Abstract
Seven distinct sequence variants of the Epstein-Barr virus latent membrane protein 1 (LMP1) have been identified by distinguishing amino acid changes in the carboxy-terminal domain. In this study the transmembrane domains are shown to segregate identically with the distinct carboxy-terminal amino acid sequences. Since strains of LMP1 have been shown to differ in abundance between blood and throat washes, nasopharyngeal carcinomas (NPCs) from areas of endemicity and nonendemicity with matching blood were analyzed by using a heteroduplex tracking assay to distinguish LMP1 variants. Striking differences were found between the compartments with the Ch1 strain prevalent in the NPCs from areas of endemicity and nonendemicity and the B958 strain prevalent in the blood of the endemic samples, whereas multiple strains of LMP1 were prevalent in the blood of the nonendemic samples. The possible selection against the B958 strain appearing in the tumor was highly significant (P < 0.0001). Sequence analysis of the full-length LMP1 variants revealed changes in many of the known and computer-predicted HLA-restricted epitopes with changes in key positions in multiple, potential epitopes for the specific HLA of the patients. These amino acid substitutions at key positions in the LMP1 epitopes may result in a reduced cytotoxic-T-lymphocyte response. These data indicate that strains with specific variants of LMP1 are more likely to be found in NPC. The predominance of specific LMP1 variants in NPC could reflect differences in the biologic or molecular properties of the distinct forms of LMP1 or possible immune selection.
The Epstein-Barr virus (EBV) is a prevalent human gammaherpesvirus that establishes a persistent latent infection in over 90% of the world's population (76). EBV can avoid recognition by the immune system in part through minimal viral gene expression in resting memory B lymphocytes, alteration and modification of antigen expression machinery, and alteration of the immune response to infected cells through the release of anti-inflammatory cytokines (33, 79, 89).
Primary infection is usually asymptomatic, but may result in infectious mononucleosis, a benign lymphoproliferative disease whose pathogenesis partially results from the immune response to EBV infection of B lymphocytes (67). After the development of the major histocompatibility complex class I (MHC-I)-restricted T-cell response to EBV antigens, the number of EBV-infected B cells declines sharply and is eventually controlled by a functional immune system (11, 46, 66). Although the majority of infections are asymptomatic, EBV infection is also strongly associated with the pathogenesis of several malignancies including Burkitt's lymphoma, nasopharyngeal carcinoma (NPC), Hodgkin's disease (HD), and AIDS-associated and posttransplant lymphomas (74). NPC is particularly significant in that it is one of the leading causes of cancer death in the southern region of China (36).
Virus-specific T lymphocytes are critical for controlling latent EBV infection in healthy individuals, and both CD8+ and CD4+ cells have been shown to respond to latently expressed EBV proteins (54, 63, 64, 77). The strongest cytotoxic T lymphocyte, CTL, response is directed against the EBV nuclear antigens (EBNAs) 3A, -B, and -C to 6, and a minor response to the latent membrane proteins, LMP2 and LMP1, has been detected (42, 77). The EBNAs and LMP1 are not expressed in the persistently infected memory B cells, and only EBNA1 and LMP2 are thought to be expressed (89).
Under conditions of immunosuppression, EBV expression may activate to a transforming infection with expansion of the EBV-infected cells and uncontrolled B-cell proliferation, resulting in the development of posttransplant lymphomas and AIDS-associated lymphomas (2, 13, 82). In posttransplant lymphomas, all of the latent EBV proteins are expressed, including EBNA1 and -2, the immunodominant EBNA3A to -C, LMP2, and LMP1 (28, 93). Restoration of the immune response by reduction of immune suppressive drugs or transfusion of EBV-specific cytotoxic T lymphocytes (CTL) has been shown to be effective therapy (34, 78, 86).
In NPC and HD, only the less-immunogenic proteins EBNA1, LMP1, and LMP2 are expressed (7, 10, 32, 72, 73). Patients have elevated titers of antibodies to multiple EBV proteins, and T cells infiltrate the tumor without apparent cytotoxicity (29, 30, 50, 59, 68, 88). It is possible that this immune evasion may be due to changes in CTL-recognized viral epitopes, since MHC-I and transporter-associated proteins (TAP-1 and -2) are still expressed within tumors and may be upregulated by LMP1 (44, 51, 65).
Several HLA-restricted epitopes have been identified in EBNA1, LMP1, and LMP2 of the prototype B958 virus (4, 43, 47, 53, 58, 65, 70). Variants of EBV have also been identified by polymorphisms in the viral genome, including the EBNA3 and LMP1 genes that have led to changes in the known HLA-restricted epitopes (14, 40). LMP1 variants have been shown to differ between the blood and tumor tissue in NPC and HD (18, 31, 57, 87). Variants in the EBNA1 and LMP2 genes have also been described; however, the same EBNA1 variant was detected in both the tumor and the blood, whereas possible differences in LMP2 between tumor and the blood have not been investigated (9, 50, 92).
Work from our laboratory has identified seven distinct LMP1 variants based on signature changes in the carboxy terminus, and our recent studies indicate that the EBV strains with distinct forms of LMP1 differ in abundance in blood and throat washings (18, 84). To determine whether the LMP1 variants were also distinguishable in the transmembrane domain of LMP1, the full-length sequences of the seven strains of LMP1 were determined. Strain-distinguishing changes were identified in the transmembrane domains, and phylogenetic analysis revealed that the transmembrane sequences segregated identically to the strains distinguished by the carboxy-terminal sequence of LMP1 (18).
To determine whether strain differences exist between tumor tissue and blood based on LMP1, samples of NPC and matching blood were analyzed from both the southern region where NPC is endemic and the northern region of China where NPC is not endemic by using a heteroduplex tracking assay (HTA) that can distinguish all of the LMP1 variants. Many of the LMP1 variants were detected in the region where NPC is not endemic, whereas fewer variants were detected in the region where NPC is endemic. In addition, clear differences were detected between the tumor and the peripheral blood lymphocytes (PBL). The LMP1 variant most frequently detected in tumor tissue had changes in many of the known and computer-predicted HLA-restricted epitopes and these changes were predicted to result in a reduced CTL response. The known HLA-restricted LMP2 epitopes were also analyzed and, although changes were identified, in most cases these changes were present in the strains prevalent in tumors and those prevalent in the blood. These data indicate that strains with specific variants of LMP1 are more likely to be found in NPC. The predominance of specific LMP1 variants in NPC could reflect differences in the biologic or molecular properties of the distinct forms of LMP1 or possible decreased immune recognition of the LMP1 prevalent in NPC.
MATERIALS AND METHODS
Patient tissue specimens.
The NPC tissue biopsies and matching PBL from regions where NPC is endemic were obtained from the Guangxi Regional Hospital in Nanning, Guangxi Autonomous Region (specimens GX1 to -6 and 138 to 141). The specimens from regions where NPC is not endemic were obtained from the Cancer Hospital in Beijing (specimens 108, 509, and 614) and the Bai Qui En Medical University Hospital in Changchun, Jilin Province (specimens 15 to 19 and 22) (87). NPCs C15, C17, 4, 36, 38, 39, 13, 23, 27, 24, 60, as well as pOT, PTL1, and HLP 11, have been previously described (18). Mono samples 43, 72, 80, 81, 82, 84, 85, 86, and 87 were cell lines derived from throat wash and/or PBL of European infectious mononucleosis patients. Sample N2 TW (throat wash) was from a healthy American.
The tumor biopsies from areas of endemicity and nonendemicity were processed as previously described (87). Briefly, tissues were Dounce homogenized on ice in a buffer containing 15 mM NaCl, 15 mM Tris-HCl (pH 8), and 1 mM EDTA; subjected to four freeze-thaw cycles; and digested with proteinase K-sodium dodecyl sulfate for 4 h at 56°C. The samples were subjected to phenol-chloroform extraction, and the DNA was ethanol precipitated. PBL were purified over lymphocyte separation medium (Organon Teknika) and processed the same as the biopsies obtained from areas of endemicity and nonendemicity described above. The remaining frozen tumors were processed as previously described (75). Tumor specimens were pulverized in a microdismembrator and suspended in 4 M guanidine isothiocyanate. After centrifugation through a cesium chloride step gradient, the DNA was dialyzed, proteinase K digested, phenol-chloroform extracted, and ethanol precipitated. The throat wash sample was purified as previously described (83).
DNA sequencing.
The DNA sequence corresponding to LMP1 was determined by amplifying 0.3 μg of DNA with the PCR by using Taq polymerase (Promega, Madison, Wis.). The transmembrane domain of LMP1 was amplified by using primers 168658R (EBV coordinates 168658 to 168677; 5′-CTCGTTGGAGTTAGAGTCAG-3′) and 169251L (EBV coordinates 169251 to 169233; 5′-ACCTTCTCTGTCCACTTGGA-3′). The PCR product was sequenced with primer 168658R.
Full-length LMP1 was amplified by using the primers LMP3UT (EBV coordinates 168017 to 168036; 5′-ATCACGAGGAATTCAATGTGGCTTTTCAGCCTAG-3′) and LMPEco (EBV coordinates169627 to 169607; 5′-ATCACGAGGAATTCCCCGTACTGCCTCCGGCAGAC-3′). A second round of PCR with 6 μl of template from the full-length reaction was performed with the primers FUE (EBV coordinates 168163 to 168183; 5′-GTCATAGTAGCTTAGCTGAAC-3′) and 168808L (EBV coordinates 168830 to 168808; 5′-GTGGACTCTATTGGTTGATCTC-3′) for the 3′ end of LMP1 and the primers LMPExc (EBV coordinates 168813 to 168833; 5′-CAACCAATAGAGTCCACCAGT-3′) and 169584Ld [EBV coordinates 169584 to 169565; 5′-CATCC(A/C)AGAAACACGCGTT-3′] for the 5′ end of LMP1. Primers 168808L and 169584Ld were used for sequencing. LMP1 sequence of several samples has been previously described and was reconfirmed (3, 18, 61, 87).
LMP2 was amplified from tumor samples by using the primers 166538R (5′-GTTTTGCAGTCGCTGCYGCA-3′) and 167039L (5′-GACCTGTTGTCCCTGAGATG-3′) for exon 1, primers −24R (5′-GGTCGGATTTCGCCCTTATT-3′) and 691L (5′-CACAGTTACAGCTCCAAGGA-3′) for exons 2 and 3, primers 362R (5′-TTTGCAATTTGCCTAACATG-3′) and 1015L (5′-GATGCCAAGTTAGAGCTGCGA-3′) for exons 4 and 5, and primers 995R (5′-TCGCAGCTCTAACTTGGCATC-3′) and 1677L (5′-TACAGTGTTGCGATATGG-3′) for exons 6 to 8. LMP2 was amplified from PBL by using primers 362R and 1677L, followed by nested reactions with primers 362R and 1015L and primers 995R and 1677L.
The samples were sequenced at the University of North Carolina-Chapel Hill Automated DNA Sequencing Facility on a model 3100 genetic analyzer (Applied Biosystems Division, Perkin-Elmer Cetus, Norwalk, Conn.) by using the ABI Prism dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (Applied Biosystems Division, Perkin-Elmer Cetus).
Phylogenetic analysis.
To determine the phylogenetic relationship within the various EBV amino acid sequences of the transmembrane domain of LMP1 and the carboxy-terminal amino acid sequence of LMP1, the amino acid sequences were aligned by using the CLUSTAL W multiple alignment, distance matrices were calculated by using the two-parameter model (45), and phylogenetic trees were inferred from the calculated distances by using neighbor joining (80) with the Vector NTI software program (Invitrogen). Consensus sequences of each strain were determined with the Vector NTI program by the alignment of the samples which grouped on the phylogenetic trees.
HTA.
Amplification of LMP1 and HTA analysis were performed as previously described (85). Briefly, LMP1 was amplified from the samples by nested PCR with 300 ng of DNA template in duplicate independent reactions with Taq polymerase (Promega, Madison, Wis.). The first round of amplification with primers LMP3UT and FUC-Hind3 (EBV coordinates 168427 to 168408; 5′-ATCAGAGAGCTTTGACAATGGCCCACATGACC-3′) yielded a 411- or a 381-bp product, depending on the presence or absence of the 30-bp deletion. Nested PCR with 6 μl of template from the initial reaction was performed with the primers FUE-Eco (EBV coordinates 168163 to 168183; 5′-ATCACGAGGAATTCGTCATAGTAGCTTAGCTGAAC-3′) and FUC-Hind3, which yielded a 264- or a 234-bp product.
The phylogenetically distinct LMP1 variants that have been identified include the prototypic undeleted B958 and other undeleted strains (Ch2, AL, NC, and Med-), as well as the 30-bp-deletion strains Ch1 and Med+. Clones of the carboxy terminus were prepared for each strain and used as positive controls for the HTA to determine the migration position of each strain. Each sample was analyzed with both an undeleted Ch2 and deleted Med+ probe at least twice from independent PCRs. The probes were made as previously described (85).
The heteroduplex formation reaction was performed with 8 μl of PCR product of a strain control or patient sample, 1 μl of annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.5], 20 mM EDTA), and 1 μl of 35S-radiolabeled probe. The mixtures were denatured for 5 min at 100°C, allowed to reanneal for 4 min at 4°C, separated on nondenaturing 10% polyacrylamide gels (Hoefer apparatus; Pharmacia Biotech, San Francisco, Calif.), dried in a gel dryer (Bio-Rad, Hercules, Calif.), and exposed to a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
HLA typing.
The samples were HLA typed by using the micro-SSP HLA class I DNA typing tray (One Lambda, Inc., Canoga Park, Calif.).
HLA epitope prediction.
HLA epitopes were predicted for each consensus LMP1 strain, as well as the full-length LMP1 sequence of the tumor biopsies, by using the HLA Peptide Binding Prediction of BioInformatics and Molecular Analysis Section available at (http://bimas.dcrt.nih.gov/molbio/hta_bind/).
Statistical methods.
For data that was in the paired format, the statistical method used was McNemar's test of equality of paired proportions. For data categorized into two by two contingency tables, the Fisher exact test was used to detect differences between proportions. Statistical analyses were performed with SAS statistical software (version 8.2; SAS Institute, Inc., Cary, N.C.).
Nucleotide sequence accession numbers.
The complete LMP1 nucleotide sequences of the different strains are available under the following GenBank accession numbers: AY337721, AY337722, AY337723, AY337724, AY337725, and AY337726. The strains are reported with four of the 33-bp repeats, although a range of two to six repeats was detected, with four or five being the most common.
RESULTS
Analysis of the transmembrane domain sequence of LMP1.
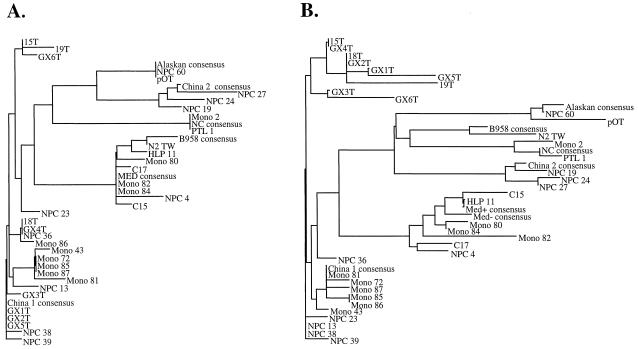
Seven distinct strains of LMP1 that can be clearly distinguished by characteristic base pair changes in the carboxy terminus have previously been described (18). Strain changes in the amino terminus of LMP1 also correlate but do not distinguish the strains as clearly (18). In the present study the transmembrane domain of LMP1 comprising amino acids (aa) 25 to 187 was sequenced to determine whether distinguishing changes between strains were present in the transmembrane domain. Samples sequenced over the transmembrane domain of LMP1 include one B958 strain (sample N2 TW); three previously described Med strain samples (NPCs C15 and C17 and HLP 11), as well as NPC 4 and Mono 80, 82, and 84; five previously described Ch1 strain samples (NPCs 13, 23, 36a, 38, and 39), as well as GX1-6T, -15T, -18T, and -19T and Mono 43, 72, 81, 85, and 87; three previously described Ch2 strain samples (NPCs 19, 24, and 27); two previously described AL strain samples (NPC 60 and pOT); and two previously described NC strain sample (PTL1 and Mono 2) (18).
The consensus sequence of each strain was determined by alignment with the Vector NTI computer program of the samples that grouped together on the phylogenetic tree drawn from the alignment of the 35 samples. Many of the strains share common coding and noncoding base pair changes in the transmembrane domain (Fig. 1). The Med strain samples had the least number of base pair changes compared to B958 in the coding region of LMP1. The Ch1 strain shared changes with the Med strain but also had additional changes. Unique base pair changes in the Ch1 strain resulted in amino acid changes 150D→A and 178L→M, with silent changes at aa 93, 114, and 186. Base pair changes unique to the Ch2 strain were detected at aa 33, 84, 91, 95, 124, and 144. A unique silent change was found at aa 60. The AL strain samples also shared common base pair changes with the Ch1 and Ch2 strains, but the changes at aa 29, 63, 82, and 101 were unique to the AL strain along with the silent change at aa 48. The NC strain had similar base pair changes to the Ch1, Ch2, and AL strains; however, this strain had numerous unique changes in the coding region of the transmembrane domain of LMP1 that included the changes at aa 26, 37, 43, 101, 109, 148, 155, and 179. Silent changes were found at aa 56 and 100 (Fig. 1).
FIG. 1.
Sequence variation and corresponding amino acid changes in the computer-derived consensus sequence of the transmembrane domain of the six strains of LMP1. Numbers across the top row correspond to the EBV genome coordinates; names in the left column refer to the six identified LMP1 strains. The base pair and the amino acid changes are in boldface.
Phylogenetic analysis of the consensus transmembrane and carboxy-terminal LMP1 amino acid sequences.
Previous work has shown that seven LMP1 strains can be distinguished by distinct changes in the carboxy terminus (18). Since the transmembrane domain also showed common and distinct aa changes among the LMP1 strains, it was of interest to determine whether the transmembrane domain also distinguished EBV strains and, if so, whether there was a similar correlation among the strains. Distant matrices were calculated from the amino acid sequences of the transmembrane domain of the samples, along with the computer-derived consensus for each strain and phylogenetic trees, were inferred by using neighbor joining (45, 80). The unrooted tree produced had six branches, since the Med+/− strains could not be distinguished since they differ only in the presence or absence of the 30-bp deletion, and all samples of a strain grouped with their consensus sequence (Fig. 2A). The B958 and Med strains were similar to each other, as were the Ch2 and AL strains (Fig. 2A). The Ch1 strain divided into subgroups by the absence of a consensus change or the presence of an additional change. Samples 15T and GX6T are missing the change at aa 144; 18T, GX4T, NPC 36, and Mono 86 are missing the characteristic change at aa 46. In contrast, Mono 43, 72, 85, 87, and 81 have an R→G mutation at aa 132. The other Ch1 samples (GX3T; NPCs 13, 38, 39, and 23; and 19T) contain sporadic but different changes (data not shown).
FIG. 2.
Identification of the LMP1 strains by phylogenetic analysis. (A) Phylogenetic tree drawn from amino acid sequences of the transmembrane domain of the computer-derived consensus sequences of the seven (the Med+ and Med− strains are identical in the transmembrane domain) previously identified LMP1 strains and the 35 samples. (B) Phylogenetic tree drawn from the consensus carboxy-terminal amino acid sequences of the seven previously identified LMP1 strains and the 35 samples.
The phylogenetic tree inferred from the distances matrices calculated from the carboxy-terminal amino acid sequences of the samples and the computer-derived consensus sequence for each of the seven LMP1 strains revealed results similar to those obtained from analysis with the transmembrane domain (Fig. 2B). All samples again grouped with their consensus sequence. The Ch1 strain again had two branches with the lower branch representing samples with a sporadic change or a D at 335, whereas many in the upper branch shared an L at aa 195 and a D at aa 335 (data not shown and Fig. 2B). The trees derived from the LMP1 sequences further strengthen the classification of these patterns as true phylogenetically distinct strains.
Full-length LMP1 sequence and mapping of known and computer-predicted HLA-restricted epitopes.
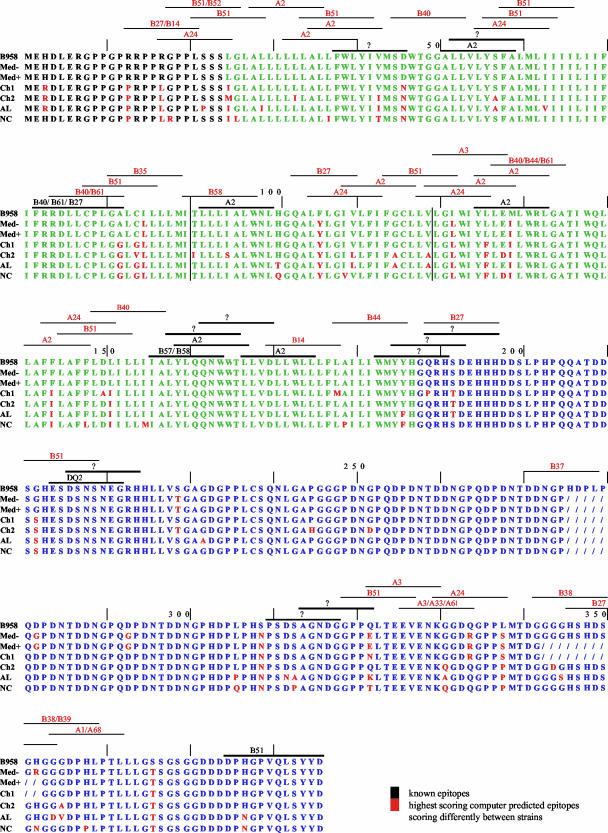
LMP1 and LMP2 are among the few viral proteins expressed in latent EBV infections in NPC and HD (7, 32, 72). Due to this restricted protein expression, the LMP1 and LMP2 proteins remain the only clear candidates for CTL-based therapy. Several functional epitopes have been described for LMP1, with the majority restricted to HLA-A2, so use of computer prediction models may allow for the identification of additional epitopes presented by haplotypes found in NPCs (17, 43, 58, 70). Due to the sequence variation in LMP1, the computer prediction model can also predict possible epitope functionality differences between the strains of LMP1. The majority of the known HLA-restricted epitopes scored in the top 10 of the epitopes predicted for an HLA-restriction by computer, suggesting congruency between the known and predicted epitopes. The majority of the computer-predicted epitopes discussed in this analysis of strains were in the top six of the strong scorers of the HLA restriction.
Previously identified restricted epitopes were identified within these strains of LMP1 (Fig. 3). Several changes in amino acid sequences of the known epitopes were detected in the different strains, including three of the known A2-restricted epitopes at positions 51, 92, and 125 (Fig. 3) (17, 43). All of the strains except B958 have changes in the A2-restricted epitope at 125 (YLL) (Fig. 3). The Ch2 and AL strains have changes in the epitope at position 51 (ALL) and Ch2 also has a change in the epitope at 92 (LLL) (Fig. 3). The Ch1, Ch2, AL, and NC strains have changes in the B40/B61/B27-restricted epitope at position 72 (FRR), and the AL and NC strains have changes in the B51-restricted epitope at position 375 (DPH) (Fig. 3) (58, 70).
FIG. 3.
Full-length consensus amino acid sequence of the LMP1 strains with previously described and computer-predicted HLA epitopes. The amino terminus is depicted in black; the transmembrane domain is depicted in green; and the carboxy terminus is depicted in blue. Amino acid changes from B958 consensus are shown in red, and deletions are shown with a forward slash (/). Vertical lines separate the exons. Previously described functional HLA epitopes are shown in black with the HLA restriction; a “?” refers to functional epitopes of unknown restriction. Computer-predicted epitopes that score differentially among the strains are shown in red.
Several other functionally identified epitopes of unknown HLA restriction also differed among the LMP1 strains, including the epitopes at positions 38 (FWL), 52 (LLV), 183 (WMY), 185 (YYH), 189 (QRH), 310 (PSD), and 314 (AGN) (Fig. 3) (17, 58). To determine whether these and other possible epitopes differed between the LMP1 strains, the LMP1 consensus sequence of each strain was analyzed for HLA-restricted epitopes by using a computer based prediction model. Numerous predicted HLA-restricted epitopes scored markedly different among the LMP1 strains. The most notable was the A2-restricted epitope at position 125 (YLL) that had the highest output score by the computer, indicating a possible strong reaction in vivo. This epitope has been previously identified to be a functional epitope (43). This A2-restricted epitope was only present in the B958 and Med strains and when actual strain variants of this epitope have been tested, a much reduced response has been observed supporting the use of the computer prediction model (17). The HLA-B14 and -B27 predicted epitope at position 13 (RRP) also scored only or much higher in the B958 and Med strains than the Ch1, Ch2, AL, and NC strains. These predicted differences in reactivity may contribute to evasion of the immune response.
HTA analysis of NPC and matching blood from areas of endemicity and nonendemicity.
The HTA based on strain-defining changes in the carboxy terminus of LMP1 has proved to be a powerful tool in determining the LMP1 strains of EBV in a sample (84, 85). The seven strains migrate uniquely when screened with both a deleted (Med+) and undeleted (Ch2) probe (Fig. 4) (85). In previous studies, compartmental differences in strain prevalence were detected between the oral cavity and PBL in asymptomatic carriers and infectious mononucleosis patients (83, 84).
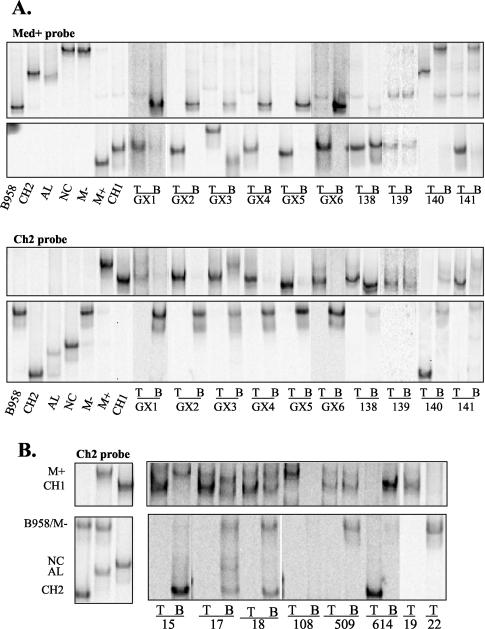
FIG. 4.
(A) Strain profile of EBV in NPC and matching blood of patients from the region of China where NPC is endemic, as determined by LMP1 HTA with Med+ and Ch2 probes. The first seven lanes correspond to control lanes for migration of the different strains. NPC tumor (lanes T) and peripheral blood (lanes B) strains were identified based on their migration in relation to the controls. (B) Strain profile of EBV in NPC and matching blood of patients from the region of China where NPC is not endemic, as determined by LMP1 HTA with Ch2 probe. The first three lanes correspond to control lanes for the migration of the different strains. NPC tumor (lanes T) and peripheral blood (lanes B) strains were identified based on their migration in relation to the controls.
To determine whether strains differed between tumor and matching PBL, samples were screened by using the LMP1 HTA. Ten NPCs from the southern region of China were analyzed by HTA where NPC is endemic. Nine of the ten NPCs (GX1, GX2, GX3, GX4, GX5, GX6, 138, 139, and 141) had the Ch1 strain present in the tumor, as shown by the migration on the HTA with both the Med+ and Ch2 probes (Fig. 4A). It has been previously shown that strains that contain additional sequence changes from the known controls have a slightly altered migration (Fig. 4A). Thus, the Ch1 band in tumor sample GX3 ran higher with the Med+ probe due to two additional base pair changes at positions 168239 and 168300 and the Ch1 band in GX6 due to changes at positions 168246 and 168300 (Fig. 4A). The final endemic sample, NPC 140, had the Ch2 strain (Fig. 4A). The identification of specific strains by HTA was confirmed by using the Ch2 probe (Fig. 4A). These data revealed that a single strain was usually detected in the tumors.
The strain profile of the matching PBL was quite different than the tumor. Interestingly, the matching PBL from the region where NPC is endemic had predominantly or only the B958 strain (Fig. 4A). In PBL samples from GX1, GX2, GX4, GX5, and GX6, the predominant strain detected was B958 (Fig. 4A). GX3 PBL had both the B958 and Med+ strain (Fig. 4A). The Ch1 strain was prevalent in the PBL and tumor of sample 138 (Fig. 4A). Sample 139 contained the Ch1 strain in both the tumor and PBL (Fig. 4A). In sample 141, the Ch1 strain was detected in the tumor and Med− in the PBL (Fig. 4A). Sample 140 had the Ch2 strain in the tumor and the Med− and Ch1 strains in the PBL (Fig. 4A). In most cases (9 of 10) the LMP1 variants detected in the tumor were also faintly detected in the PBL after long exposure of the film (data not shown). Using the McNemar's test for equality with the paired endemic samples, we found it statistically significant that the Ch1 strain was consistently in the tumor but not the predominant strain in the PBL (P = 0.0156). Combining all of the HTA data from the samples in the present study and other samples analyzed in the lab for a total of 26 endemic tumor samples and 11 endemic PBL samples, the sample size was sufficient to be able to detect an association between the presence (or absence) of a particular strain in either the endemic tumors or the endemic PBL by using the Fisher exact test (data not shown). It was highly statistically significant that the B958 strain that was consistently detected in the blood (7 of 11 samples) was never detected in the tumors of the endemic NPCs (0 of 26 samples) (P < 0.0001), and the Med− strain present in blood (4 of 11 samples) was not detected in the endemic tumors (0 of 26 samples) (P = 0.005) (Table 1). These compartmental differences suggest a possible selection against these strains in NPC.
TABLE 1.
Prevalence of LMP1 strain in Chinese NPC and PBLa
| Strain | Region of endemicity (no. of positive samples/total no.)
|
P | Region of nonendemicity (no. of positive samples/total no.)
|
P | ||
|---|---|---|---|---|---|---|
| T | PBL | T | PBL | |||
| B958 | 0/26 | 7/11 | <0.0001 | 0/26 | 3/7 | 0.006 |
| Med− | 0/26 | 4/11 | 0.005 | 3/26 | 6/7 | 0.0005 |
| Med+ | 0/26 | 1/11 | 0.3 | 0/26 | 3/7 | 0.006 |
| Ch1 | 21/26 | 8/11 | 0.67 | 17/26 | 7/7 | 0.15 |
| Ch2 | 5/26 | 1/11 | 0.65 | 6/26 | 3/7 | 0.36 |
| NC | 0/26 | 1/11 | 0.3 | 0/26 | 4/7 | 0.0009 |
| AL | 0/26 | 0/11 | 0/26 | 0/7 | ||
P values were calculated by using the Fisher exact test, analyzing for the presence of strain versus location (i.e., tumor [T] or PBL).
Eight NPCs from the northern region of China where NPC was not endemic, six with matching blood, were screened by HTA. Six of the nonendemic NPC samples contained the Ch1 strain (i.e., samples 15, 17, 18, 108, 509, and 19) (Fig. 4B). NPC 614 contained the Ch2 strain, and NPC 22 contained the Med− strain with two additional base pair changes at positions 168246 and 168348 (Fig. 4B and data not shown). Although sample 15 had Ch1 in the tumor, the PBL had Ch2, Med+, and Med− strains (Fig. 4B). NPCs 17 and 18 had multiple strains in the blood, including Ch2, NC, B958, Med−, Med+, and Ch1 (Fig. 4B). The tumor of subject 108 revealed the Ch1 strain (with additional changes), whereas the PBL contained predominantly Med− (Fig. 4B). Sample 509 had strains B958, Med−, and Ch1 in the PBL and Ch1 in the tumor (Fig. 4B). Sample 614 had Ch1 and Med− strains in the PBL, with the Ch2 strain in the tumor (Fig. 4B). With long exposure, sample 19 was found to have the Ch1 strain in the PBL and, in most cases (6 of 7 samples), the strain detected in the other nonendemic NPCs was faintly detected in the PBL (data not shown). All strains detected with the Ch2 probe were confirmed with the Med+ probe (data not shown). Although the Ch1 strain was prevalent in the NPCs from both regions of endemicity and nonendemicity, the B958 strain was prevalent in the PBL of patients from areas where NPC was endemic, and multiple LMP1 strains were detected in patients from areas where NPC was not endemic (Fig. 4). Importantly, the strain in the tumor was usually not the prevalent strain in the blood in the majority of cases (12 of 16 samples). Combining all HTA data from the samples in the present study and others analyzed in the lab for a total of 26 nonendemic tumors and 7 nonendemic PBL samples, a Fisher exact test to determine associations between the presence or absence of a particular strain in nonendemic tumor or PBL indicated that it was statistically significant that (i) the Med− strain, which was detected in six of seven PBL samples, was present in only 3 of 26 tumors (P = 0.0005); (ii) the NC strain, which was detected in four of seven PBL samples, was not present in the tumors (0 of 26) (P = 0.0009); (iii) the Med+ strain detected in three of seven PBL samples was not detected in the nonendemic tumors (0 of 26) (P = 0.006); and (iv) the B958 strain present in three of seven PBL samples was not detected in the tumors (0 of 26) (P = 0.006) (Table 1 and data not shown). The striking absence of certain LMP1 strains in the tumors again suggests a possible selection against these strains in NPC.
HLA typing and epitope sequencing of samples from areas of endemicity and nonendemicity.
Specific HLA haplotypes, notably A2 and B46, have been shown to be prevalent in NPC patients (35, 50). The data presented here indicate that in the Ch1 strain several A2-restricted epitopes are altered, suggesting that changes in HLA-restricted epitopes in LMP1 variants may result in reduced immune recognition. Since there was a striking difference in the LMP1 strain detected in the tumors compared to the PBL, several of the samples from areas of endemicity and nonendemicity were HLA typed (Table 2). The majority of samples were HLA-A24 or -A2 restricted.
TABLE 2.
HLA typing of NPC samples from regions of endemicity and nonendemicity
| Origin and sample | LMP1 strain
|
HLA-A
|
HLA-B
|
|||
|---|---|---|---|---|---|---|
| Tumor | PBL | 1 | 2 | 1 | 2 | |
| Endemic | ||||||
| GX1 | Ch1 | B958 | A24 | B35 | B15 | |
| GX2 | Ch1 | B958 | A24 | B13 | B51 | |
| GX3 | Ch1 | B958 | A2 | A24 | B40 | B40 |
| GX4 | Ch1 | B958 | A11 | A24 | B27 | B40 |
| GX5 | Ch1 | B958 | A24 | A33 | B51 | B58 |
| GX6 | Ch1 | B958 | A2 | A24 | B46 | |
| Nonendemic | ||||||
| 15 | Ch1 | Multiple | A2 | A11 | B13 | B46 |
| 18 | Ch1 | Multiple | A2 | B51 | ||
| 19 | Ch1 | Ch1 | A1 | A2 | B7 | B15 |
| 22 | Med− | NDa | A2 | A33 | B44 | B15 |
ND, not determined.
To determine whether possible changes were present in HLA-restricted epitopes and to confirm that the strain identified by HTA of the carboxy terminus was consistent throughout the gene, the full-length sequence of LMP1 of these tumor samples was determined, and HLA-restricted epitopes were predicted for each protein sequence by computer analysis. All 10 tumor samples were found to have the characteristic strain changes throughout the full-length LMP1, as predicted by the HTA strain, although some had minor base pair change differences (data not shown). For example, GX1T was predicted by HTA to contain the Ch1 strain in the carboxy terminus, and analysis of the amino terminus and transmembrane domains confirmed the corresponding characteristic Ch1 base pair changes in these regions.
When the scores of the computer-predicted epitopes from the full-length LMP1 sequences of the tumors were compared to B958 sequence, which was not present in the tumors, marked differences could be found. Although changes were not found in the known A2-restricted epitopes ALLVLYSA (ALL), LLLIALWNL (LLL), and YLQQNWWTL (YLQ), the known epitope YLLEMLWRL (YLL) was predicted not to be recognized in the Ch1, Ch2, AL, and NC strains (Table 3) (43). This epitope scores the highest of all LMP1 epitopes predicted by the computer, and functional studies have shown that the Ch1 changes in this epitope render it less responsive in vitro (17, 40). Four of the A2-restricted tumors (GX6, -15, -18, and -19) had the characteristic Ch1 changes at positions 126 and 129, and one tumor (GX3) had a different change at position 126 (Table 3). Most interestingly, the A2-restricted 22T that had the Med− strain by HTA and full-length sequencing had an additional nonconsensus change at position 130 that rendered this epitope less responsive by computer prediction (Table 3). All five Ch1 strain samples (GX3T, GX6T, 15T, 18T, and 19T) were predicted by computer to not react to the predicted A2 epitope at position 118, LVL, due to amino acid changes at positions 122 and 126 (Table 3). These changes in the A2 epitopes at positions 118 and 125 were changed in the 2 or 9 position, known to be critical for binding to MHC-I (71). NPC 19T had additional mutations in aa 172, 173, and 154 that rendered the known A2 epitope at position 167, LLV, nonreactive and the predicted epitope at position 152, ILL, fourfold less reactive than B958 (Table 3) (43).
TABLE 3.
Known and predicted HLA-restricted epitopes in LMP1 and the variants found in NPC that showed a marked decrease in reactivity as determined by computer compared to B958-like epitopes
| HLA | Positiona | B958-like epitope [sample(s)]b | Variant epitope(s) in Ch1 and Med− strains [sample(s)]b |
|---|---|---|---|
| A2 | 51* | ALLVLYSFA (GX3T, GX6T, 15T, 18T, 19T, 22T) | |
| 92* | LLLIALWNL (GX3T,GX6T,15T,18T,19T, 22T) | ||
| 125* | YLLEMLWRL | YFLEILWRL (GX6T, 15T, 18T, 19T), YILEILWRL (GX3T), YFLEIFWRL (22T) | |
| 159* | YLQQNWWTL (GX3T, GX6T, 15T, 18T, 19T, 22T) | ||
| 167* | LLVDLLWLL (GX3T, GX6T, 15T, 18T, 22T) | LLVDLVRLL (19T) | |
| 118 | LVLGIWIYL (22T) | LVLGLWIYF (GX6T, 15T, 18T, 19T), LVLGLWIYI (GX3T) | |
| 152 | ILLIIALYL (GX3T, GX6T, 15T, 18T, 22T) | ILVIIALYL (19T) | |
| A24 | 143 | FFLAFFLDL | FILAFFLAI (GX1T, GX2T, GX3T, GX4T, GX5T), FFLAFFLAI (GX6T) |
| 17 | RGPPLSSSL | LGPPLSSSI (GX1T, GX2T, GX3T, GX4T, GX5T, GX6T) | |
| 330 | KGGDQGPPL | KGGRDGPPS (GX1T, GX2T, GX3T, GX4T, GX5T, GX6T) | |
| 118 | LVLGIWIYL | LVLGLWIYF (GX2T, GX4T, GX5T, GX6T), LVLGLWIYI (GX1T, GX3T) | |
| A33 | 65 | IILIIFIFR (GX5T) | ILLIIFIFR (22T) |
| B35 | 81 | GALCILLLM | GGLGLLLLM (GX1T) |
| B51 | 375* | DPHGPVQLSYYD (GX2T, GX5T, 18T) | |
| 114 | FGCLLVLGI | FGCLLVLGL (GX2T, GX5T, 18T) | |
| 81 | GALCILLLM | GGLGLLLLM (GX2T, GX5T, 18T) | |
| B40 | 74* | RDLLCPLGA | RDLLCPLGG (GX3T, GX4T) |
| 45 | SDWTGGALL | SNWTGGALL (GX3T, GX4T) | |
| 149 | LDLILLIIA | LAIILLIIA (GX3T, GX4T) | |
| B27 | 72* | FRRDLLCPL (GX4T) | |
| 13 | RRPPRGPPL | PRPPLGPPL (GX4T) | |
| 189 | QRHSDEHHH | PRHTDELHH (GX4T) | |
| B58 | 156* | IALYLQQNW (GX5T) | |
| 218 | NSNEGRHHL | NSNDGRHLL (GX5T) |
The position corresponds to the B958 amino acid sequence of LMP1. Known LMP1 epitopes are indicated by an asterisk.
T, tumor. Mutations in the epitopes are indicated in boldface.
Since functional A24-restricted epitopes have not been identified and many of the NPC samples were A24 restricted, the computer-predicted epitopes were identified. Four HLA-A24-restricted epitopes identified in the B958 sequence were predicted to be nonfunctional in the Ch1 sequence. All six of the A24-restricted samples had changes in all four epitopes. GX1, GX2, GX3, GX4, and GX5 had three changes in the A24-restricted 143 epitope, FFL, due to the characteristic Ch1 strain changes in LMP1 found in the tumor, and one sample (GX6) had two of the changes but lacked the characteristic Ch1 change at aa 144 (Table 3). All six A24 typed samples (GX1 to -6) had two changes in the predicted epitope at position 17, RGP, three changes in the predicted epitope at 330, KGG, as well as two changes in the predicted epitope at 118, LVL, due to the characteristic Ch1 strain changes in the tumors (Table 3).
The HLA-A33-restricted sample 22 also responded two fold less to the A33 computer-predicted epitope at position 65, IIL. This change in sample 22T at aa 66 is not a consensus Med- sequence change (Table 3). The HLA-B35-restricted epitope predicted at position 81, GAL, did not score with the characteristic Ch1 strain changes at aa 82, 84, and 85 found in GX1T (Table 3).
Changes were not detected in the Ch1 strain in the known B51-restricted epitope at position 375, DPH, possibly since this epitope encompasses the PxQxS motif that is important for signaling (Fig. 3) (58, 70). However, the three HLA-B51-typed samples (GX2, GX5, and 18) had a much weaker predicted response to the B51 predicted epitopes at position 114, FGC, and at position 81, GAL, due to the characteristic Ch1 changes at positions 122, 82, 84, and 85 (4- and 12-fold, respectively) (Table 3).
Similarly, the B40 typed samples (GX3 and GX4) were predicted not to react to the known epitope at 74, RDL, and the predicted epitope at position 45, SDW, due to the characteristic Ch1 changes at aa 82 and 46 and displayed a much weaker response to the predicted epitope at 149, LDL, due to the amino acid changes at positions 150 and 151 (Table 3) (58, 70). Possible anchoring amino acids in the epitopes were again changed.
Although the HLA-B27-typed GX4 did not have changes in the known B27 epitope at position 72, FRR, the computer-derived epitope at position 13, RRP, was predicted to be 30-fold less reactive due to characteristic Ch1 amino acid changes at positions 13 and 17 (58, 70). Similarly, the epitope predicted at aa 189, QRH, in the B95 strain was not identified due to Ch1 changes at aa 189 and 192 in the tumor (Table 3) (58). The QRH epitope has been previously described, but the HLA restriction is unknown, although it is thought not to be HLA-B27 restricted (17).
GX5, HLA-B58 typed, did not have changes in the known epitope at position 156, IAL, but due to two additional changes from the Ch1 strain LMP1 sequence at aa 221 and 225 had a weaker predicted response to a predicted epitope at position 218, NSN (Fig. 3 and Table 3) (17). The NSN epitope has been described as a functional epitope, but the restriction is unknown (17).
These data indicate that the strains detected in NPC had changes in several of the potential epitopes of LMP1 presented by a specific HLA of the patient. The prevalence of specific strains in tumors with changes in these epitopes may contribute to reduced immune recognition of virus-infected cells. Computer predictions for the other HLA haplotypes identified in these samples (B13, B15, and B46) are not available.
LMP2 sequence of known HLA-restricted epitopes.
LMP2 is expressed in NPC tumors and also in latently infected peripheral blood memory B cells and has been shown to induce a stronger CTL response than LMP1 (77). Consequently, LMP2 sequence was determined from the two compartments to evaluate possible epitope changes that could also contribute to the difference in the prevalence of one strain in the tumor versus the strains detected in the blood. LMP2 sequences from the tumor samples of nine patients (GX1, GX2, GX3, GX4, GX5, GX6, 18, 19, and 22) and the blood samples from four patients (GX1, GX2, GX5, and 19) were determined. Variation was detected in LMP2 from the Chinese samples, but in most cases the same changes were found in the tumor and blood. Of the six known HLA-A2-restricted epitopes, a change was only identified in the known A2 epitope at position 426, CLG. This epitope was changed in GX6T, 18T, 19B, and 22T but not in GX3T or 19T (Table 4) (52). This change in this epitope has been shown not to affect reactivity, and the computer also predicted no change in reactivity (52). The remaining tumor samples did not have changes in the known A2 epitopes, suggesting that the tumors and PBL would be equally recognized by LMP2-specific CTL (Table 4) (49).
TABLE 4.
Sequence of known and predicted LMP2 epitopes from NPC and PBL from regions of endemicity and nonendemicity
| HLA | Positiona | Consensus (B958) epitope [sample(s)]b | Variant epitopes [sample(s)]b |
|---|---|---|---|
| A2 | 292* | GGLGTLGAAL (GX6T, 18T, 19T and -B, 22T) | |
| 329* | LLWTLVVLL (19T) | ||
| 356* | FLYALALLLLAS (GX3T, GX6T, 18T, 19T and -B, 22T) | ||
| 426* | CLGGLLTMVA (GX3T, 19T) | SLGGLLTMVA (GX6T, 18T, 19B, 22T) | |
| 447* | LLSAWILTAGF (GX3T, GX6T, 18T, 19T and -B, 22T) | ||
| 453* | LTAGFLIFLI (GX3T, GX6T, 18T, 19T, and -B, 22T) | ||
| A24 | 222* | IYVLVMLVL (GX1T and -B, GX2T and -B, GX3T, GX4T, GX5T and -B, GX6T) | |
| 419* | TYGPVFMCL | TYGPVFMSL (GX1T and -B, GX2T and -B, GX3T, GX4T, GX5T and -B, GX6T) | |
| 170 | SYAAAQRKL | SSAAAQRKL (GX1T and -B, GX2T and -B, GX3T, GX4T, GX5T and -B, GX6T) | |
| A11 | 341* | SCSSCPLSK (GX4T) | |
| B27 | 236* | RRRWRRLTV | LRRWRRLTV (GX4T) |
| B40 | 200* | IEDPPFNSLLF | IEDPPFNSILF (GX3T) |
| ?c | 61 | EDPYWGNGDRHSDYQ (GX2T, GX3T, GX4T, GX6T) | EDLDWGNGDRHSDYQ (19T, 22T), EDLYWGNGDRHSDYQ (GX5T) |
| 121 | NPVCLPVIVAPYLF (GX2T, GX3T, GX5T, 18T, 19T, 22T) | LPLCLRVIVAPYLF (GX4T) | |
| 141 | ASCFTASVSTVVTAT (19T) | ASCFTASVSTVVSAT (GX2T, GX3T, GX5T, 18T, 22T) | |
| 249 | MFLACVLVLIVDAV (19T) | MFLACLVVLIVDAV (GX1T and -B, GX2T and -B, GX4T, GX5T and -B, GX6T 18T, 19B, 22T) |
The position corresponds to B958 amino acid sequence of LMP2. Known LMP2 epitopes are indicated by an asterisk.
T, tumor; B, PBL. Mutations in the epitopes are indicated in boldface.
?, epitope(s) of unknown HLA restriction.
Of the six A24-restricted patients (GX1, GX2, GX3, GX4, GX5, and GX6), tumor and blood samples were unchanged in the known epitope at aa 222, whereas all had changes in the known A24 epitope at position 419, TYG (Table 4) (58). However, this change was also found in the PBL from GX1, GX2, and GX5. Similarly, the computer-predicted A24-restricted epitope at 170 (SYAAAQRKL) was changed in all of the A24 samples (SSA), but this change was also found in the matched PBL samples.
The known A11-restricted epitope at position 341, SCS, was unchanged in GX4T; however, the known B27 epitope at position 236, RRR, had a change and by computer was predicted to be threefold less reactive (58). The known B40-restricted epitope at position 200, IED, was changed in GX3T and, as determined by computer prediction, is fivefold less reactive. Again, possible anchoring positions of the epitope were changed (Table 4) (49, 58).
However, several of the described epitopes of unknown restriction were changed in some samples (Table 4) (58). The epitope at position 61, EDP, had two changes in two of the samples (19T and 22T) from regions where NPC is not endemic and one change in a sample (GX5T) from a region where NPC is endemic, but it was unchanged in samples GX2T, GX3T, GX4T, and GX6T from areas of endemicity (Table 4) (58). GX4T had three changes in the epitope at position 121, NPV; however, six of the tumor samples did not have changes (Table 4) (58). The epitope at position 141, ASC, was unchanged in 19T, whereas the tumor samples from GX2, GX3, GX5, 18, and 22 had one amino acid change (Table 4) (58). Sample 19T did not have a change in the known epitope at position 249, MFL, but two changes were detected in GX1T and PBL, GX2T and PBL, GX4T, GX5T and PBL, GX6T, 18T, 19PBL, and 22T (Table 4) (58). This described epitope overlaps a predicted A2 epitope (IMFLACVLV) that is predicted to be much less reactive with the amino acid changes (MMMFLACLVV) seen in the samples. Only sample 19, which had the same Ch1 LMP1 strain in the tumor and PBL, had a different LMP2 sequence in the tumor compared to the blood, with the blood LMP2 being more variant. The changes that were found in these potential epitopes of LMP2 may render the virus less susceptible to immune surveillance; however, in all but one example the LMP2 was the same in the blood and tumor and would be equally recognized by the immune system. The fact that a common LMP2 strain was found in the Chinese population regardless of the LMP1 strain suggests that immune recognition of LMP2 likely does not account for the difference in strain prevalence between the blood and tumor samples.
DISCUSSION
The data presented here further reveals that the seven phylogenetically distinct strains of LMP1 previously identified by amino acid changes in the carboxy terminus are also distinguished by amino acid changes throughout the protein (18). Interestingly, identification of LMP1 variants by using HTA indicates that there are striking differences in relative abundance of the variants between the samples of NPCs and PBL. The frequent detection of the Ch1 strain in NPC has been suggested to reflect the high prevalence of Ch1 in the general population. However, the data presented here reveal that the B958 strain is also prevalent in peripheral blood samples from the region of China where NPC is endemic, although the B958 strain is rarely found in NPC. In the majority of samples from this region, a single different strain was detected in the NPC and PBL samples, whereas multiple LMP1 variants were detected in the PBL samples from the region of China where NPC is not endemic. The absence of some of these variants within the tumor is highly statistically significant and suggests a selection against their presence within the tumors. Importantly, the strains detected in all examples of tumors with a given HLA restriction had changes in several of the known and predicted HLA epitopes within LMP1 in comparison with the strains detected in the PBL that retained the epitope sequence. It is possible that the changes that are characteristic of the Ch1 strain in several HLA-restricted epitopes may result in decreased binding and presentation by MHC-I and/or less-efficient recognition by the T-cell receptor.
The HLA-A11 haplotype is a dominant allele in China; however, the present study and previous studies have determined that most NPC patients are HLA-A2 or -A24 restricted (35, 50, 53). Although A2 and A24 alleles are prevalent in the general population of China, they are found more frequently in NPC patients, which suggests a possible relationship between HLA type and the development of NPC (12). The majority of the identified epitopes in LMP1 and LMP2 are restricted through HLA-A2 (58). Since A24-restricted epitopes have not been identified in LMP1 and since the majority of the NPC samples analyzed were A24 restricted, computer prediction enabled additional analysis. The data presented here reveal that many of the A2- and A24-restricted LMP1 epitopes are changed in the Ch1 and Ch2 strains that were predominant in the tumors in comparison with some of the strains found in the PBL of the same patient. Many of these changes were in anchoring positions within the epitopes and could possibly interfere with MHC-I binding (71). An example is the known A2-restricted YLL epitope, which has been shown to induce a much weaker CTL response in a chromium release assay when the amino acid changes characteristic of Ch1 were tested (17, 40).
Although many of the known and predicted A2-restricted epitopes had potentially significant changes, not all of the A2 epitopes are altered in the Ch1 strain, and the functional epitopes at aa 51, 92, and 156 should be presented in A2-restricted individuals with the Ch1 strain. Intriguingly, several studies to identify LMP1 epitopes have shown that in Asian populations A2-restricted healthy individuals respond with much less frequency than A2-restricted healthy Caucasians to the identified epitopes at 51 and 159 spanning LMP1 (17, 50). It has been shown that multiple epitopes of a common HLA restriction may exist within a single protein and, within the EBNA3B protein, multiple A11 epitopes have been identified (25). A screen of individual responses revealed that most individuals responded to one strong epitope but that the degree of response to other epitopes was variable in presence and strength (25). Considering the multiple known and predicted A2 epitopes in LMP1, it is possible that in Asian populations the presentation of the epitopes at 51, 92, or 159 is apparently nonexistent or undetectable. Thus, the Ch1 consensus sequence changes within the strong epitope at 125 could eliminate presentation of LMP1 in Asian A2-restricted individuals.
LMP1 is not expressed in latently infected PBL but is detected in a considerable proportion of NPC and HD tumors and is thought to be expressed in all cells in early, preinvasive examples of NPC (73). Malignant cells that express LMP1 but are not recognized due to epitope changes in LMP1 would persist, and cells whose LMP1 epitopes are recognized would be eliminated. This negative selection of the immune system on strains detected in the blood would be reflected in the striking predominance of the Ch1 strain in the tumors.
It is of interest that many of the LMP1 variants had changes in predicted HLA epitopes of various restrictions. This variation in potential immune recognition could contribute to the development of EBV-associated diseases in distinct populations and individuals. Changes in HLA epitopes in the EBNA proteins have also been identified within isolated populations and likely contribute to the prevalence and persistence of specific EBV strains in populations (14). The strongest CTL response to EBV has been shown to be directed against EBNA3A, -B, and -C (40, 77). Interestingly, an escape mutant of EBV in a posttransplant patient treated with CTL therapy developed into a lethal lymphoma due to a deletion in EBNA3B that encompassed two immunodominant CTL epitopes (27). The sporadic, nonconsensus mutation in the Med− strain in the 22T sample, an A2-restricted sample, suggests that sporadic mutations may also occur in LMP1 that could enable immune escape.
In contrast to LMP1, the LMP2 sequences did not distinguish the strains in the tumor and PBL. Several A2- and A24-restricted epitopes have been described for LMP2, and most were unchanged in the tumors and PBL in the present study (58). Many of these LMP2 epitopes, including the A2-restricted FLY, CLG, and LLW and the A24-restricted TYG and IYV, are presented in a TAP-independent manner (41, 47, 48). The hydrophobic A2-restricted epitopes, LLS and CTA, may also be presented in a TAP-independent manner and did not have changes. It has been proposed that upregulation of TAP1 and TAP2 by LMP1 may lead to more efficient presentation of TAP-dependent epitopes, so TAP-independent epitopes (like LMP2) may evade recognition by being diluted on the surface of the cell (69). The decreased presentation of LMP2 on the cell surface would decrease the selective pressure for epitope changes, as well as enable the persistence of both tumor cells and PBL expressing LMP2 (7, 10, 89). However, several predicted epitopes of LMP2 were changed and may contribute to possible immune evasion in the peripheral blood and tumor.
Although multiple potential CTL epitopes are changed among the variant LMP1, several strains have some changes in common with the Ch1 and Ch2 strains and yet are not detected in the tumors. This may indicate that differences in the biologic properties of LMP1 variants are also a factor in the development of cancer. LMP1 is a transmembrane protein that functions as a constitutively activated member of the tumor necrosis factor receptor family (62). The protein contains three distinct domains. The short 24-aa cytoplasmic amino terminus anchors the protein in the membrane and regulates the turnover of the protein by ubiquitin-proteasome degradation pathway (1, 91). The six transmembrane domains induce aggregation of LMP1 in the membrane (24). The carboxy terminus has two activation domains, carboxy-terminal activating region 1 (CTAR1) and CTAR2, responsible for activation of NF-κB, JAK/STAT, and JNK (20, 26). All LMP1 strains retain the PxQxT motif in CTAR1 and the PxQxS motif in CTAR2, suggesting that all strains can interact equally with the TNF receptor associated factors leading to NF-κB activation (6, 15, 19, 37, 38, 81). One notable change among the strains of LMP1 is at aa 129, where only the B958 strain retains the methionine for the lytic LMP1; consequently, only the B958 strain can produce lyLMP1. Lytic LMP1 has been suggested to negatively regulate LMP1 activation of NF-κB (21). Increased AP1 and NF-κB activation has been reported with the Ch1 and NC strains (using our nomenclature), and increased activation of NF-κB and upregulation of epidermal growth factor receptor has been observed with the Med+ strain in comparison with B958 (22, 60). These differences in signaling map to the amino terminus and the transmembrane domains of LMP1 (5, 39, 60). There are potential sequence differences in the amino and transmembrane domains that may be responsible. Decreased NF-κB and AP1 activation have been described for a substrain of Ch1 that lacks the first four amino acid changes in the amino terminus characteristic of Ch1 (aa 3, 13, 17, and 25) (22). This Ch1 substrain has been detected in several NPCs and is the prevalent Ch1 strain in European and American asymptomatic carriers and infectious mononucleosis subjects (data not shown) (18). These same changes in the four amino-terminal amino acids are also found in the Ch2 strain and may also contribute to functional differences (18, 87). The Ch1 strain found in the endemic NPCs was also found to frequently carry a change at aa 335 (G→D), an amino acid change previously shown prevalent in NPCs, but not in healthy Chinese subjects (data not shown) (90). Secretion of LMP1 has been suggested to have direct immunosuppressive effects on tumor-infiltrating lymphocytes (16, 23, 56). This immunosuppressive ability maps to aa 34 to 40 of LMP1, a region where only the NC strain has a change (16). Perhaps the absence of the NC strain in NPC reflects an inability of the NC LMP1 to inhibit T-cell proliferation and natural killer cytotoxicity. These potential differences in signaling and biologic properties of the LMP1 variants may also contribute to differences in pathogenicity.
Multiple studies have suggested that the immune recognition of EBV-infected cells may be affected through several mechanisms. In latently infected PBL, EBV expression is restricted to EBNA1 and LMP2 (89). EBNA1 is not thought to be presented by MHC-I and the TAP-independent epitopes within LMP2 may not be efficiently presented (4, 55, 69). Latently infected lymphocytes that shift into a latent, proliferating mode of infection are eliminated due to CTL recognition of the EBNA2 and EBNA3 proteins (77). Interestingly, epitope changes have been identified in these proteins that may facilitate persistence in populations with predominant HLA types (8, 14). During the development of NPC, LMP1 is expressed in the absence of the EBNAs, the major viral CTL targets. The data presented here suggest the strain-specific changes in HLA epitopes in LMP1 may enable its expression in the tumors without recognition by LMP1-specific CTL. In summary, the striking consistent sequence variation of LMP1 strains may contribute to the transformation of epithelial cells in NPC through reduced immune recognition and also through differences in molecular and biologic properties that are yet to be elucidated.
Acknowledgments
We thank John Schmitz of the Department of Pathology and Laboratory Medicine, UNC Hospital, for the HLA typing of the samples. We thank Nancy Sung, Yi Zeng, Alan Rickinson, and Rajadura Pathmanathan for samples.
This study was supported by grants CA32979 and DE 11644 from the National Institutes of Health.
REFERENCES
- 1.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway: targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275:23491-23499. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, G. J., L. L. Decker, R. B. Freeman, and D. A. Thorley-Lawson. 1999. Epstein-Barr virus-infected resting memory B cells, not proliferating lymphoblasts, accumulate in the peripheral blood of immunosuppressed patients. J. Exp. Med. 190:567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 4.Blake, N., S. Lee, I. Redchenko, W. Thomas, N. Steven, A. Leese, P. Steigerwald-Mullen, M. G. Kurilla, L. Frappier, and A. Rickinson. 1997. Human CD8+ T-cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity 7:791-802. [DOI] [PubMed] [Google Scholar]
- 5.Blake, S. M., A. G. Eliopoulos, C. W. Dawson, and L. S. Young. 2001. The transmembrane domains of the EBV-encoded latent membrane protein 1 (LMP1) variant CAO regulate enhanced signalling activity. Virology 282:278-287. [DOI] [PubMed] [Google Scholar]
- 6.Brodeur, S. R., G. Cheng, D. Baltimore, and D. A. Thorley-Lawson. 1997. Localization of the major NF-κB-activating site and the sole TRAF3 binding site of LMP-1 defines two distinct signaling motifs. J. Biol. Chem. 272:19777-19784. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, L., Q. Y. Yao, A. B. Rickinson, and L. S. Young. 1992. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J. Virol. 66:2689-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows, J. M., S. R. Burrows, L. M. Poulsen, T. B. Sculley, D. J. Moss, and R. Khanna. 1996. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. J. Virol. 70:2490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busson, P., R. H. Edwards, T. Tursz, and N. Raab-Traub. 1995. Sequence polymorphism in the Epstein-Barr virus latent membrane protein (LMP)-2 gene. J. Gen. Virol. 76:139-145. [DOI] [PubMed] [Google Scholar]
- 10.Busson, P., R. McCoy, R. Sadler, K. Gilligan, T. Tursz, and N. Raab-Traub. 1992. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J. Virol. 66:3257-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callan, M. F., N. Steven, P. Krausa, J. D. Wilson, P. A. Moss, G. M. Gillespie, J. I. Bell, A. B. Rickinson, and A. J. McMichael. 1996. Large clonal expansions of CD8+ T cells in acute infectious mononucleosis. Nat. Med. 2:906-911. [DOI] [PubMed] [Google Scholar]
- 12.Cao, K., J. Hollenbach, X. Shi, W. Shi, M. Chopek, and M. A. Fernandez-Vina. 2001. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum. Immunol. 62:1009-1030. [DOI] [PubMed] [Google Scholar]
- 13.d'Amore, E. S., J. C. Manivel, K. J. Gajl-Peczalska, C. E. Litz, C. M. Copenhaver, R. S. Shapiro, and J. G. Strickler. 1991. B-cell lymphoproliferative disorders after bone marrow transplant: an analysis of ten cases with emphasis on Epstein-Barr virus detection by in situ hybridization. Cancer 68:1285-1295. [DOI] [PubMed] [Google Scholar]
- 14.de Campos-Lima, P. O., V. Levitsky, J. Brooks, S. P. Lee, L. F. Hu, A. B. Rickinson, and M. G. Masucci. 1994. T-cell responses and virus evolution: loss of HLA A11-restricted CTL epitopes in Epstein-Barr virus isolates from highly A11-positive populations by selective mutation of anchor residues. J. Exp. Med. 179:1297-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devergne, O., E. Hatzivassiliou, K. M. Izumi, K. M. Kaye, M. F. Kleijnen, E. Kieff, and G. Mosialos. 1996. Association of TRAF1, TRAF2, and TRAF3 with an Epstein-Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-κB activation. Mol. Cell. Biol. 16:7098-7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dukers, D. F., P. Meij, M. B. Vervoort, W. Vos, R. J. Scheper, C. J. Meijer, E. Bloemena, and J. M. Middeldorp. 2000. Direct immunosuppressive effects of EBV-encoded latent membrane protein 1. J. Immunol. 165:663-670. [DOI] [PubMed] [Google Scholar]
- 17.Duraiswamy, J., J. M. Burrows, M. Bharadwaj, S. R. Burrows, L. Cooper, N. Pimtanothai, and R. Khanna. 2003. Ex vivo analysis of T-cell responses to Epstein-Barr virus-encoded oncogene latent membrane protein 1 reveals highly conserved epitope sequences in virus isolates from diverse geographic regions. J. Virol. 77:7401-7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards, R. H., F. Seillier-Moiseiwitsch, and N. Raab-Traub. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79-95. [DOI] [PubMed] [Google Scholar]
- 19.Eliopoulos, A. G., E. R. Waites, S. M. Blake, C. Davies, P. Murray, and L. S. Young. 2003. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 77:1316-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliopoulos, A. G., and L. S. Young. 2001. LMP1 structure and signal transduction. Semin. Cancer Biol. 11:435-444. [DOI] [PubMed] [Google Scholar]
- 21.Erickson, K. D., and J. M. Martin. 2000. The late lytic LMP-1 protein of Epstein-Barr virus can negatively regulate LMP-1 signaling. J. Virol. 74:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fielding, C. A., K. Sandvej, A. Mehl, P. Brennan, M. Jones, and M. Rowe. 2001. Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 75:9129-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flanagan, J., J. Middeldorp, and T. Sculley. 2003. Localization of the Epstein-Barr virus protein LMP1 to exosomes. J. Gen. Virol. 84:1871-1879. [DOI] [PubMed] [Google Scholar]
- 24.Floettmann, J. E., and M. Rowe. 1997. Epstein-BArr virus latent membrane protein-1 (LMP1) C terminus activation region 2 (CTAR2) maps to the far C terminus and requires oligomerisation for NF-κB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 25.Gavioli, R., M. G. Kurilla, P. O. de Campos-Lima, L. E. Wallace, R. Dolcetti, R. J. Murray, A. B. Rickinson, and M. G. Masucci. 1993. Multiple HLA A11-restricted cytotoxic T-lymphocyte epitopes of different immunogenicities in the Epstein-Barr virus-encoded nuclear antigen 4. J. Virol. 67:1572-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gires, O., F. Kohlhuber, E. Kilger, M. Baumann, A. Kieser, C. Kaiser, R. Zeidler, B. Scheffer, M. Ueffing, and W. Hammerschmidt. 1999. Latent membrane protein 1 of Epstein-Barr virus interacts with JAK3 and activates STAT proteins. EMBO J. 18:3064-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottschalk, S., C. Y. Ng, M. Perez, C. A. Smith, C. Sample, M. K. Brenner, H. E. Heslop, and C. M. Rooney. 2001. An Epstein-Barr virus deletion mutant associated with fatal lymphoproliferative disease unresponsive to therapy with virus-specific CTLs. Blood 97:835-843. [DOI] [PubMed] [Google Scholar]
- 28.Gratama, J. W., M. M. Zutter, J. Minarovits, M. A. Oosterveer, E. D. Thomas, G. Klein, and I. Ernberg. 1991. Expression of Epstein-Barr virus-encoded growth-transformation-associated proteins in lymphoproliferations of bone-marrow transplant recipients. Int. J. Cancer 47:188-192. [DOI] [PubMed] [Google Scholar]
- 29.Henle, G., and W. Henle. 1976. Epstein-Barr virus-specific IgA serum antibodies as an outstanding feature of nasopharyngeal carcinoma. Int. J. Cancer 17:1-7. [DOI] [PubMed] [Google Scholar]
- 30.Henle, W., G. Henle, H. C. Ho, P. Burtin, Y. Cachin, P. Clifford, A. de Schryver, G. de-The, V. Diehl, and G. Klein. 1970. Antibodies to Epstein-Barr virus in nasopharyngeal carcinoma, other head and neck neoplasms, and control groups. J. Natl. Cancer Inst. 44:225-231. [PubMed] [Google Scholar]
- 31.Henry, S., C. Sacaze, L. Berrajah, H. Karray, M. Drira, A. Hammami, J. Icart, and B. Mariame. 2001. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int. J. Cancer 91:698-704. [DOI] [PubMed] [Google Scholar]
- 32.Herbst, H., F. Dallenbach, M. Hummel, G. Niedobitek, S. Pileri, N. Muller-Lantzsch, and H. Stein. 1991. Epstein-Barr virus latent membrane protein expression in Hodgkin and Reed-Sternberg cells. Proc. Natl. Acad. Sci. USA 88:4766-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbst, H., H. D. Foss, J. Samol, I. Araujo, H. Klotzbach, H. Krause, A. Agathanggelou, G. Niedobitek, and H. Stein. 1996. Frequent expression of interleukin-10 by Epstein-Barr virus-harboring tumor cells of Hodgkin's disease. Blood 87:2918-2929. [PubMed] [Google Scholar]
- 34.Heslop, H. E., and C. M. Rooney. 1997. Adoptive cellular immunotherapy for EBV lymphoproliferative disease. Immunol. Rev. 157:217-222. [DOI] [PubMed] [Google Scholar]
- 35.Hildesheim, A., R. J. Apple, C. J. Chen, S. S. Wang, Y. J. Cheng, W. Klitz, S. J. Mack, I. H. Chen, M. M. Hsu, C. S. Yang, L. A. Brinton, P. H. Levine, and H. A. Erlich. 2002. Association of HLA class I and II alleles and extended haplotypes with nasopharyngeal carcinoma in Taiwan. J. Natl. Cancer Inst. 94:1780-1789. [DOI] [PubMed] [Google Scholar]
- 36.Ho, J. H. 1972. Nasopharyngeal carcinoma (NPC). Adv. Cancer Res. 15:57-92. [DOI] [PubMed] [Google Scholar]
- 37.Izumi, K. M., E. D. Cahir McFarland, A. T. Ting, E. A. Riley, B. Seed, and E. D. Kieff. 1999. The Epstein-Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-κB activation. Mol. Cell. Biol. 19:5759-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson, R. J., M. Stack, S. A. Hazlewood, M. Jones, C. G. Blackmore, L. F. Hu, and M. Rowe. 1998. The 30-base-pair deletion in Chinese variants of the Epstein-Barr virus LMP1 gene is not the major effector of functional differences between variant LMP1 genes in human lymphocytes. J. Virol. 72:4038-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khanna, R., and S. R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19-48. [DOI] [PubMed] [Google Scholar]
- 41.Khanna, R., S. R. Burrows, D. J. Moss, and S. L. Silins. 1996. Peptide transporter (TAP-1 and TAP-2)-independent endogenous processing of Epstein-Barr virus (EBV) latent membrane protein 2A: implications for cytotoxic T-lymphocyte control of EBV-associated malignancies. J. Virol. 70:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khanna, R., S. R. Burrows, A. Neisig, J. Neefjes, D. J. Moss, and S. L. Silins. 1997. Hierarchy of Epstein-Barr virus-specific cytotoxic T-cell responses in individuals carrying different subtypes of an HLA allele: implications for epitope-based antiviral vaccines. J. Virol. 71:7429-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khanna, R., S. R. Burrows, J. Nicholls, and L. M. Poulsen. 1998. Identification of cytotoxic T-cell epitopes within Epstein-Barr virus (EBV) oncogene latent membrane protein 1 (LMP1): evidence for HLA A2 supertype-restricted immune recognition of EBV-infected cells by LMP1-specific cytotoxic T lymphocytes. Eur. J. Immunol. 28:451-458. [DOI] [PubMed] [Google Scholar]
- 44.Khanna, R., P. Busson, S. R. Burrows, C. Raffoux, D. J. Moss, J. M. Nicholls, and L. Cooper. 1998. Molecular characterization of antigen-processing function in nasopharyngeal carcinoma (NPC): evidence for efficient presentation of Epstein-Barr virus cytotoxic T-cell epitopes by NPC cells. Cancer Res. 58:310-314. [PubMed] [Google Scholar]
- 45.Kimura, M. 1983. The neutral theory of molecular evolution. Cambridge University Press, New York, N.Y.
- 46.Klein, E. 1998. The complexity of the Epstein-Barr virus infection in humans. Pathol. Oncol. Res. 4:3-7. [DOI] [PubMed] [Google Scholar]
- 47.Kuzushima, K., N. Hayashi, A. Kudoh, Y. Akatsuka, K. Tsujimura, Y. Morishima, and T. Tsurumi. 2003. Tetramer-assisted identification and characterization of epitopes recognized by HLA A*2402-restricted Epstein-Barr virus-specific CD8+ T cells. Blood 101:1460-1468. [DOI] [PubMed] [Google Scholar]
- 48.Lautscham, G., T. Haigh, S. Mayrhofer, G. Taylor, D. Croom-Carter, A. Leese, S. Gadola, V. Cerundolo, A. Rickinson, and N. Blake. 2003. Identification of a TAP-independent, immunoproteasome-dependent CD8+ T-cell epitope in Epstein-Barr virus latent membrane protein 2. J. Virol. 77:2757-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lautscham, G., S. Mayrhofer, G. Taylor, T. Haigh, A. Leese, A. Rickinson, and N. Blake. 2001. Processing of a multiple membrane spanning Epstein-Barr virus protein for CD8+ T-cell recognition reveals a proteasome-dependent, transporter associated with antigen processing-independent pathway. J. Exp. Med. 194:1053-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee, S. P., A. T. Chan, S. T. Cheung, W. A. Thomas, D. CroomCarter, C. W. Dawson, C. H. Tsai, S. F. Leung, P. J. Johnson, and D. P. Huang. 2000. CTL control of EBV in nasopharyngeal carcinoma (NPC): EBV-specific CTL responses in the blood and tumors of NPC patients and the antigen-processing function of the tumor cells. J. Immunol. 165:573-582. [DOI] [PubMed] [Google Scholar]
- 51.Lee, S. P., C. M. Constandinou, W. A. Thomas, D. Croom-Carter, N. W. Blake, P. G. Murray, J. Crocker, and A. B. Rickinson. 1998. Antigen-presenting phenotype of Hodgkin Reed-Sternberg cells: analysis of the HLA class I processing pathway and the effects of interleukin-10 on Epstein-Barr virus-specific cytotoxic T-cell recognition. Blood 92:1020-1030. [PubMed] [Google Scholar]
- 52.Lee, S. P., W. A. Thomas, R. J. Murray, F. Khanim, S. Kaur, L. S. Young, M. Rowe, M. Kurilla, and A. B. Rickinson. 1993. HLA A2.1-restricted cytotoxic T cells recognizing a range of Epstein-Barr virus isolates through a defined epitope in latent membrane protein LMP2. J. Virol. 67:7428-7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee, S. P., R. J. Tierney, W. A. Thomas, J. M. Brooks, and A. B. Rickinson. 1997. Conserved CTL epitopes within EBV latent membrane protein 2: a potential target for CTL-based tumor therapy. J. Immunol. 158:3325-3334. [PubMed] [Google Scholar]
- 54.Leen, A., P. Meij, I. Redchenko, J. Middeldorp, E. Bloemena, A. Rickinson, and N. Blake. 2001. Differential immunogenicity of Epstein-Barr virus latent-cycle proteins for human CD4+ T-helper 1 responses. J. Virol. 75:8649-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald-Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375:685-688. [DOI] [PubMed] [Google Scholar]
- 56.Marshall, N. A., M. A. Vickers, and R. N. Barker. 2003. Regulatory T cells secreting IL-10 dominate the immune response to EBV latent membrane protein 1. J. Immunol. 170:6183-6189. [DOI] [PubMed] [Google Scholar]
- 57.Meggetto, F., P. Brousset, J. Selves, G. Delsol, and B. Mariame. 1997. Reed-Sternberg cells and “bystander” lymphocytes in lymph nodes affected by Hodgkin's disease are infected with different strains of Epstein-Barr virus. J. Virol. 71:2547-2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meij, P., A. Leen, A. B. Rickinson, S. Verkoeijen, M. B. Vervoort, E. Bloemena, and J. M. Middeldorp. 2002. Identification and prevalence of CD8+ T-cell responses directed against Epstein-Barr virus-encoded latent membrane protein 1 and latent membrane protein 2. Int. J. Cancer 99:93-99. [DOI] [PubMed] [Google Scholar]
- 59.Meij, P., M. B. Vervoort, J. Aarbiou, P. van Dissel, A. Brink, E. Bloemena, C. J. Meijer, and J. M. Middeldorp. 1999. Restricted low-level human antibody responses against Epstein-Barr virus (EBV)-encoded latent membrane protein 1 in a subgroup of patients with EBV-associated diseases. J. Infect. Dis. 179:1108-1115. [DOI] [PubMed] [Google Scholar]
- 60.Miller, W. E., J. L. Cheshire, A. S. Baldwin, Jr., and N. Raab-Traub. 1998. The NPC derived C15 LMP1 protein confers enhanced activation of NF-κB and induction of the EGFR in epithelial cells. Oncogene 16:1869-1877. [DOI] [PubMed] [Google Scholar]
- 61.Miller, W. E., R. H. Edwards, D. M. Walling, and N. Raab-Traub. 1994. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J. Gen. Virol. 75:2729-2740. [DOI] [PubMed] [Google Scholar]
- 62.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signalling proteins for the tumor necrosis factor receptor family. Cell 80:389-399. [DOI] [PubMed] [Google Scholar]
- 63.Moss, D. J., A. B. Rickinson, and J. H. Pope. 1978. Long-term T-cell-mediated immunity to Epstein-Barr virus in man. I. Complete regression of virus-induced transformation in cultures of seropositive donor leukocytes. Int. J. Cancer 22:662-668. [DOI] [PubMed] [Google Scholar]
- 64.Moss, D. J., A. B. Rickinson, and J. H. Pope. 1979. Long-term T-cell-mediated immunity to Epstein-Barr virus in man. III. Activation of cytotoxic T cells in virus-infected leukocyte cultures. Int. J. Cancer 23:618-625. [DOI] [PubMed] [Google Scholar]
- 65.Murray, P. G., C. M. Constandinou, J. Crocker, L. S. Young, and R. F. Ambinder. 1998. Analysis of major histocompatibility complex class I, TAP expression, and LMP2 epitope sequence in Epstein-Barr virus-positive Hodgkin's disease. Blood 92:2477-2483. [PubMed] [Google Scholar]
- 66.Murray, R. J., M. G. Kurilla, J. M. Brooks, W. A. Thomas, M. Rowe, E. Kieff, and A. B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T-cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niederman, J. C., A. S. Evans, L. Subrahmanyan, and R. W. McCollum. 1970. Prevalence, incidence and persistence of EB virus antibody in young adults. N. Engl. J. Med. 282:361-365. [DOI] [PubMed] [Google Scholar]
- 68.Oudejans, J. J., N. M. Jiwa, J. A. Kummer, A. Horstman, W. Vos, J. P. Baak, P. M. Kluin, P. van der Valk, J. M. Walboomers, and C. J. Meijer. 1996. Analysis of major histocompatibility complex class I expression on Reed-Sternberg cells in relation to the cytotoxic T-cell response in Epstein-Barr virus-positive and -negative Hodgkin's disease. Blood 87:3844-3851. [PubMed] [Google Scholar]
- 69.Pai, S., and R. Khanna. 2001. Role of LMP1 in immune control of EBV infection. Semin. Cancer Biol. 11:455-460. [DOI] [PubMed] [Google Scholar]
- 70.Parker, K. C., M. A. Bednarek, and J. E. Coligan. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152:163-175. [PubMed] [Google Scholar]
- 71.Parker, K. C., M. A. Bednarek, L. K. Hull, U. Utz, B. Cunningham, H. J. Zweerink, W. E. Biddison, and J. E. Coligan. 1992. Sequence motifs important for peptide binding to the human MHC class 1 molecule, HLA-A2. J. Immunol. 149:3580-3587. [PubMed] [Google Scholar]
- 72.Pathmanathan, R., U. Prasad, G. Chandrika, R. Sadler, K. Flynn, and N. Raab-Traub. 1995. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx: variants of Epstein-Barr virus-infected neoplasia. Am. J. Pathol. 146:1355-1367. [PMC free article] [PubMed] [Google Scholar]
- 73.Pathmanathan, R., U. Prasad, R. Sadler, K. Flynn, and N. Raab-Traub. 1995. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N. Engl. J. Med. 333:693-698. [DOI] [PubMed] [Google Scholar]
- 74.Raab-Traub, N. 1996. Pathogenesis of Epstein-Barr virus and its associated malignancies. Semin. Virol. 7:315-323. [Google Scholar]
- 75.Raab-Traub, N., R. Hood, C. S. Yang, B. Henry II, and J. S. Pagano. 1983. Epstein-Barr virus transcription in nasopharyngeal carcinoma. J. Virol. 48:580-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rickinson, A., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 77.Rickinson, A. B., and D. J. Moss. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405-431. [DOI] [PubMed] [Google Scholar]
- 78.Rooney, C. M., C. A. Smith, C. Ng, S. K. Loftin, C. Li, R. A. Krance, et al. 1995. Use of gene-modified virus-specific T lymphocytes to control Epstein-Barr virus-related lymphoproliferation. Lancet 345:9-13. [DOI] [PubMed] [Google Scholar]
- 79.Rowe, M., R. Khanna, C. A. Jacob, V. Argaet, A. Kelly, S. Powis, M. Belich, D. Croom-Carter, S. Lee, S. R. Burrows, et al. 1995. Restoration of endogenous antigen processing in Burkitt's lymphoma cells by Epstein-Barr virus latent membrane protein-1: coordinate upregulation of peptide transporters and HLA-class I antigen expression. Eur. J. Immunol. 25:1374-1384. [DOI] [PubMed] [Google Scholar]
- 80.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 81.Sandberg, M. L., A. Kaykas, and B. Sugden. 2000. Latent membrane protein 1 of Epstein-Barr virus inhibits as well as stimulates gene expression. J. Virol. 74:9755-9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shapiro, R. S., K. McClain, G. Frizzera, K. J. Gajl-Peczalska, J. H. Kersey, B. R. Blazar, D. C. Arthur, D. F. Patton, J. S. Greenberg, B. Burke, et al. 1988. Epstein-Barr virus associated B-cell lymphoproliferative disorders following bone marrow transplantation. Blood 71:1234-1243. [PubMed] [Google Scholar]
- 83.Sitki-Green, D., M. Covington, and N. Raab-Traub. The biology of the Epstein-Barr virus during infectious mononucleosis. J. Infect. Dis., in press. [DOI] [PubMed]
- 84.Sitki-Green, D., M. Covington, and N. Raab-Traub. 2003. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J. Virol. 77:1840-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sitki-Green, D., R. H. Edwards, J. Webster-Cyriaque, and N. Raab-Traub. 2002. Identification of Epstein-Barr virus strain variants in hairy leukoplakia and peripheral blood by use of a heteroduplex tracking assay. J. Virol. 76:9645-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Starzl, T. E., M. A. Nalesnik, K. A. Porter, M. Ho, S. Iwatsuki, B. P. Griffith, J. T. Rosenthal, T. R. Hakala, B. W. Shaw, Jr., R. L. Hardesty, et al. 1984. Reversibility of lymphomas and lymphoproliferative lesions developing under cyclosporin-steroid therapy. Lancet i:583-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sung, N. S., R. H. Edwards, F. Seillier-Moiseiwitsch, A. G. Perkins, Y. Zeng, and N. Raab-Traub. 1998. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int. J. Cancer 76:207-215. [DOI] [PubMed] [Google Scholar]
- 88.Thomas, J. A., V. Iliescu, D. H. Crawford, R. Ellouz, M. Cammoun, and G. de-The. 1984. Expression of HLA-DR antigens in nasopharyngeal carcinoma: an immunohistological analysis of the tumour cells and infiltrating lymphocytes. Int. J. Cancer 33:813-819. [DOI] [PubMed] [Google Scholar]
- 89.Tierney, R. J., N. Steven, L. S. Young, and A. B. Rickinson. 1994. Epstein-Barr virus latency in blood mononuclear cells: analysis of viral gene transcription during primary infection and in the carrier state. J. Virol. 68:7374-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsao, S. W., G. Tramoutanis, C. W. Dawson, A. K. F. Lo, and D. P. Huang. 2002. The significance of LMP1 expression in nasopharyngeal carcinoma. Semin. Cancer Biol. 12:473-487. [DOI] [PubMed] [Google Scholar]
- 91.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, W. Y., Y. C. Chien, J. S. Jan, C. M. Chueh, and J. C. Lin. 2002. Consistent sequence variation of Epstein-Barr virus nuclear antigen 1 in primary tumor and peripheral blood cells of patients with nasopharyngeal carcinoma. Clin. Cancer Res. 8:2586-2590. [PubMed] [Google Scholar]
- 93.Young, L., C. Alfieri, K. Hennessy, H. Evans, C. O'Hara, K. C. Anderson, J. Ritz, R. S. Shapiro, A. Rickinson, E. Kieff, et al. 1989. Expression of Epstein-Barr virus transformation-associated genes in tissues of patients with EBV lymphoproliferative disease. N. Engl. J. Med. 321:1080-1085. [DOI] [PubMed] [Google Scholar]