS-2 were revealed by using specific anti-NS-1 (D) and anti-NS-2 (E) antibodies.
DISCUSSION
The high level of sequence conservation of NS-3 protein among the members of the genus Densovirus (Fig. 2B) suggests that the biological function(s) of this protein is essential for the accomplishment of their life cycle. In the present study, we unambiguously demonstrated that the knockout of the JcDNV NS-3 gene either by site-directed mutagenesis of its ATG initiation codon or partial or complete deletion of its coding sequence prevented the accomplishment of a productive infectious cycle both in cell culture and in larvae. Further analysis clearly showed the involvement of NS-3 in the viral DNA replication since in the absence of NS-3 expression, no RF could be detected in cells transfected with constructs having partial or complete deletions of the NS-3 coding sequence (Fig. 5A and C). Definite confirmation of the essential role played by NS-3 in DNA replication was brought about by transcomplementation experiments showing that cotransfection of cells with an NS-3 deletion mutant plasmid and a plasmid expressing in trans homologous (JcDNV) or heterologous (MlDNV) NS-3 polypeptide restored this function (Fig. 5C). As expected for such a function, time course analysis showed that the NS-3 protein expression occurred early in the virus cycle. The increasing amount of NS-3 detected from day 1 to day 3 posttransfection (Fig. 6C) correlated well with the increase of viral RF DNA detected in pBRJ-transfected cells during the same period of time (Fig. 5B). Preliminary results of time course analysis indicated that the 2.4-kb mRNA responsible for NS-3 synthesis is always significantly less abundant than the spliced 1.7-kb mRNA encoding NS-1 and NS-2 (unpublished data). Thus, the amount of NS-3 compared to that of NS-1 and NS-2 appears to be regulated essentially by the ratio of unspliced to spliced NS mRNAs in an infected cell. This situation likely explains the difficulties we encountered in detecting NS-3 in pBRJ-transfected cells. The fact that the presence or absence of NS-3 does not affect the level of NS-1 and NS-2 expression indicates that this protein is not involved in the regulation of the two others.
Our results constitute a first step toward the understanding of NS-3 function(s) during the JcDNV life cycle. However, the precise level at which this polypeptide acts and its specific biological function(s) remain to be elucidated. The lack of significant homology between the NS-3 amino acid sequence and proteins of the data banks, including NS proteins of parvoviruses, implies that its function is either specific to this type of virus, i.e., of the genus Densovirus, or that it is somehow related to function(s) of the host cell necessary for their replication. At present, it is not possible to decide in favor of one of these hypotheses, which are not exclusive of one another. The two representatives of the genus Iteravirus, B. mori DNV and Casphalia extranea DNV, both of which lack NS-3 (14, 21), share with the members of the genus Densovirus the property of having as natural hosts exclusively lepidopteran insects (13). The essential difference between these two groups of viruses at the biological level lies in their host range and tissue tropism. Whereas B. mori DNV and C. extranea DNV have a restricted host range (B. mori and C. extranea, respectively) and replicate almost exclusively in the midgut epithelial cells (12, 29), members of the genus Densovirus have usually a wide host spectrum and, more important, replicate in most larval tissues, with the exception of midgut (2, 3, 28). As recently clearly demonstrated for vertebrate parvoviruses, these differences might reflect specific requirements for cellular proteins functionning in parternship with NS and (or) VP proteins in order to achieve the viral replicative cycle, as has been recently demonstrated for vertebrate parvoviruses (6, 7, 22, 23, 24, 30, 31).
The hypothesis that the role of NS-3 reflects intrinsic properties of the JcDNV genomic structure is supported by the unique property of the bona fide members of the genus Densovirus to possess a long (>500-nt) ITR. The folding and annealing of complementary sequences within the ITR is able to generate numerous complex stem-loop structures (10, 11). The folding of the terminal 96 nt forms a typical T-shaped hairpin structure assumed to function as the origin of replication. Demonstration of the in vitro binding of JcDNV NS-1 to a specific sequence in the A′ region of this structure, of strand- and site-specific nicking, and of an ATP-dependent DNA helicase activities of this polypeptide similar to those described for vertebrate parvoviruses, provided good evidence for a replicative model of this virus (10). The cis-acting function(s) of the 420 to 430 nt of the ITR downstream of the terminal hairpin is still unknown, and it is tempting to speculate that NS-3 could somehow interact either alone or in synergy with NS-1 or NS-2 or a cellular partner(s) involved in DNA replication by binding to one of the stem-loop structures downstream of the terminal hairpin. Investigations to verify these hypotheses are under way.
Acknowledgments
We thank Christine Lopez for skillful technical assistance.
A.A.-A. was a recipient of a doctoral fellowship from the Egyptian government.
REFERENCES
- 1.Bando, H., J. Kusuda, T. Gojobori, T. Maruyama, and S. Kawase. 1987. Organization and nucleotide sequence of a densovirus genome imply a host-dependent evolution of the parvoviruses. J. Virol. 61:553-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergoin, M., and N. Bres. 1968. Lésions tissulaires chez la larve du lepidiptère Galleria mellonela L. atteinte du virus de la densonucléose. Bull. Apicole. 11:5-12. [Google Scholar]
- 3.Bergoin, M., and P. Tijssen. 1998. Biological and molecular properties of densoviruses and their use in protein expression and biological control, p. 141-169. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing, Inc., New York, N.Y.
- 4.Bergoin, M., and P. Tijssen. 2000. Molecular biology of Densovirinae, p. 12-32. In S. Faisst and J. Rommelaere (ed.), Parvoviruses from molecular biology to pathology and therapeutic uses. Karger, Basel, Switzerland. [DOI] [PubMed]
- 5.Berns, K. I., M. Bergoin, M. Bloom, M. Lederman, N. Muzyczka, G. Siegl, J. Tal, and P. Tattersall. 2000. Family Parvoviridae, p. 311-323. In M. H. V. Van Regenmortel, C. M. Fauquet, D. H. L. Bishop, et al., Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, Inc., San Diego, Calif.
- 6.Bodendorf, U., C. Cziepluch, J. C. Jauniaux, J. Rommelaere, and N. Salome. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulon-Bublex, M. N. Mounier, P. Couble, and J. C. Prudhomme. 1993. Cytoplasmic actin A3-gene promoter injected as supercoiled plasmid is transiently active in Bombyx mori embryonic vitellophages. Rouxs Arch. Dev. Biol. 202:123-127. [DOI] [PubMed] [Google Scholar]
- 10.Ding, C., M. Urabe, M. Bergoin, and R. M. Kotin. 2002. Biochemical characterization of Junonia coenia densovirus nonstructural protein NS-1. J. Virol. 76:338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumas, B., M. Jourdan, A. M. Pascaud, and M. Bergoin. 1992. Complete nucleotide sequence of the cloned infectious genome of Junonia coenia densovirus reveals an organization unique among parvoviruses. Virology 191:202-222. [DOI] [PubMed] [Google Scholar]
- 12.Fédière, G. 1983. Recherches sur des viroses épizootiques de lépidoptères Limacodidae ravageurs de palmacées. Ph.D. dissertation. Universite ScienceTechnologie, Languedoc, Montpellier, France.
- 13.Fédière, G. 2000. Epidemiology and pathology of Densovirinae, p. 1-11. In S. Faisst and J. Rommelaere (ed.), Parvoviruses from molecular biology to pathology and therapeutic uses. Karger, Basel, Switzerland.
- 14.Fédière, G., Y. Li, Z. Zadori, J. Szelei, and P. Tijssen. 2002. Genome organization of Casphalia extranea densovirus, a new iteravirus. Virology 292:299-308. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin, R. H., J. R. Adams, and M. Shapiro. 1990. Replication of the entomopoxvirus from Amsacta moorei in serum-free cultures of a gypsy moth cell line. J. Invertebr. Pathol. 56:190-205. [Google Scholar]
- 17.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R., R. G. Meidinger, and K. Iatrou. 1992. A cellular promoter-based expression cassette for generating recombinant baculoviruses directing rapid expression of passenger genes in infected insects. Virology 190:815-823. [DOI] [PubMed] [Google Scholar]
- 19.Jourdan, M., F. X. Jousset, M. Gervais, S. Skory, M. Bergoin, and B. Dumas. 1990. Cloning of the genome of a densovirus and rescue of infectious virions from recombinant plasmid in the insect host Spodoptera littoralis. Virology 179:403-409. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., F. X. Jousset, C. Giraud, F. Rolling, J. M. Quiot, and M. Bergoin. 1996. A titration procedure of the Junonia coenia densovirus and quantitation of transfection by its cloned genomic DNA in four lepidopteran cell lines. J. Virol. Methods 57:47-60. [DOI] [PubMed] [Google Scholar]
- 21.Li, Y., Z. Zadori, H. Bando, R. Dubuc, G. Fediere, J. Szelei, and P. Tijssen. 2001. Genome organization of the densovirus from Bombyx mori (BmDNV-1) and enzyme activity of its capsid. J. Gen. Virol. 82:2821-2825. [DOI] [PubMed] [Google Scholar]
- 22.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuesch, J. P., S. Lachmann, R. Corbau, and J. Rommelaere. 2003. Regulation of minute virus of mice NS1 replicative functions by atypical PKCλ in vivo. J. Virol. 77:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raab, U., K. Beckenlehner, T. Lowin, H. H. Niller, S. Doyle, and S. Modrow. 2002. NS1 protein of parvovirus B19 interacts directly with DNA sequences of the p6 promoter and with the cellular transcription factors Sp1/Sp3. Virology 293:86-93. [DOI] [PubMed] [Google Scholar]
- 25.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 26.Tijssen, P., and M. Bergoin. 1995. Densonucleosis viruses constitute an increasingly diversified subfamily among the parvoviruses. Semin. Virol. 6:347-355. [Google Scholar]
- 27.Tijssen, P., J. van den Hurk, and E. Kurstak. 1976. Biochemical, biophysical, and biological properties of densonucleosis virus. I. Structural proteins. J. Virol. 17:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vago, C., J.-L. Duthoit, and F. Delahaye. 1966. Les lésions nucléaires de la “virose à noyaux denses” du lépidoptère Galleria mellonella. Arch. Gesamte Virusforsch. 18:344-349. [Google Scholar]
- 29.Watanabe, H., S. Maeda, M. Matsui, and T. Shimizu. 1976. Histopathology of the midgut epithelium of the silkworm, Bombyx mori, infected with a newly isolated virus from the flacherie diseased larvae. J. Sericult. Sci. Jpn. 45:29-34. [Google Scholar]
- 30.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 76:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice small nonstructural protein NS2 interacts and colocalizes with the Smn protein. J. Virol. 76:6364-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]