Abstract
Cytomegalovirus (CMV) is known to rapidly induce activation of nuclear factor κB (NF-κB) after infection of fibroblast and macrophage cells. NF-κB response elements are present in the enhancer region of the CMV major immediate-early promoter (MIEP), and activity of the MIEP is strongly upregulated by NF-κB in transient-transfection assays. Here we investigate whether the NF-κB-dependent pathway is required for initiating or potentiating human and murine CMV replication in vitro. We show that expression of a dominant negative mutant of the inhibitor of NF-κB-alpha (IκBαM) does not alter the replication kinetics of human or mouse CMV in cultured cells. In addition, mouse embryo fibroblasts genetically deficient for p65/RelA actually showed elevated levels of MCMV replication. Mutation of all NF-κB response elements within the enhancer of the MIEP in a recombinant mouse CMV containing the human MIEP (hMCMV-ES), which we have previously shown to replicate in murine fibroblasts with kinetics equivalent to that of wild-type mouse CMV, did not negatively affect replication in fibroblasts. Taken together, these data show that, for CMV replication in cultured fibroblasts activation of the canonical NF-κB pathway and binding of NF-κB to the MIEP are dispensable, and in the case of p65 may even interfere, thus uncovering a previously unrecognized level of complexity in the host regulatory network governing MIE gene expression in the context of a viral infection.
Cytomegalovirus (CMV), the prototypic betaherpesvirus, shows sequential regulation of gene expression, as is the case for all the members of family Herpesviridae (35). CMV replication begins with expression of the immediate-early (IE/α) proteins shortly after infection and is dependent upon the host RNA polymerase. Expression of the CMV ie genes leads to expression of viral transactivators that promote induction of CMV early (β) genes and eventually late (γ) genes, which are expressed after DNA replication has been initiated. Transcription of the major ie genes is controlled by the major immediate-early promoter (MIEP). The enhancer of the MIEP is a highly complex region, and its activity depends on a diverse number of positive and negative cis-acting elements. Although the MIEP enhancers of mouse and human CMV (MCMV and HCMV, respectively) show limited homology at the nucleotide level, they share many common functional regulatory elements (6, 13). The MIE proteins in HCMV and MCMV (IE1/IE2 and IE1/IE3, respectively) have been shown to transactivate viral and host gene expression as well as to regulate their own expression through binding the MIEP (24, 30, 35, 46-48). The MIEP enhancer of MCMV has been shown to play an important yet dispensable role in initiation and potentiation of viral replication in tissue culture (2) and more recently has been found to be absolutely essential for in vivo growth (14). Replacement of the MIEP enhancer of MCMV with the paralogous region from HCMV does not restrict replication of this MCMV “enhancer swap” virus in tissue culture (2) or in vivo (20). Directed mutagenesis of the HCMV MIEP has identified the distal region of the enhancer as important for optimal expression of IE genes and viral replication at low multiplicity of infection (32, 33). Accordingly, expression of the MIE proteins is required for efficient CMV growth (1, 17, 32, 34, 36), and significant work has been done to delineate what specific host factors are required for the initiation of ie gene expression.
The nuclear factor κB (NF-κB) family of transcription factors (including c-Rel, p65/RelA, RelB, p50/NF-κB1, and p52/NF-κB2) is critical for mounting both innate and adaptive host immune responses to pathogen infection through induction of various “inflammatory” genes (15). Stimulation of cells with agonists (including microbial products, viruses, or host cytokines) can trigger various signal transduction pathways, ultimately leading to activation of the inhibitor of NF-κB kinase (IKK) complex (25). The “classical” IKK complex is composed of IKKα, IKKβ, and IKKγ, and the different roles of these proteins in regulating the various NF-κB family members when responding to specific extracellular and intracellular signals is a topic of heated study. In the case of the canonical NF-κB pathway (p50/p65 dependent) (15), the IKK complex can directly phosphorylate the inhibitor of NF-κB-alpha (IκBα) on two serine residues (S32 and S36), resulting in ubiquitin-mediated degradation of IκBα, translocation of p50/p65 to the nucleus, and initiation of transcription through binding to NF-κB response elements. Importantly, although the majority of studies in the literature to date have concentrated on IKKαβγ-mediated activation of the canonical NF-κB pathway, newly identified homologues of the IKKs (40) and more detailed elucidation of alternative or noncanonical NF-κB signaling pathways activated by tumor necrosis factor (TNF) family cytokines (11, 26) highlights the enormous degree of complexity, which ultimately results in the ability of this pathway to translate the input of multiple stimuli into specific downstream effects.
Infection of target cells by HCMV results in the modulation of hundreds of cellular genes (7, 53), and increased numbers of genes are upregulated when cells are exposed to replication-inactivated HCMV (7). This result indicates that infection itself, in the absence of viral gene expression, activates multiple cellular signaling pathways which lead to transcriptional induction of host and viral genes. It has been shown by several groups that HCMV infection of primary fibroblasts and monocyte-derived cell lines induces rapid (within 15 to 30 min) translocation of NF-κB to the nucleus and activates NF-κB-dependent transcription (9, 27, 43, 45, 53, 54). NF-κB-responsive elements present within 18-bp repeat sequences in the MIEP enhancer bind NF-κB and activate transcription of reporter genes (43, 45). IE1 and IE2 have been shown to transactivate both the p50 and p65 promoters, resulting in increased levels of these proteins at later times (24 to 48 h) in the HCMV replication cycle (55). Finally, it has recently been reported that the overexpression of a constitutively active form of the mitogen-activated protein kinase kinase kinase-1 results in increased expression from the MIEP and that this condition depends upon the NF-κB binding elements in the 18-bp repeat sequences (49).
Less is known regarding the activation and regulation of NF-κB in MCMV-infected cells. The MIEP of MCMV shows high-level constitutive activity similar to its counterpart in HCMV (13), and multiple consensus NF-κB response elements are contained in its sequence (13, 44). Infection of murine fibroblasts with MCMV has been shown to result in translocation of NF-κB (p50/p65) to the nucleus, similar to that seen for HCMV (18). Taken together, these data suggest a potentially important role for NF-κB in regulating both human and murine CMV ie gene expression and replication.
On the basis of these studies, it has become accepted that activation of NF-κB upon a primary CMV infection “kick starts” CMV replication at the immediate-early phase, and transcriptional induction by CMV IE proteins of the p50/p65 subunits of NF-κB may help to promote viral replication through the early and late phases. Here we provide direct lines of experimental evidence showing that mutating the NF-κB response elements in the MIEP and inhibition of the canonical NF-κB-dependent pathway does not hinder transcription-replication of HCMV or MCMV in cultured fibroblasts. Instead, our results clearly demonstrate the regulatory robustness of the CMV enhancer and suggest a potential p65-dependent suppressive role of the NF-κB pathway, thereby providing a new framework for understanding the host regulatory network of ie gene expression.
MATERIALS AND METHODS
Cells and virus.
Normal human dermal fibroblasts (NHDF) from neonatal foreskin (Clonetics, San Diego, Calif.) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco/BRL) supplemented with 10% fetal bovine serum, 5 μg of insulin per ml, and 1 ng of basic fibroblast growth factor (Sigma, St. Louis, Mo.) per ml. Mouse NIH 3T3 cells (ATCC CRL1658) were grown in DMEM supplemented with 10% newborn calf serum, and J774A.1 cells (ATCC TIB 67) and immortalized p65+/+ and p65−/− murine embryonic fibroblasts (MEFs) (kind gift of A. Beg) (3) were grown in DMEM supplemented with 10% fetal calf serum. MCMV (Smith strain, ATCC VR-1399) and bacterial artificial chromosome (BAC)-derived MCMVs were propagated on NIH 3T3 cells for generating viral stocks, and titers were determined by standard plaque assays in NIH 3T3 cells. HCMV strain AD169-ATCC was acquired from the American Type Culture Collection, and the Toledo strain was a kind gift from S. Starr (passage 10). HCMV stocks were prepared by low-multiplicity-of-infection (MOI) challenge of NHDF, combining sedimented extracellular virus (25,000 × g) with cell-associated virus (generated by cell sonication and removal of cell debris by low-speed centrifugation). To avoid contamination of viral preparations by soluble cellular factors, HCMV was then pelleted (25,000 × g) through a 20% sorbitol cushion. HCMV titers were determined by limiting-dilution plaque assays in NHDF.
Generation of MCMV mutants.
Plasmid pIE111H carries the MCMV sequences from nucleotide (nt) 177008 (44) (EcoRI) to 187889 (HindIII) with a replacement of the MCMV enhancer sequences (MCMV nt 182940 to 184086; −48 to −1191 relative to the MCMV ie1/ie3 transcription start site) by the corresponding sequences of the HCMV enhancer (−52 to −667 relative to the HCMV ie1/ie2 transcription start site) (2). In order to introduce the HCMV enhancer sequences into the MCMV BAC pSM3fr, the 1.0-kbp MluI fragment of plasmid pEnhDel (2) was replaced with the 2.1-kbp MluI fragment of plasmid pIE111H, and an 8.6-kbp SalI fragment of the resulting plasmid was then transferred to the shuttle plasmid pST76-ASacB.
The four putative NF-κB binding sites in the 613-bp HCMV enhancer sequence of plasmid pIE111H-NF-κB were destroyed by site-directed mutagenesis. Unique restriction sites were introduced during mutagenesis (−98 to −103 StuI, −161 to −166 KpnI, and −265 to −270 and −416 to −421 BglII), and mutated sequences are illustrated in Fig. 1A. A 2.1-kbp MluI- fragment carrying the mutated NF-κB response elements was transferred into pEnhDel and finally into pST76-ASacB exactly as described for the corresponding DNA fragment of pIE111H.
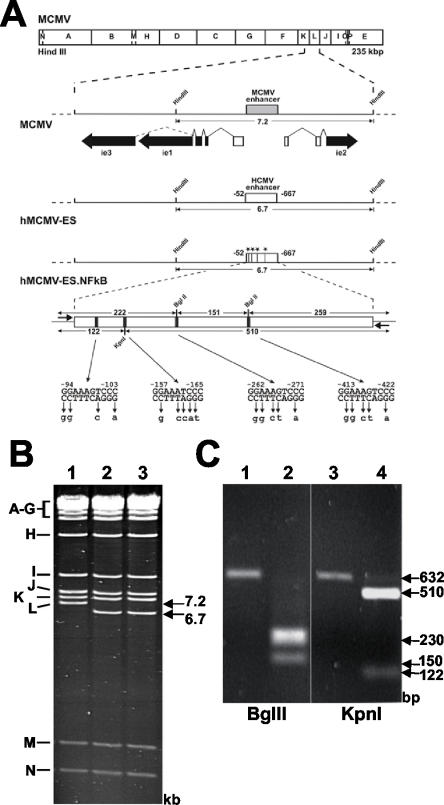
FIG. 1.
Mutagenesis of the HCMV-MIEP NF-κB binding sites in hMCMV-ES. (A) Organization of the MIE regions of the constructed MCMV BAC genomes. The HindIII map of the MCMV genome is given at the top. The expanded map below represents the MIE region of MCMV with the enhancer (gray box) and the exon-intron structure of the ie1, ie2, and ie3 genes indicated. Coding and noncoding exons are depicted as black and white boxes, respectively. The BAC plasmids hMCMV-ES and hMCMV-ES.NF-κB carry a 616-bp fragment (white rectangle) from the HCMV MIE promoter in replacement of the MCMV enhancer. In hMCMV-ES.NF-κB, the NF-κB binding sites were mutated, introducing unique restriction sites (−98 to −103 StuI, −161 to −166 KpnI, and −265 to −270 and −416 to −421 BglII), which are indicated by asterisks or black bars. The sizes of the expected HindIII L fragments of the MCMV BAC genomes and the sizes of the expected BglII and KpnI fragments of the PCR-amplified HCMV enhancer (with primers shown with short arrows flanking the enhancer) are indicated by the arrows. Coordinates are given relative to the MCMV ie1/ie2 transcription start site. The diagrams are not drawn to scale. (B) Ethidium bromide-stained agarose gel of HindIII-digested MCMV BAC plasmid (1), hMCMV-ES (2), and hMCMV-ES.NF-κB (3). The names and sizes of the HindIII fragments of the MCMV genome are indicated in the margins. (C) To verify successful mutagenesis of NF-κB sites, enhancer sequences were amplified from MCMV-ES (lanes 1 and 3), and hMCMV-ES.NF-κB (lanes 2 and 4)BACs by PCR. The amplified products were digested with restriction enzyme BglII (lanes 1 and 2) or KpnI (lanes 3 and 4), and the resulting DNA fragments were separated by gel electrophoresis.
Mutagenesis of the full-length MCMV BAC plasmid pSM3fr (51) was performed in the Escherichia coli strain CBTS by using a two-step replacement method (34, 38) with the modifications recently described (2, 5, 51). BAC DNA was prepared from 10-ml overnight cultures by the alkaline lysis procedure (31) and subjected to treatment with appropriate restriction endonucleases followed by agarose gel electrophoresis. Primers dN (5′-GCG GCC GCT GCA TAC GTT GTA TCC ATA TC-3′) and dBlp (5′-CGC TCA GCC CGC CCA TTT GCG TCA ATG GGG-3′) were used to amplify the authentic or mutant 616-bp HCMV enhancer fragments that were inserted into the BACs MCMV-ES and MCMV-ES.NF-κB, respectively. Ten nanograms of the BAC DNA was used as the template, and PCR conditions were as follows: 1 cycle at 96°C for 3 min; 30 cycles of 30 s at 96°C, 45 s at 60°C, and 30 s at 72°C; and 1 cycle at 72°C for 10 min. Amplified DNA products were isolated by using a PCR purification kit (Qiagen, Hilden, Germany), and 1/10 of the eluate was subjected to digestion with endonuclease BglII or KpnI, followed by electrophoretic separation in a 1.2% low-melting-point agarose gel. BACs were transfected into MEFs by using the calcium phosphate transfection technique. Six hours posttransfection, cells were treated with glycerol (15% glycerol in HEPES-buffered saline) for 3 min as described previously (38). The progeny virus that originated from these cultures was harvested when 100% cytopathic effect was observed. BAC-derived viruses were amplified, subjected to three rounds of plaque purification, and used for the preparation of high-titer stocks. The integrity of the viruses generated was confirmed by restriction enzyme analysis of the corresponding genomes and sequencing of the enhancer regions (data not shown). Enhancer sequences were as expected, except for a one-base deletion in MCMV-ES.NF-κB corresponding to nt position 424 (relative to the transcription start site of the HCMV MIEP).
Construction of IκBαM cell lines.
NHDF IκBαM cells were described previously (4). NIH 3T3 and J774 IκBαM cells were generated by multiple rounds of transduction with IκBαM-expressing retrovirus, a gift from I. Verma (50), and stable cell pools were selected with G418 (0.8 mg/ml) as described previously (4) in order to reduce the possibility of artifacts arising from clonal populations.
Determination of CMV replication kinetics.
Cell lines were infected at ∼80% confluency (∼105 cells in a 12-well dish) with CMV for 2 h (350-μl volume). Virus was aspirated, and cells were then washed twice and incubated in growth medium. Cell supernatants were collected at various times postinfection, centrifuged briefly to pellet any cell debris, and frozen at −80°C. Viral titers were then determined by limiting-dilution plaque assay on NHDF (HCMV) or NIH 3T3 cells (MCMV) by standard methods.
Analysis of NF-κB activation.
For analysis of ICAM-1 upregulation on the cell surface, NHDF, NIH 3T3, or J774 LXSN or IκBαM cells (∼105) were treated with 1 nM human TNF (gift of J. Browning; Biogen, Boston, Mass.) or lymphotoxin alpha (LTα) for ∼24 h. Cells were then dislodged with 5 mM EDTA in phosphate-buffered saline and analyzed for cell surface expression of human ICAM-1 (NHDF, mAB2146; Chemicon, Temecula, Calif.) or murine ICAM-1 (NIH 3T3 or J774 cells, clone 3E2; BD Biosciences, San Diego, Calif.) by flow cytometry as described previously (4).
For electrophoretic mobility shift assay (EMSA) analysis of p50/p65 NF-κB activation, cells (∼3 × 106) were treated with TNF, LTα, or CMV at the indicated concentrations or MOI for 2 h. Cells were harvested by scraping and lysed in 500 μl of cytoplasmic extraction buffer A (20 mM HEPES [pH 7.55], 10 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol, 1 mM orthovanadate, 1 mM phenylmethylsulfonyl fluoride, leupeptin, and aprotinin) on ice for 15 min. NP-40 was added to a final concentration of 0.5%, and the cell nuclei were pelleted by centrifugation at 18,000 × g for 1 min in a microcentrifuge. The nuclear pellet was lysed in 50 μl of nuclear lysis buffer B (20 mM HEPES [pH 7.55], 400 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, leupeptin, and aprotinin) and vortexed gently for ∼1 h at 4°C, and lysates were cleared by centrifugation for 5 min at 18,000 × g.
Fifteen microliters of buffer B containing 50% glycerol was added to the cleared lysate, and protein concentrations were determined by using the bovine serum albumin protein assay (Pierce, Rockford, Ill.) following the manufacturer's protocol. Ten micrograms of total protein for each sample was incubated with 32P-labeled (polynucleotide kinase) double-stranded oligonucleotides (20,000 cpm/sample) corresponding to the NF-κB binding site in the immunoglobulin κ light-chain enhancer (sense, 5-ATGTGAGGGGACTTTCC CAGGC-3′; antisense, 5′-GCCTGGGAAAGTCCCCTCAACT-3′) and 100 ng of poly(dI:C) per ml and then analyzed by nondenaturing polyacrylamide gel electrophoresis (5% polyacrylamide gel). The dried gel was exposed to film, and the bands were visualized by autoradiography. Unlabeled oligonucleotides (200 ng) were used as a competitor to show binding specificity. Supershifts were performed with both anti-p65 (Santa Cruz Biotech, Santa Cruz, Calif.) and anti-p50 (Upstate, Lake Placid, N.Y.) antibodies to verify the identity of the p50/p65 heterodimer.
Analysis of CMV IE expression.
For analysis of ie1 expression, cells were infected with HCMV (AD169-ATCC) or MCMV and harvested at various times postinfection for isolation of RNA and subsequent analysis by real-time PCR as described previously (4). Primer sequences designed to amplify within exon 4 of ie1 were as follows: HCMV, 5′-CATCCACACTAGGAGAGCAGACTC-3′ and 5′-GCATGAAGGTCTTTGCCCAG-3′; MCMV, 5′-AGCTGTTGGTGGTGTCACTCAA-3′ and 5′-GGCTGGGACTCATCTTCTTCAG-3′. For MCMV ie3 (exon 5), the primer sequences used were 5′-GCCCCAACCTGTCCATCTC-3′ and 5′-CCTGCTGTGGTTAATGGGCT-3′. For analysis of IE1 protein levels, infected NHDF (∼105) were harvested at various time points and subjected to Western blot analysis using anti-IE1 monoclonal antibodies (mix of clones 63 and 27; kind gift of W. Britt) as described previously (4).
RESULTS
NF-κB response elements in the MIEP are not required for replication of MCMV in fibroblasts.
Previous studies have shown that infection of fibroblasts with MCMV results in activation of NF-κB (18, 19), similar to that seen for HCMV, and raises the question of whether this activation is required for efficient MCMV replication. In the following set of experiments, we first sought to test whether the NF-κB binding sites in the MIEP contribute to MCMV replication in fibroblasts by introducing point mutations that abolish the ability of NF-κB to bind to the enhancer. The MCMV enhancer has over 12 consensus NF-κB response elements contained within a highly repetitive sequence that is prohibitive for generating site-directed mutations. For this and other reasons, we previously generated a recombinant MCMV in which the native MIEP enhancer has been replaced by the less repetitive HCMV enhancer (2). The HCMV enhancer, unlike its mouse counterpart, contains only four NF-κB binding sites. Importantly, this recombinant “enhancer swap” virus (hMCMV-ES) exhibits infectious kinetics similar to that of wild-type MCMV upon infection of murine fibroblasts (2). To directly investigate the role of the NF-κB binding sites present in the HCMV enhancer on viral replication, an hMCMV-ES mutant was generated (hMCMV-ES.NF-κB) that contained mutations known to abolish NF-κB binding within all four sites. The viral mutant was constructed by using the previously described CMV BAC system (34), and a schematic representation of the parental and recombinant viruses is shown in Fig. 1A.
Unique restriction sites were introduced when mutating the NF-κB binding sites and were used to confirm the successful construction of hMCMV-ES.NF-κB by restriction digestion. To verify the genomic structure of the recombinant BACs used in the study, their HindIII restriction patterns were analyzed. As expected (Fig. 1B), the natural 7.2-kbp MCMV HindIII L fragment of the wild-type MCMV BAC plasmid was replaced by a new 6.7-kbp HindIII L fragment in the recombinant hMCMV-ES and hMCMV-ES.NF-κB plasmids (compare lane 1 with lanes 2 and 3). To confirm that the intended mutations were introduced in the BAC plasmid hMCMV-ES.NF-κB, a 632-bp fragment corresponding to the HCMV enhancer regions of hMCMV-ES and hMCMV-ES.NF-κB was PCR amplified and subsequently digested with BglII or KpnI (Fig. 1C) (see Materials and Methods for details). When the PCR-amplified product from hMCMV-ES.NF-κB was digested with BglII, two major bands of 150 and around 230 bp (the latter corresponding to the predicted 222- and 259-bp digested fragments) were detected. Similarly, digestion of the PCR-amplified product from hMCMV-ES.NF-κB with KpnI resulted in the two expected fragments of 122 and 510 bp. In contrast, the PCR-amplified product from hMCMV-ES was not altered after digestion with restriction enzymes BglII or KpnI. Mutations were also confirmed by sequencing the entire enhancer and flanking regions of the BAC (data not shown). Altogether, these results indicate the integrity of the BAC constructs and the successful disruption of the NF-κB binding sites within the enhancer region of the hMCMV-ES.NF-κB BAC.
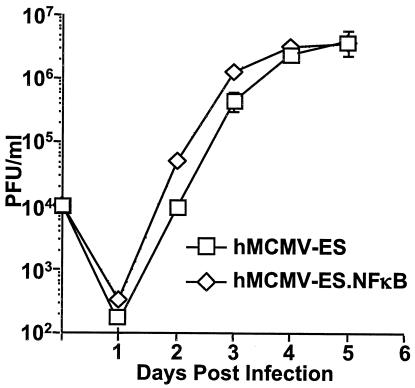
Recombinant viruses were generated by transfection of the constructed BACs into NIH 3T3 cells, collection of viral supernatants, and three subsequent rounds of plaque purification. hMCMV-ES.NF-κB-derived virus was then tested to determine whether the mutations introduced in the NF-κB binding sites altered the ability of the virus to grow in cell culture. For this purpose, NIH 3T3 cells were infected with the parental hMCMV-ES and hMCMV-ES.NF-κB at a low MOI of 0.1, and viral titers in the supernatants of the cultures were determined at various times postinfection. hMCMV-ES.NF-κB exhibited no growth defects when compared to the parental virus (Fig. 2). Since the genetic abolition of all four consensus NF-κB sites within the enhancer region had no negative effect on CMV growth, we conclude that the NF-κB binding sites in the enhancer are nonessential for potentiating MCMV replication in cultured fibroblasts.
FIG. 2.
The MIEP NF-κB binding sites are not required for hMCMV-ES replication in NIH 3T3 cells. NIH 3T3 cells were infected at an MOI of 0.1 with either hMCMV-ES or hMCMV-ES.NF-κB recombinant virus. Cell supernatants were collected at various days postinfection, and titers (PFU/ml) were analyzed in triplicate by plaque assays on NIH 3T3 cells. Error bars represent standard errors of the means.
The canonical NF-κB pathway is not required for replication of murine CMV in fibroblast- or macrophage-derived cell lines.
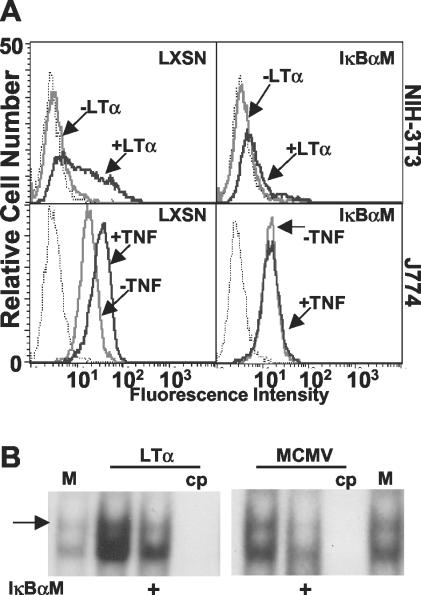
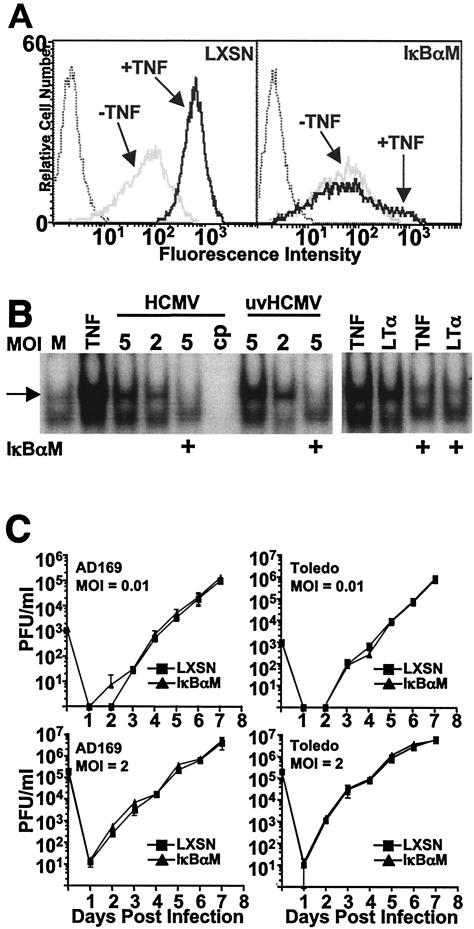
In addition to mutating the NF-κB response elements in the MIEP, a second approach, which avoids causing mutations in the virus, was also taken to address the role of NF-κB in MCMV replication. In order to determine whether NF-κB activation was directly or indirectly required for MCMV replication, a dominant negative version of the inhibitor of NF-κB-alpha (IκBα) that had been mutated at two critical serine residues (S32 and S36) to alanine (IκBαM) was utilized (50). An IκBαM-expressing retroviral vector, or control vector (LXSN), was used to transduce both NIH 3T3 and J774 cells, murine fibroblast- and macrophage-derived cell lines, respectively. Stable cell pools expressing IκBαM were selected after transduction to avoid possible artifacts arising from single-cell clones. NF-κB-mediated transcription was shown to be inhibited by the use of a flow cytometry-based assay examining the ability of the NF-κB-responsive gene ICAM-1 (21) to be upregulated in response to treatment of cells with the TNF family ligand, LTα (Fig. 3A). LTα binds to TNF receptor 1 (TNFR-1) (52), which is expressed on the surface of most cells at low levels, resulting in the TNFR-associated factor 2-dependent activation of NF-κB (p50/p65). Both TNF and LTα bind TNFR-1 and activate the canonical NF-κB pathway through an IKK-dependent mechanism. These ligands can also induce apoptosis in many cell lines, but TNF oftentimes exhibits more potent apoptotic activity in vitro than does LTα (52). NIH 3T3 cells are highly sensitive to apoptosis induced by TNF, as opposed to J774 cells, which are largely resistant, necessitating the use of LTα as an agonist in these cells to allow detection of ICAM-1 after a 24-h incubation in cytokine (Fig. 3A). As expected, based on the observed inhibition of ICAM-1 upregulation, the translocation of p50/p65 NF-κB to the nucleus after treatment of 3T3-IκBαM cells with LTα was inhibited compared to control cells, and this was also the case after infection with MCMV (Fig. 3B).
FIG. 3.
IκBαM overexpression inhibits p50/p65 NF-κB activation in NIH 3T3 and J774 cells. NIH 3T3 or J774 cells were transduced with either control (LXSN) or IκBαM retroviral vector. (A) To verify that the IκBαM-expressing cells were inhibited for p50/p65 NF-κB-mediated transcription, J774 and 3T3 cells were incubated with ±1 nM TNF or LTα for 24 h, respectively, and cell surface levels of ICAM-1 (a NF-κB responsive gene) were analyzed by flow cytometry. Dotted histograms represent binding of isotype control antibody, gray histograms are cells untreated with cytokine, and black histograms are cytokine-treated cells. (B) Translocation of p50/p65 NF-κB to the nucleus is inhibited in IκBαM-expressing cells. NIH 3T3 LXSN or IκBαM (+) cells were treated with LTα (1 nM) or MCMV (MOI = 2) for 2 h; nuclear extracts were prepared and subjected to EMSA. The arrow identifies the p50/p65 heterodimer of NF-κB. M, mock treated; cp, addition of unlabeled competitor, NF-κB-binding oligonucleotide probe.
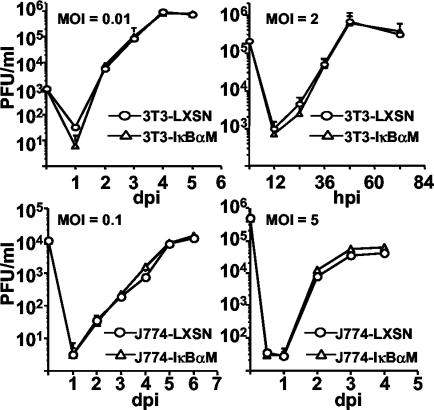
After p50/p65 NF-κB-mediated nuclear translocation and transcription was verified to be inhibited, cells were infected with MCMV (Smith strain) at various MOIs, and the kinetics of viral replication was compared to that of control cells (Fig. 4). Infections at different MOIs were performed due to previous results indicating that disruption of ie1 expression shows no phenotype at high MOI, but replication deficiencies are unmasked at low MOI (14, 17, 32, 33). No differences in the ability of MCMV to replicate were seen when IκBαM was used to inhibit activation of the canonical NF-κB pathway in either of these cell lines, even at low MOI (Fig. 4).
FIG. 4.
Replication of MCMV is normal in cells expressing a dominant negative inhibitor of IκBα. NIH 3T3 (upper two panels) and J774 (lower two panels) control (LXSN) and IκBαM cells were infected at either high or low MOI with MCMV, and supernatant was collected at different times (days or hours) postinfection (dpi and hpi, respectively). MCMV titers (PFU/ml) were determined in triplicate by limit dilution in NIH 3T3 cells. Error bars represent standard errors of the means.
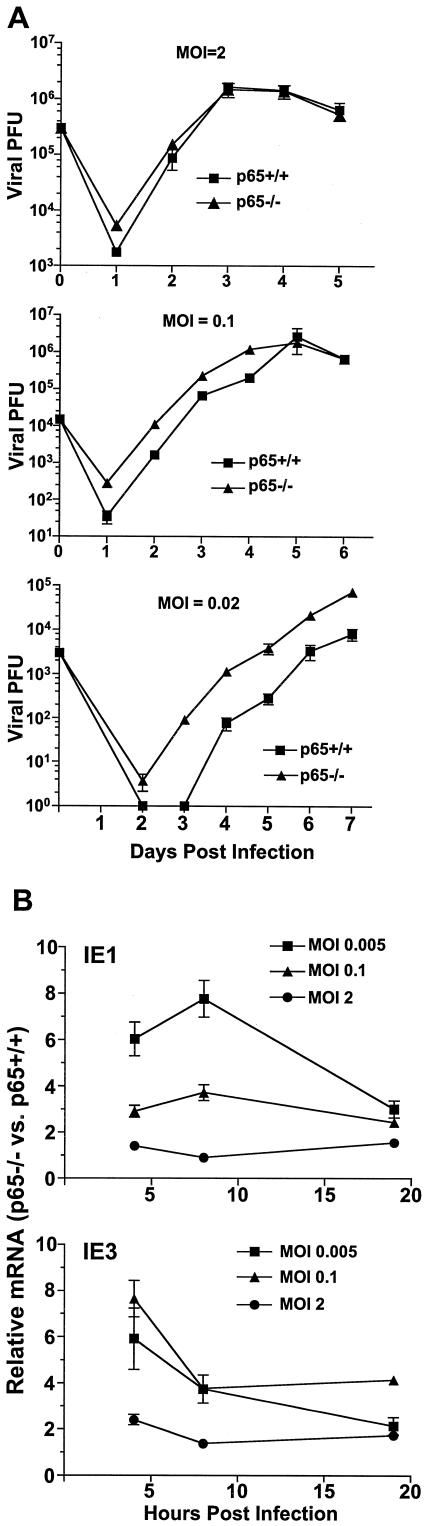
As an alternative approach to assess the contribution of the canonical NF-κB pathway to MCMV infection, replication of MCMV was examined in p65/RelA-deficient immortalized MEFs (p65−/− MEFs). Infection was performed at both high and low MOI, and replication of MCMV in p65−/− MEFs was compared to that in p65+/+ MEFs (Fig. 5A). No restriction of MCMV replication was observed in p65−/− MEFs at any MOI. Alternatively, increased levels of MCMV replication were observed at low MOI compared to those of p65+/+ MEFs derived from wild-type littermate control mice (3). Consistent with these results, significantly higher levels of MCMV ie1 and ie3 expression were detected at low MOI in p65−/− MEFs, and these differences were less marked as the MOI was increased (Fig. 5B). This observation suggests a potentially p65-dependent but IκBα-independent suppression of MCMV replication.
FIG.5.
Mouse CMV replication is increased in p65-deficient fibroblasts. (A) p65-deficient (p65−/−) and control (p65+/+) immortalized MEFs were infected with MCMV at several different MOIs, and supernatant was collected at various days postinfection for analysis of viral PFU produced. Titers were determined in triplicate in NIH 3T3 cells, and error bars represent standard errors of the means. (B) The relative level of MCMV ie1 and ie3 expression in p65−/− MEFs was compared to that of p65+/+ MEFs at various times postinfection at several different MOI by real-time PCR. Values are representative of three independent experiments, and error bars represent the standard errors of the means.
Taken together, these data unequivocally show that activation of the canonical NF-κB pathway is nonessential for initiating and promoting MCMV replication or expression from the MIEP in immortalized fibroblasts or a macrophage cell line.
Inhibiting the canonical NF-κB pathway has no effect on human CMV replication in fibroblasts.
Numerous studies have shown that human CMV infection induces activation of NF-κB (9, 10, 45, 54), and the NF-κB-responsive elements present in the MIEP (Fig. 1A) bind NF-κB and are responsive to NF-κB-mediated transcriptional enhancement in transient-transfection reporter-based expression assays (45). To investigate the role of the canonical NF-κB pathway for HCMV replication, we determined growth kinetics of laboratory and clinical strains upon infection of an NHDF line which expresses IκBαM (4). This cell line (NHDF-IκBαM) shows severely reduced levels of IκBα degradation (4) and ICAM-1 upregulation in response to treatment of cells with TNF when compared to that of a control cell line (NHDF-LXSN) (Fig. 6A). Importantly, NHDF-IκBαM cells show no translocation of p50/p65 NF-κB to the nucleus after treatment with TNF or LTα or after infection with HCMV or UV light-inactivated HCMV (Fig. 6B).
FIG. 6.
The canonical NF-κB pathway is not required for replication of human CMV in fibroblasts. (A) NHDF were transduced with control (LXSN) or IκBαM retroviral vector. Cells were examined for upregulation of ICAM-1 by flow cytometry after incubation with ±1 nM TNF. Dotted histogram represents binding of isotype control antibody. (B) Nuclear extracts from NHDF-LXSN or -IκBαM (+) cells treated with TNF (1 nM) or LTα (1 nM) or infected with HCMV or UV light-inactivated HCMV (uvHCMV; MOI, 5 or 2) for 2 h were subjected to EMSA to analyze the level of p50/p65 NF-κB (indicated by the arrow) translocation to the nucleus. (C) NHDF-LXSN or -IκBαM cells were infected at an MOI of 0.01 or 2 with HCMV strain AD169 or Toledo. Cell supernatant was collected at various days postinfection, and viral titers (PFU/ml) were determined in triplicate by plaque assays in NHDF. Error bars represent standard errors of the means.
NHDF-IκBαM cells were compared to NHDF-LXSN cells for their ability to support replication of HCMV. Infections were performed at both high and low MOI with two distinct strains of HCMV, AD169-ATCC and Toledo (Fig. 6C). No differences were detected in the ability of AD169 to replicate in NHDF-IκBαM cells compared to that of control fibroblasts when infected at either high or low MOI. The Toledo strain of HCMV was then analyzed because it harbors at least 19 open reading frames which are not present in AD169 (8), resembling viral clinical isolates. As was seen for AD169, no differences in the replication of Toledo were detected when comparing the two NHDF lines (Fig. 6B). Based on these results, we conclude that the blockade of the canonical NF-κB pathway by overexpressing the IκBαM in NHDF does not alter viral replication.
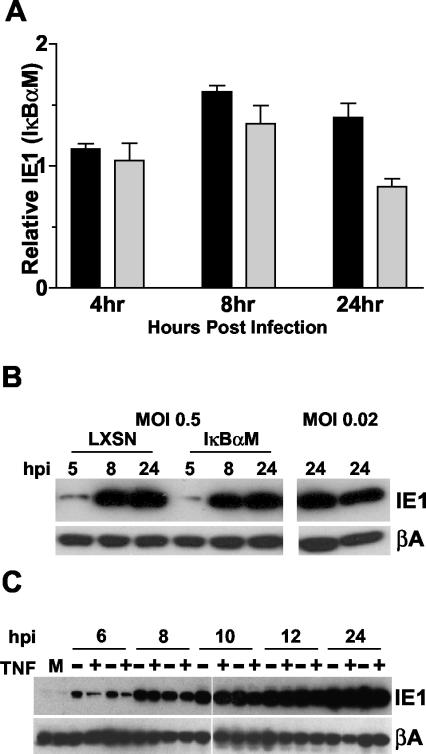
Expression of HCMV IE1 in infected cells does not require NF-κB.
Although HCMV replicates as well in NHDF-IκBαM cells as it does in control fibroblasts, previous reports suggest that the levels of ie1 expression might be lower in cells deficient in NF-κB activation (9, 43, 45, 49). Therefore, the levels of IE1 message and protein were compared in NHDF-IκBαM and -LXSN cells infected with HCMV at a low MOI (0.005 and 0.1, respectively). IE1 message levels were roughly equivalent in NHDF-IκBαM and NHDF-LXSN cells as determined by real-time PCR at various times postinfection, indicating that the canonical NF-κB pathway is not required for transactivation of the ie1 promoter-enhancer during viral replication in fibroblasts (Fig. 7A). IE1 protein levels were also compared in HCMV-infected NHDF-IκBαM and -LXSN cells. In order to detect IE1 at early times postinfection (5 or 8 h) fibroblasts needed to be infected at a relatively high MOI (0.5), taking into consideration that infecting at higher MOI is not ideal for analyzing effects on MIEP activity. When IE1 levels were compared in IκBαM- versus LXSN-infected cells, no significant differences were seen (Fig. 7B), consistent with the results obtained when analyzing ie1 mRNA (Fig. 7A). At lower MOI (0.02), similar levels of IE1 were also seen when analyzed at later times (24 h postinfection) (Fig. 7B). Finally, treatment of HCMV-infected IκBαM and LXSN fibroblasts with TNF, a strong activator of NF-κB, showed no differential effects on IE1 expression, although slightly lower levels of IE1 were seen at 6 h postinfection in both cell lines compared to those found in untreated samples (Fig. 7C). These results further confirm that inhibition of the canonical NF-κB pathway does not restrict HCMV gene expression in cultured fibroblasts.
FIG. 7.
Inhibition of the canonical NF-κB pathway does not inhibit expression of HCMV IE1. (A) NHDF-LXSN or -IκBαM cells were infected at an MOI of 0.005 (dark bars) or 0.1 (light bars) and were harvested at various hours postinfection (hpi) for analysis of ie1 expression by real-time PCR. Results represent the relative amount of ie1 expression in IκBαM cells compared to that of control cells (LXSN) at the various times. Values are averaged data from three independent experiments, and error bars represent the standard errors of the means. (B) NHDF-LXSN or -IκBαM cells were infected with HCMV at an MOI of 0.5 or 0.02. Cells were harvested at various hpi for analysis of IE1 protein levels by Western blotting. For samples treated at an MOI of 0.02, LXSN-infected cells are shown in the left lane, and IκBαM cells are shown in the right. (C) NHDF-LXSN (first two lanes of each time point) or -IκBαM cells (second two lanes of each time point) were infected with HCMV at an MOI of 2, and cells were harvested at various hpi for analysis of IE1 protein levels by Western blot. For each time point, cells were either treated with (+) 1 nM TNF or were not (−).
DISCUSSION
The role of NF-κB during viral infection is critical for mounting antiviral immune responses through activation of various host inflammatory genes (15). In this regard, it would seem deleterious for viruses to trigger NF-κB upon infection of host cells. However, several viruses, including CMV (45, 55), herpes simplex virus type 1 (39), human immunodeficiency virus (12), and others have been reported to utilize NF-κB to their advantage by incorporating NF-κB response elements into their genomes. In the case of CMV, the activation of NF-κB upon infection has been proposed to be important for modulating expression of viral immediate-early genes (45, 55). Although it is well established that NF-κB is activated by CMV upon infection of cells in culture, little has been done to address the importance of this key transcription factor in viral replication. In this study we have used several different approaches to test directly whether NF-κB positively contributes to the CMV transcription-replication cycle in vitro. We present multiple lines of evidence that show a remarkably neutral engagement of the pathway, and in certain experiments a moderate negative contribution to CMV replication can be observed. These findings are more consistent with the notion that CMV effectively buffers itself against a strong innate immune response to an acute infection.
In hMCMV-ES, the mutation of the NF-κB binding sites in the HCMV MIEP did not restrict viral replication in NIH 3T3 cells. In this connection, it is worth noting that consensus NF-κB binding sites are present only in the MIEP enhancer region of the MCMV genome (44) (P. Ghazal, unpublished observation). We further analyzed the contribution of the canonical, or classical, NF-κB pathway by using dominant negative inhibitors of IκBα and immortalized p65/RelA-deficient fibroblasts. Infection of fibroblasts by CMV has been shown to activate the translocation of transcriptionally active p50/p65 to the nucleus (10, 19, 27) and to induce activation of the p50 and p65 promoters by IE1 (19, 54, 55). Overexpression of HCMV IE1 has also recently been reported to induce transcription of the NF-κB family member RelB (23). Our results support a neutral or redundant role for the canonical NF-κB pathway for CMV replication in cultured cells. In fact, MCMV replicated significantly better in fibroblasts lacking p65 when infection was performed at low MOI. Expression of ie1 and ie3 was also significantly higher in p65−/− MEFs infected at low MOI, consistent with the increases seen in viral titers. In this regard, it appears that p65 plays an inhibitory role for MCMV replication in fibroblasts, and this negative effect is amplified at lower MOIs. One possible explanation for this result is that innate cellular defenses to MCMV infection are compromised in p65-deficient fibroblasts. We found that induction of beta interferon (IFN-β) by MCMV is ∼40-fold lower in p65−/− fibroblasts; however, the addition of neutralizing IFN-β antibody to the supernatant had no effect (C. Benedict, unpublished observation). Our inability to detect a role for the lower alpha/beta IFN levels being responsible for the increased CMV replication in p65-deficient cells is consistent with our observations that overexpression of IκBαM also inhibits induction of IFN-β (4), but no significant increases in CMV replication are observed in these cell lines. Taken together, these data suggest a potential dominant role for p65 in suppressing CMV replication, possibly independent of IκBα, and studies to further elucidate this mechanism are currently under way.
It is worthwhile to note that we cannot rule out the possibility that during the process of immortalization, p65−/− MEFs may have compensated for the lack of p65 by hyperinduction of other NF-κB family members and that this may contribute to the increased levels of CMV replication and gene expression. However, we did observe increased sensitivity to TNF-mediated apoptosis of immortalized p65−/− MEFs compared to that of p65+/+ MEFs as originally reported by Beg and Baltimore (3) as well as for 3T3-IκBαM cells (C. Benedict, unpublished observations), indicating an inhibition of cell survival gene induction by NF-κB in these cell lines. Also noteworthy is the fact that although cells inhibited for activation of the canonical NF-κB pathway often show increased sensitivity to apoptosis mediated by TNF or other stimuli, MCMV replication is not inhibited. In fact, significantly increased replication is seen in p65−/− MEFs at low MOI, suggesting that induction of apoptosis is not a rate limiting factor for replication of MCMV in fibroblasts as has been reported for herpes simplex virus (16).
HCMV engages direct interactions with the cellular transcriptional regulatory network through various regulatory elements (reviewed in references 35 and 37). The consortium of host transcription factors which bind the MIEP is likely to vary based on the cell type, differentiation state, and activation status, to name a few. Our present study has concentrated on assessing the contribution of NF-κB to CMV replication as opposed to its role in modulating expression from plasmids containing MIEP sequences. Our data demonstrate that, in the context of an infection, the contribution of NF-κB is redundant in cultured cells, revealing a highly robust regulatory network determining the expression state of CMV, and these findings should be considered when forming conclusions based on results achieved in more isolated systems.
Although NF-κB does not appear to be required for replication of human or murine CMV in fibroblasts, it is premature to extrapolate from these data or draw conclusions regarding the contribution of NF-κB to CMV pathogenesis in vivo. The life cycle of CMV, as for all herpesviruses, may be subdivided into two major phases. First, there is the initial, acute infection which lasts only a few weeks and allows CMV to seed various host organs; then comes the lifelong, latent phase during which a competent immune system keeps viral replication largely in check with sporadic periods of reactivation in select tissues (28, 29). HCMV and MCMV have been reported to establish latency in cells of the monocyte lineage (22, 41), and activation of NF-κB in these cells may contribute to reactivation of viral replication under certain circumstances of inflammation. Because the intracellular milieu of host factors during active CMV infection is not likely to be similar in latently infected cells in which active viral replication is not occurring, it is not unreasonable to speculate that the factors which regulate ie gene expression would be different in these two situations. In addition, the differentiation state of infected cells is also likely to play a role in NF-κB's ability to regulate CMV ie expression due to the presence of specific inhibitory factors (42). Importantly, the hMCMV-ES recombinant virus system will allow us to directly address these questions in vivo.
Through this connection, we have recently shown that the enhancer is absolutely essential for in vivo growth (14), indicating that the cellular network of transcriptional regulators of the MIEP has a crucial role in establishing an infection. The present contribution provides new insights into the nature and behavior of this network. The striking observation that the CMV enhancer is insensitive to elimination of NF-κB underscores the significance of the density of regulatory interactions within the network and thus its inherent robustness. In evolutionary terms, robustness of the CMV enhancer would confer functional flexibility, helping to make the virus resistant to the deleterious effects of mutations in an essential control region; consequently, the virus is highly adaptive. This finding may provide an explanation for the relatively high sequence divergence between the MIEPs among the different species members of the CMV family. At the functional level, and in the context of an acute infection, a robust enhancer would allow flexibility to operate efficiently under various cellular conditions, possibly providing a higher degree of autonomy from cellular factor constraints upon infection.
Acknowledgments
We thank A. Yurochko and E. Dejardin for helpful discussions and W. Britt and A. Beg for providing reagents.
This work was funded in part by grants from the National Institutes of Health (grant numbers AI33068, CA69831, AI48073 [C.F.W.], AI44851 [A.A.], and AI30627 [P.G.]); the Wilhelm-Roux Program of the Medical Faculty of the University of Halle-Wittenberg (M.M.); BBSRC and Wellcome Trust Programme grant GR066784 (P.G.); the Ministerio de Ciencia y Tecnología (SAF 2002-00270) (A.A.); and the American Heart Association 0330064N (C.A.B.) A.A. is a fellow from the Ramón y Cajal program.
REFERENCES
- 1.Angulo, A., P. Ghazal, and M. Messerle. 2000. The major immediate-early gene ie3 of mouse cytomegalovirus is essential for viral growth. J. Virol. 74:11129-11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo, A., M. Messerle, U. H. Koszinowski, and P. Ghazal. 1998. Enhancer requirement for murine cytomegalovirus growth and genetic complementation by the human cytomegalovirus enhancer. J. Virol. 72:8502-8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beg, A. A., and D. Baltimore. 1996. An essential role for NF-κB in preventing TNF-α-induced cell death. Science 274:782-784. [DOI] [PubMed] [Google Scholar]
- 4.Benedict, C. A., T. A. Banks, L. Senderowicz, M. Ko, W. J. Britt, A. Angulo, P. Ghazal, and C. F. Ware. 2001. Lymphotoxins and cytomegalovirus cooperatively induce interferon-β, establishing host-virus détente. Immunity 15:617-626. [DOI] [PubMed] [Google Scholar]
- 5.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boshart, M., F. Weber, G. Jahn, K. Dorsch-Hasler, B. Fleckenstein, and W. Schaffner. 1985. A very strong enhancer is located upstream of an immediate-early gene of human cytomegalovirus. Cell 41:521-530. [DOI] [PubMed] [Google Scholar]
- 7.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha, T.-A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherrington, J. M., and E. S. Mocarski. 1989. Human cytomegalovirus ie1 transactivates the β promoter-enhancer via an 18-base-pair repeat element. J. Virol. 63:1435-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cinatl, J. J., S. Margraf, J. U. Vogel, M. Scholz, J. Cinatl, and H. W. Doerr. 2001. Human cytomegalovirus circumvents NF-κB dependence in retinal pigment epithelial cells. J. Immunol. 167:1900-1908. [DOI] [PubMed] [Google Scholar]
- 11.Dejardin, E., N. M. Droin, M. Delhase, E. Haas, Y. Cao, C. Makris, Z. W. Li, M. Karin, C. F. Ware, and D. R. Green. 2002. The Lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17:525-535. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca, C., H. Kwon, R. Lin, M. Wainberg, and J. Hiscott. 1999. NF-κB activation and HIV-1-induced apoptosis. Cytokine Growth Factor Rev. 10:235-253. [DOI] [PubMed] [Google Scholar]
- 13.Dorsch-Hasler, K., G. M. Keil, F. Weber, M. Jasin, W. Schaffner, and U. H. Koszinowski. 1985. A long and complex enhancer activates transcription of the gene coding for the highly abundant immediate-early mRNA in murine cytomegalovirus. Proc. Natl. Acad. Sci. USA 82:8325-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghazal, P., M. Messerle, K. Osborn, and A. Angulo. 2003. An essential role of the enhancer for murine cytomegalovirus in vivo growth and pathogenesis. J. Virol. 77:3217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed] [Google Scholar]
- 16.Goodkin, M. L., A. T. Ting, and J. A. Blaho. 2003. NF-κB is required for apoptosis prevention during herpes simplex virus type 1 infection. J. Virol. 77:7261-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gribaudo, G., S. Ravaglia, M. Gaboli, M. Gariglio, R. Cavallo, and S. Landolfo. 1995. Interferon-α inhibits the murine cytomegalovirus immediate-early gene expression by down-regulating NF-κB activity. Virology 211:251-260. [DOI] [PubMed] [Google Scholar]
- 19.Gribaudo, G., S. Ravaglia, L. Guandalini, R. Cavallo, M. Gariglio, and S. Landolfo. 1996. The murine cytomegalovirus immediate-early 1 protein stimulates NF-κB activity by transactivating the NF-κB p105/p50 promoter. Virus Res. 45:15-27. [DOI] [PubMed] [Google Scholar]
- 20.Grzimek, N. K., J. Podlech, H. P. Steffens, R. Holtappels, S. Schmalz, and M. J. Reddehase. 1999. In vivo replication of recombinant murine cytomegalovirus driven by the paralogous major immediate-early promoter-enhancer of human cytomegalovirus. J. Virol. 73:5043-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou, J., V. Baichwal, and Z. Cao. 1994. Regulatory elements and transcription factors controlling basal and cytokine-induced expression of the gene encoding intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. USA 91:11641-11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis, M., and J. Nelson. 2002. Human cytomegalovirus persistence and latency in endothelial cells and macrophages. Curr. Opin. Microbiol. 5:403-407. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, H. Y., C. Petrovas, and G. E. Sonenshein. 2002. RelB-p50 NF-κB complexes are selectively induced by cytomegalovirus immediate-early protein 1: differential regulation of Bcl-xL promoter activity by NF-κB family members. J. Virol. 76:5737-5747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 67:7539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 26.Kayagaki, N., M. Yan, D. Seshasayee, H. Wang, W. Lee, D. M. French, I. S. Grewal, A. G. Cochran, N. C. Gordon, J. Yin, M. A. Starovasnik, and V. M. Dixit. 2002. BAFF/BLyS receptor 3 binds the B-cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-κB2. Immunity 17:515-524. [DOI] [PubMed] [Google Scholar]
- 27.Kowalik, T. F., B. Wing, J. S. Haskill, J. C. Azizkhan, A. S. Baldwin, Jr., and E. S. Huang. 1993. Multiple mechanisms are implicated in the regulation of NF-κB activity during human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 90:1107-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurz, S. K., M. Rapp, H. P. Steffens, N. K. Grzimek, S. Schmalz, and M. J. Reddehase. 1999. Focal transcriptional activity of murine cytomegalovirus during latency in the lungs. J. Virol. 73:482-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurz, S. K., and M. J. Reddehase. 1999. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 73:8612-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Meier, J. L., M. J. Keller, and J. J. McCoy. 2002. Requirement of multiple cis-acting elements in the human cytomegalovirus major immediate-early distal enhancer for viral gene expression and replication. J. Virol. 76:313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier, J. L., and J. A. Pruessner. 2000. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J. Virol. 74:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. Lippincott, Williams and Wilkins, Philadelphia, Pa.
- 36.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding IE1491aa is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson, J. A., J. W. Gnann, Jr., and P. Ghazal. 1990. Regulation and tissue-specific expression of human cytomegalovirus. Curr. Top. Microbiol. Immunol. 154:75-100. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 244:1307-1312. [DOI] [PubMed] [Google Scholar]
- 39.Patel, A., J. Hanson, T. I. McLean, J. Olgiate, M. Hilton, W. E. Miller, and S. L. Bachenheimer. 1998. Herpes simplex type 1 induction of persistent NF-κB nuclear translocation increases the efficiency of virus replication. Virology 247:212-222. [DOI] [PubMed] [Google Scholar]
- 40.Peters, R. T., and T. Maniatis. 2001. A new family of IKK-related kinases may function as IκB kinase kinases. Biochim. Biophys. Acta 1471:M57-M62. [DOI] [PubMed] [Google Scholar]
- 41.Pollock, J. L., R. M. Presti, S. Paetzold, and H. W. Virgin. 1997. Latent murine cytomegalovirus infection in macrophages. Virology 227:168-179. [DOI] [PubMed] [Google Scholar]
- 42.Prosch, S., A. K. Heine, H. D. Volk, and D. H. Kruger. 2001. CCAAT/enhancer-binding proteins α and β negatively influence the capacity of tumor necrosis factor α to up-regulate the human cytomegalovirus IE1/2 enhancer/promoter by nuclear factor κB during monocyte differentiation. J. Biol. Chem. 276:40712-40720. [DOI] [PubMed] [Google Scholar]
- 43.Prosch, S., K. Staak, J. Stein, C. Liebenthal, T. Stamminger, H. D. Volk, and D. H. Kruger. 1995. Stimulation of the human cytomegalovirus IE enhancer/promoter in HL-60 cells by TNF-α is mediated via induction of NF-κB. Virology 208:197-206. [DOI] [PubMed] [Google Scholar]
- 44.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambucetti, L. C., J. M. Cherrington, G. W. Wilkinson, and E. S. Mocarski. 1989. NF-κB activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T-cell stimulation. EMBO J. 8:4251-4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz, R., B. Helmich, and D. H. Spector. 1996. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J. Virol. 70:6955-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stenberg, R. M. 1996. The human cytomegalovirus major immediate-early gene. Intervirology 39:343-349. [DOI] [PubMed] [Google Scholar]
- 48.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, B., G. Harrowe, C. Reinhard, C. Yoshihara, K. Chu, and S. Zhuo. 2001. Modulation of human cytomegalovirus immediate-early gene enhancer by mitogen-activated protein kinase kinase kinase-1. J. Cell. Biochem. 83:563-573. [DOI] [PubMed] [Google Scholar]
- 50.Van Antwerp, D. J., S. J. Martin, T. Kafri, D. R. Green, and I. M. Verma. 1996. Suppression of TNF-α-induced apoptosis by NF-κB. Science 274:787-789. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ware, C. F., S. Santee, and A. Glass. 1998. Tumor necrosis factor-related ligands and receptors, p. 549-592. In A. Thompson (ed.), The cytokine handbook, 3rd ed. Academic Press, San Diego, Calif.
- 53.Yurochko, A. D., and E. S. Huang. 1999. Human cytomegalovirus binding to human monocytes induces immunoregulatory gene expression. J. Immunol. 162:4806-4816. [PubMed] [Google Scholar]
- 54.Yurochko, A. D., E. S. Hwang, L. Rasmussen, S. Keay, L. Pereira, and E. S. Huang. 1997. The human cytomegalovirus UL55 (gB) and UL75 (gH) glycoprotein ligands initiate the rapid activation of Sp1 and NF-κB during infection. J. Virol. 71:5051-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yurochko, A. D., T. F. Kowalik, S. M. Huong, and E. S. Huang. 1995. Human cytomegalovirus upregulates NF-κB activity by transactivating the NF-κB p105/p50 and p65 promoters. J. Virol. 69:5391-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]