Abstract
Repressor activator protein 1 (RAP1) is the most highly conserved telomere protein. It is involved in protecting chromosome ends in fission yeast, promoting gene silencing in Saccharomyces cerevisiae while in Kluyveromyces lactis it is required to repress homology directed recombination (HDR) at telomeres. Since mammalian RAP1 requires TRF2 for stable expression, its role in telomere function has remained obscure. To understand how RAP1 plays such diverse functions at telomeres, we solved the crystal or solution structures of the C-terminal RCT domains of RAP1 from multiple organisms in complex with their respective protein-binding partners. Our comparative structural analysis establishes the RCT domain of RAP1 as an evolutionarily conserved protein-protein interaction module. In mammalian and fission yeast cells, this module interacts with TRF2 and Taz1, respectively, targeting RAP1 to chromosome ends for telomere end protection. While RAP1 repress NHEJ at fission yeast telomeres, at mammalian telomeres it is required to repress HDR. In contrast, S. cerevisiae RAP1 utilizes the RCT domain to recruit Sir3 to telomeres to mediate gene silencing. Together, our results reveal that depending on the organism, the evolutionarily conserved RAP1 RCT motif plays diverse functional roles at telomeres.
Telomeres, the natural ends of linear eukaryotic chromosomes, are essential for cell viability and genome integrity1. In most organisms, telomeric DNA consists of short repetitive sequences that terminates in 3’ single-stranded overhangs. Both the double stranded repeats and the 3’ overhangs of mammalian telomeres are bound by shelterin, a six-protein complex that exclusively associates with telomeres and protects chromosome ends from aberrant DNA repair activities2,3. Telomeric proteins have undergone a rapid rate of change during evolution4. Notably, repressor activator protein 1 (RAP1) is the only telomere protein that is conserved from budding and fission yeast to mammals. RAP1 contains a BRCT domain, one or two Myb domains, and an RCT (RAP1 C-terminus) domain (Fig. 1a). Despite this relatively conserved multi-domain architecture, RAP1 proteins in different organisms appear to have acquired diversified functions during evolution.
Figure 1.
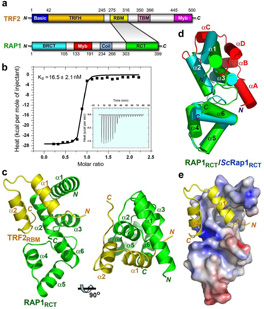
Structure of the human TRF2RBM-RAP1RCT complex. (a) Domain organization of the TRF2 and RAP1 polypeptide chains. In TRF2, the N-terminal basic region is colored in blue, the C-terminal Myb domain in magenta, the TRFH domain in orange, the RAP1-binding motif (RBM) in yellow and the TIN2-binding domain (TBM) in purple. For RAP1, the N-terminal BRCT domain is in cyan, the Myb domain in red, the coiled-coil region in slate, and the C-terminal RCT domain in green. The shaded area between TRF2 and RAP1 indicates that the TRF2-RAP1 interaction is mediated by TRF2RBM and RAP1RCT. (b) In vitro Isothermal Titration Calorimitry (ITC) measurement of the interaction between TRF2RBM with RAP1RCT. Insert is the ITC titration data. (c) Two orthogonal views of the overall structure of the TRF2RBM-RAP1RCT complex. TRF2RBM and RAP1RCT are colored in yellow and green, respectively. (d) Superposition of the crystal structure of human RAP1RCT on that of budding yeast ScRap1 RCT domain 21. Helices are shown as colored cylinders; human RAP1RCT is in green, whereas the helix-bundle core and the N-terminal extension of ScRap1RCT are in cyan and red, respectively. (e) Electrostatic surface potential of the TRF2RBM binding site of RAP1RCT. Positive potential: blue, negative potential: red.
In mammalian cells, RAP1 is the least well-understood component of shelterin. RAP1 does not directly bind to telomeric DNA. Instead, it is recruited to telomeres through interaction between its C-terminal RCT domain and TRF2, another shelterin protein that binds to the duplex region of telomeres5. TRF2 is essential in telomere end protection, since removal of TRF2 from telomeres initiates a potent DNA damage response (DDR) that activates ATM and the non-homologous end joining (NHEJ) pathway, resulting in massive end-to-end chromosome fusions6–10. Recent studies suggested that the TRF2-RAP1 subcomplex is sufficient to suppress NHEJ both in vitro and in vivo11,12. Since the stability of mammalian RAP1 is dependent upon its interaction with TRF26, it remains unclear whether TRF2, RAP1 or both proteins are required to protect telomeres.
Budding yeast Saccharomyces cerevisiae Rap1 (ScRap1) was discovered as a positive transcriptional regulator of genes for multiple growth-related genes such as the ribosomal protein genes13. Later studies revealed that ScRap1 is the major double-stranded telomeric repeat-binding protein in S. cerevisiae and plays essential roles in telomere length regulation, subtelomeric gene silencing and chromosome end protection14. While the central two Myb domains are responsible for the DNA binding activity of ScRap115, the C-terminal RCT domain mediates chromatin recruitment of two sets of proteins, the Sir proteins (Sir3 and Sir4) for transcriptional silencing16 and the Rif protein (Rif1 and Rif2) for telomere length regulation17,18. Although the crystal structure of ScRap1RCT is available (Feeser and Wolberger, 2008), how this domain recruits the Sir and the Rif proteins to telomeres still remains unknown.
Fission yeast Schizosaccharomyces pombe Rap1 (SpRap1) was identified based on its limited sequence similarity to ScRap119,20. Like mammalian RAP1 but unlike budding yeast ScRap1, fission yeast SpRap1 lacks DNA binding activity and was believed to localizes to telomeres via interactions with Taz1, an ortholog of mammalian TRF1 and TRF219,20. Deletion of fission yeast rap1 results in chromosome end-to-end fusions, telomere elongation, and derepression of telomere silencing, phenotypes reminiscent of those observed in taz1Δ cells, suggesting a close relationship between SpRap1 and Taz119,20. However, SpRap1 lacks an obvious RCT domain. Therefore, how SpRap1 interacts Taz1 remains unclear.
To address these structural and functional questions of RAP1, we solved the three-dimensional crystal or solution structures of the RCT domains of human, fission yeast, and budding yeast RAP1 in complex with their respective binding partners, TRF2, Taz1 and Sir3. Our structurally focused biochemical, cellular, and genetic analyses revealed that RAP1 contains a remarkably conserved protein-protein interaction module that is utilized by both mammalian and fission yeast RAP1 proteins to interact with a telomeric double-stranded DNA binding protein for telomere regulation and protection. In contrast, budding yeast ScRap1 uses this module to recruit Sir3 to telomeres to mediate transcriptional silencing. Together, our results reveal that an evolutionarily conserved protein interaction module in RAP1 plays diverse roles at telomeres in different organisms.
RESULTS AND DISCUSSION
Structure of the human TRF2RBM-RAP1RCT complex
TRF2 is required for the recruitment of RAP1 to telomeres. A central fragment of TRF2 (residues 123 – 366) was reported to directly bind to the RCT domain of RAP1 (residues 303 – 399)5. To further map the RAP1-binding region of TRF2, various fragments of TRF2 were evaluated for their ability to interact with RAP1. A TRF2 fragment consisting of residues 275 – 316 was necessary and sufficient for binding with RAP1RCT (Fig. 1a and Supplementary Fig. 1a). TRF2275–316 binds to RAP1RCT with an equilibrium dissociation constant (Kd) of 16.5 nM, similar to that of the full-length TRF2 protein to RAP1RCT (23.9 nM) as measured by isothermal titration calorimetry (ITC) (Fig. 1b). Hereafter, we will refer to TRF2275–316 as TRF2RBM (RAP1-binding motif) (Fig. 1a).
To reveal the structural basis of RAP1 recognition by TRF2, we crystallized the TRF2RBM-RAP1RCT complex and solved its structure by multiple-wavelength anomalous dispersion (MAD) with selenomethionine-substituted crystals at a resolution of 1.95 Å (Supplementary Table 1). The TRF2RBM-RAP1RCT complex adopts a compact globular fold, resembling a single folding unit (Fig. 1c). RAP1RCT consists of six α helices arranged into two three-helix bundles. Helices α1, α2, and α3 form the first bundle and helices α4, α5, and α6 form the second. The structure of RAP1RCT closely resembles that of the RCT domain of budding yeast S. cerevisiae Rap1 (ScRap1RCT), consistent with previous sequence alignment predictions (Fig. 1d)5,21. Indeed, an unbiased search for structurally homologous proteins using the Dali server revealed that the structure of RAP1RCT is most similar to that of ScRap1RCT22. The two RCT domains can be superimposed with a root-mean-square deviation (rmsd) of 2.3 Å for 85 equivalent Cα pairs (Fig. 1d). In addition to the structurally conserved three-helix bundles, ScRap1RCT contains an N-terminal extension covering one side of the RCT domain, which is not present in RAP1RCT (Fig. 1d).
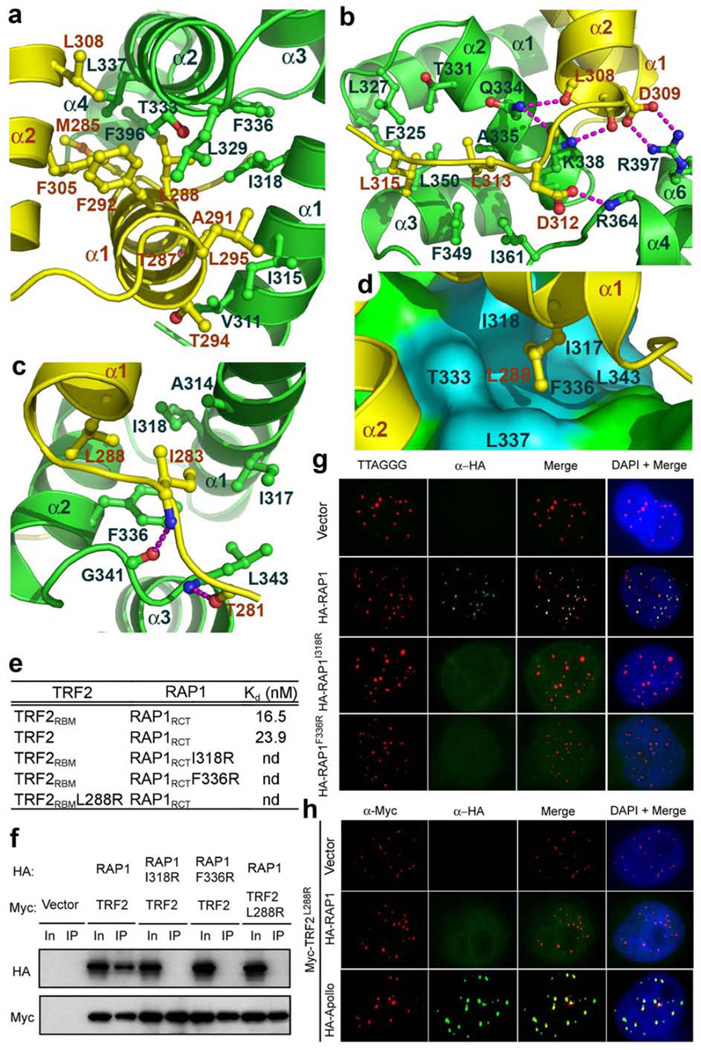
TRF2RBM is a helix-turn-helix motif that packs against helices α1 and α2 of RAP1RCT to form an intermolecular four-helix bundle (Fig. 1c). The formation of the binary complex involves an extensive set of interactions and causes the burial of 2,400 Å2 of surface area at the interface. The driving force for the binding of TRF2 to RAP1 is van der Waals interactions (Fig. 1e). Helix α1 of TRF2RBM contributes most of the hydrophobic contacts. Five hydrophobic residues of TRF2RBM (Met285, Leu288, Ala291, Phe292, and Leu295) from helix α1 make extensive contacts with the hydrophobic wedge between helices α1 and α2 of RAP1RCT (Fig. 2a). Helix α2 of TRF2RBM makes less direct hydrophobic contact with RAP1. Instead, its C-terminus mediates four intermolecular electrostatic interactions with TRF2 (Fig. 2b). In addition to helices α1 and α2, the terminal regions of TRF2RBM also contribute to the binding to RAP1. They function as the two arms of a clamp to hold helix α2 of RAP1RCT (Fig. 1c and Supplementary Fig. 1b). The N-terminal tail of TRF2RBM (residues 282–284) extends into a deep groove of RAP1 and runs antiparallel to loop L23 (residues 340–342 between helices α2 and α3) of RAP1RCT (Fig. 2c). The C-terminal tail of TRF2RBM contacts the other side of RAP1RCT (Fig. 1c and Supplementary Fig. 1b). The side chains of two leucine residues (Leu313 and Leu315) pack against a hydrophobic patch of RAP1RCT formed by residues from loop L34 and helices α2 and α3 (Fig. 2b).
Figure 2.
Crystallographic and mutational analyses of the TRF2RBM-RAP1RCT interaction. (a) Hydrophobic interactions between helix α1 of TRF2RBM and helices α1 and α2 of RAP1RCT. TRF2RBM and RAP1RCT are shown in ribbon model and colored as in Figure 1C. The interacting residues of TRF2RBM and RAP1RCT are presented as ball-and-stick models. (b) Details of the interactions around the C-terminal tail and helix α2 of TRF2RBM. Intermolecular hydrogen bonding interactions are shown as dashed magenta lines. (c) The interaction between the TRF2RBM N-terminal tail and loop L23 of RAP1RCT(d) The TRF2 Leu288 residue in the ball-and-stick model (yellow) is nested in a hydrophobic pocket of RAP1 (cyan surface). The rest of RAP1 is colored in green. (e) In vitro ITC binding data of wild-type and mutant TRF2rbm-RAP1RCT interactions (nd: not detectable by ITC). (f) Co-IP of the same sets of mutant TRF2-RAP1 interactions as in e. Lanes marked “In” represent 5% of input cell lysate used for the IPs. Co-IP data show that TRF2 mutant L288R and two RAP1 mutants I318R and F336R disrupt the TRF2-RAP1 interaction in cells. (g) Localization of retrovirally expressed HA-tagged wild type and the I318R, F336R mutants of RAP1 in HeLa cells. Telomeres were visualized by telomere peptide nucleic acid (PNA)-FISH (red). (h) Localization of transiently expressed HA-tagged RAP1 and Apollo when co-tranfected with the Myc-TRF2 L288R mutant in HeLa cells.
Mutational analyses of the TRF2RBM-RAP1RCT interaction
To corroborate our structural analysis, we examined whether missense mutations of the interface residues of TRF2RBM or RAP1RCT could weaken or disrupt the TRF2-RAP1 interaction. We focused on the hydrophobic interface between helices α1 of TRF2RBM and α1 and α2 of RAP1RCT, which are critical for stabilization of the interaction. In particular, located at the center of this interface, the side chain of Leu288 of TRF2 is nested in a pocket formed by a group of hydrophobic residues of RAP1 (Fig. 2d). Consistent with the crystal structure, substitution of TRF2 Leu288 with a positively charged and bulkier arginine residue completely abolished the interaction with RAP1 in both ITC and yeast two-hybrid assays (Fig. 2e and Supplementary Figs. 2a and 2b). Similarly, RAP1 mutations I318R and F336R on the other side of the interface also impaired the interaction (Fig. 2e and Supplementary Figs. 2a and 2b). These results indicated that a single point mutation at the hydrophobic interface is sufficient to disrupt the ability of TRF2 to bind RAP1.
To further examine the TRF2-RAP1 interaction in vivo, we next examined the interactions of mutant proteins transiently expressed in human embryonic kidney 293T cells. Consistent with the ITC and yeast two-hybrid analyses, co-immunoprecipitation (Co-IP) experiments revealed that while wild-type TRF2 and RAP1 showed the expected interaction, mutations of the conserved hydrophobic residues (Leu288 of TRF2, or Ile318 and Phe336 of RAP1) at the interface completely abolished the TRF2-RAP1 interaction in cells (Fig. 2f). To confirm that these point mutations affected only TRF2’s interaction with RAP1, we co-transfected wild-type and the L288R mutant of TRF2 with the TRF2 interacting protein Apollo in 293T cells23,24. Consistent with the previous finding that Apollo is recruited to telomeres by its interaction with the TRF homology (TRFH) domain of TRF223–25, Apollo was efficiently co-immunoprecipitated by both wild-type and the L288R mutant TRF2 (Supplementary Fig. 2c). Taken together, our mutagenesis analyses suggest that the hydrophobic interface is necessary for both in vitro and in vivo binding of RAP1 to TRF2.
To examine the role of TRF2 in targeting RAP1 to telomeres in cells, we asked whether telomeric accumulation of RAP1 dependents upon its interaction with TRF2. Indirect immunofluorescence (IF) of HeLa cells transiently transfected with HA-tagged RAP1 protein revealed that wild-type RAP1 showed a nuclear punctate staining pattern that completely co-localized with telomeric DNA (Fig. 2g). In contrast, both the RAP1 I318R and RAP1 F336R mutants distributed diffusely throughout the nucleoplasm with no obvious accumulation at telomeres (Fig. 2g and Supplementary Fig. 2d), suggesting that these residues are critical for the TRF2-RAP1 interaction. Next, we co-transfected wild-type RAP1 together with the TRF2 L288R mutant and assayed for subcellular localization of both proteins. While TRF2 L288R efficiently localizes to telomeres (Supplementary Fig. 2e), it was unable to recruit exogenous RAP1 to telomeres (Fig. 2h). This result indicates that overexpression of TRF2 L288R has a dominant negative effect on RAP1’s ability to localize to telomeres. In contrast, telomeric localization of Apollo is still retained in the presence of TRF2 L288R, consistent with the observation that Apollo is recruited to telomeres through its interaction with the TRFH domain of TRF2 (Fig. 2h). Notably, all the RAP1 proteins were overexpressed at comparable levels in cells with different combinations of TRF2 and RAP1 mutations (Supplementary Fig. 2f). Thus, these results demonstrated that telomeric localization of RAP1 depends solely on its direct interaction with TRF2.
Mammalian RAP1 is not required to repress DDR at telomeres
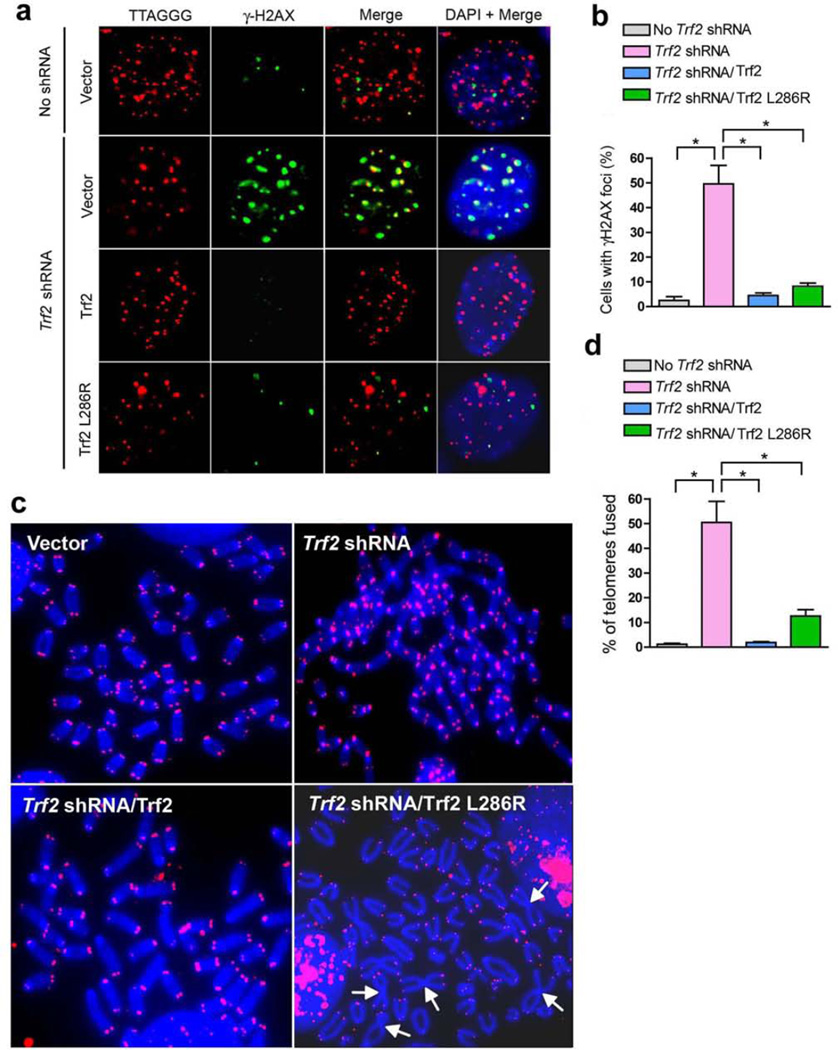
The structural information of the TRF2-RAP1 interaction provided a unique opportunity to study the in vivo function of RAP1. We first depleted endogenous mouse Trf2 using retrovirus-mediated short hairpin RNA (shRNA) to Trf2 in SV40LT immortalized mouse embryonic fibroblasts (MEFs) (Supplementary Fig. 3a)26. When Trf2 is compromised, telomeres initiate a robust ATM dependent DNA damage response, resulting in phosphorylation of Chk2 and the induction of ‘telomere dysfunction-induced foci’ (TIF) as evidenced by the telomeric accumulation of phosphorylated H2AX (γ–H2AX) in ∼50% of cells examined (Figs. 3a and 3b, and Supplementary Fig. 3a)26. Metaphase spreads collected 96 h after Trf2 shRNA treatment showed that nearly all the telomeres are joined together, resulting in long trains of fused chromosomes (Figs. 3c and 3d). This telomere deprotection phenotype was nearly completely rescued by retroviral transduction of an shRNA-resistant Trf2 cDNA in MEFs before Trf2 shRNA treatment, indicating that the observed phenotype in Trf2-shRNA-treated MEFs was caused by Trf2 deficiency and not due to off-target effects (Figs. 3a–3d).
Figure 3.
RAP1 is required for telomere end protection. (a) γ-H2AX-positive foci (green) in SV40LT immortalized MEFs expressing the indicated proteins before treatment with Trf2 shRNA or control vector. Telomeres were visualized with telomere PNA-FISH (red). Representative Trf2 L286R expressing MEFs with > 4 γ-H2AX TIFs is shown in the bottom panel. (b) Quantification of the percent of cells with > 4 γ-H2AX-positive TIFs from representative images shown in a. Error bars, s.d.; n≥ 300 nuclei analyzed per sample. * P < 0.005 calculated using a two-tailed Student’s t-test. (c) MEFs expressing the indicated proteins were treated with control vector or Trf2 shRNA for 96 h, metaphase spreads prepared and telomere fusions were visualized by telomere PNA-FISH (red) and 4,6-diamidino-2-phenylindole (DAPI; blue). Arrows point to fused chromosomes. (d) Quantification of telomere fusions from representative images shown in c. Error bars, s.d.; n >1600 telomeres analyzed per sample. * P < 0.005 calculated using a two-tailed Student’s t-test.
To examine the functional significance of Rap1 in telomere end protection, we asked whether the telomere deprotection phenotype caused by Trf2 depletion could be rescued upon introduction of an shRNA-resistant Trf2 mutant deficient in Rap1 binding (Trf2 L286R). Trf2 L286R is equivalent to human TRF2 L288R that does not bind to RAP1 (Figs. 2e and 2f). Although Trf2 L286R localized efficiently to telomeres (Supplementary Fig. 3b), endogenous Rap1 did not accumulate at telomeres when Trf2 L286R was expressed (Supplementary Fig. 3c). In fact, the protein levels of endogenous Rap1 were greatly reduced in shTrf2 treated MEFs with or without the expression of Trf2 L286R (Supplementary Fig. 3a). In contrast, expression of wild-type Trf2 in shTrf2 treated MEFs restored the protein level of endogenous Rap1 comparable to that in control cells (Supplementary Fig. 3a). Consistent with a previous study, these data indicated that the Trf2-Rap1 interaction not only was required for targeting Rap1 to telomeres, but also was critical for the stability of endogenous Rap1. Expression of Trf2 L286R in shTrf2 treated MEFs largely rescued TIF formation (Figs. 3a and 3b), suggesting that Rap1 is not required to repress the DDR at telomeres. Unlike the massive end-to-end chromosome fusions with robust telomeric signals at the sites of fusion when Trf2 is removed6,26, replacement of endogenous Trf2 with Trf2 L286R resulted in end-to-end chromosome fusions involving only ∼13% of all chromosome ends (Fig. 3c). These data suggest that Rap1 does not participate in inhibition of NHEJ-mediated fusions at telomeres. Interestingly, the chromosome fusion sites in shTrf2-treated-Trf2-L286R-expressing MEFs were largely devoid of telomeric signals (Figs. 3c and 3d). To further examine the relationship between Rap1 and this telomere loss, we fused Rap1 with shRNA-resistant Trf2 L286R and complemented this chimeric DNA into MEFs before Trf2 shRNA treatment (Supplementary Fig. 4a). As shown in Supplementary Fig. 4b, the telomere attrition phenotype was completely rescued in response to Rap1-Trf2 L286R expression, confirming that the observed telomere attrition phenotype in Trf2 L286R expressing cells was caused by the lack of Rap1 at telomeres.
Telomere loss in the absence of RAP1 is mediated by Rad51 and Exo1 dependent homologous recombination
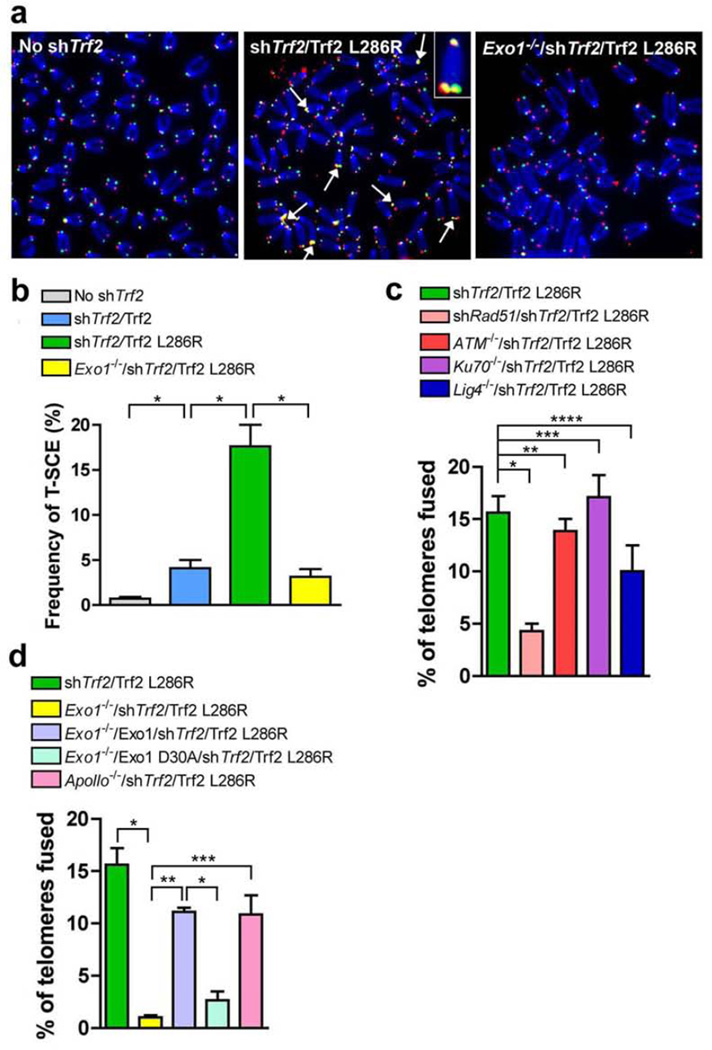
What account for the low level of chromosome fusions observed in Trf2 L286R expressing cells? One possibility is that aberrant homology-directed repair (HDR) at telomeres could result in loss of telomeric DNA, enabling end-to-end chromosome fusions with fusion sites lacking telomeric signals. To test whether Rap1 is required to repress telomere attrition due to inappropriate HDR at telomeres, we visualized both leading and lagging strand telomeres and analyzed telomere sister chromatid exchanges (T-SCEs), a marker for HDR, using chromosome-orientation FISH (CO-FISH)29. Both control metaphases and metaphases from Trf2-expressing MEFs devoid of endogenous Trf2 showed low levels of T-SCEs (Figs. 4a and 4b). In sharp contrast, metaphases from Trf2 L286R expressing cells exhibited a dramatic 4-fold increase in T-SCEs (Figs. 4a and 4b). Consistent with two recent studies30, these results suggested that Rap1 represses aberrant HDR at telomeres.
Figure 4.
Rap1 suppresses aberrant Rad51 and Exo1 dependent HDR at telomeres. (a) SV40LT immortalized MEFs expressing the indicated proteins were treated with control vector or Trf2 shRNA for 96 h, metaphase spreads prepared and telomere fusions were visualized by telomere PNA-FISH (red) and 4,6-diamidino-2-phenylindole (DAPI; blue). Arrows point to T-SCEs. (b) Quantifications of T-SCEs from representative images shown in a. Error bars, s.d.; n > 3,500 telomeres analyzed per sample. * P < 0.05 calculated using a two-tailed Student’s t-test. (c) Quantification of telomere fusions in MEFs expressing the indicated proteins and treated with Trf2 shRNA. Error bars, s.d.; n > 2,000 telomeres telomeres analyzed per sample. * P < 0.05, ** P < 0.47, *** P < 0.63, **** P < 0.20, calculated using a two-tailed Student’s t-test. (d) Quantification of telomere fusions in MEFs expressing the indicated proteins and treated with Trf2 shRNA. Error bars, s.d.; n >2700 telomeres analyzed per sample. * P < 0.01, ** P < 0.005, *** P < 0.05, calculated using a two-tailed Student’s t-test.
Recombinase Rad51 assembles onto ssDNA and mediates the pairing and shuffling of DNA sequences during HDR31. We reasoned that if HDR caused the telomere fusions observed in the absence of Rap1, deletion of Rad51 would rescue this phenotype. We expressed Trf2 L286R in MEFs where both endogenous Trf2 and Rad51 were efficiently depleted by shRNA and subsequently analyzed the metaphase spreads. Strikingly, depletion of Rad51 resulted in almost complete rescue of the telomere fusions due to Rap1 loss (Fig. 4c). Thus, Trf2 L286-induced end-to-end chromosome fusions requires functional Rad51, suggesting that HDR is involved in these events. A series of other genes involved in DNA damage signaling (e.g. ATM) and NHEJ DNA repair pathway (e.g., Ku70 and Lig4) were tested for their contribution to Rap1 loss-induced telomere attritions. Using genetically deficient MEFs, we found that deletion of none of these genes prevented chromosome fusions when Rap1 is removed from telomeres (Fig. 4c). These results further reinforce the notion that end-to-end fusions observed following Rap1 depletion from telomeres were not the result of NHEJ-mediated repair, but rather a product of HDR mediated repair of DNA ends devoid of telomeric DNA. Taken together, these data suggest that rapid telomere loss is a prerequisite for chromosome fusions in the absence of Rap1, and that these telomere-free chromosome end-to-end fusions are mediated by HDR.
A key step in HDR is the generation of ssDNA, the substrate for Rad51 binding to initiate homologous pairing and strand exchange32,33. Recent studies revealed that 5’-3’ exonuclease 1 (Exo1) plays a role in formation of telomere ssDNA overhangs in yeast cells34. Hence, we hypothesized that Exo1 might be required to generate ssDNA for Rad51 binding during HDR. In order to address the contribution of Exo1 to the HDR mediated telomere attrition in the absence of Rap1, we examined the effect of Trf2 L286 expression in Trf2 shRNA treated Exo1−/− MEFs. Surprisingly, deletion of Exo1 dramatically reduced T-SCEs due to Rap1 depletion from telomeres (Figs. 4a and 4b). Expression of Exo1 from an introduced cDNA restored the chromosome fusion phenotype to the same level as the Trf2 L286R expressing control cells (Fig. 4d). In sharp contrast, cells expressing a catalytic dead mutant Exo1 D30A (equivalent to human flap endonuclease-1 (FEN-1) D34A35) showed no sign of end-to-end chromosome fusion (Fig. 4d), suggesting that the exonuclease activity of Exo1 is required for telomere attrition after Rap1 removal from telomeres. Taken together, these data suggest that Rap1 represses aberrant HDR at telomeres mediated by both Rad51 and Exo1.
Mammalian RAP1 and TRF2 play distinct functions in telomere end protection
Our structural, biochemical, and cell biology data supports a model in which mammalian Trf2 and Rap1 play important but distinct roles in telomere end protection. Trf2 inhibits NHEJ-mediated repair of telomeres and is also required to recruit Rap1 and other telomere associated proteins to telomeres3. Rap1 is required to repress HDR-mediated telomere attrition, but is dispensable for preventing NHEJ-mediated repair of uncapped telomeres (Figs. 3c and 3d). Our data also indicate that neither Trf2 nor Rap1 alone is able to fully protect telomeres. Instead, we propose that Trf2 and Rap1 form a stable heterodimer to protect the duplex region of telomeres.
A striking consequence of loss of protective functions at telomeres following Rap1 removal from telomeres is the observation that chromosome end-to-end fusions form without telomeres at fusion sites in metaphase chromosomes when endogenous Trf2 is replaced with Trf2 L286R (Figs. 3c and 3d). This fusion phenotype is distinct from the telomere fusions observed when Trf2 is removed from telomeres, in which robust telomeric signals are abundant at fusion sites26. It is likely that loss of Trf2 from telomeres disrupts the entire protective nucleoprotein structure (for instance the ability to form t-loops) so that telomeres are subjected to NHEJ-mediated fusions immediately after Trf2 loss. In contrast, loss of Rap1 induces rapid telomere attrition through activation of telomere HDR, resulting in the induction of a DDR and subsequent repair of DNA ends devoid of telomeric sequences by HDR. While the mechanism of how Rap1 represses telomere HDR is currently unclear, we postulate that either Rap1 directly inhibits telomere HDR or that Rap1 interacts with factor(s) that are involved in repressing telomere HDR.
Our observation of increased telomere recombination when endogenous Trf2 is replaced with Trf2 L286R was consistent with two recent studies in which endogenous Rap1 was conditional deleted in MEFs (Sfeir et al., 2010, Martines et al., 2010). However, these studies did not report telomere fusion and attrition we observed in shTrf2 treated MEFs expressing Trf2 L286R. We postulate that the reason why chromosome fusions were observed in our experimental setting is due to efficient depletion of endogenous Trf2 using a robust shRNA-based approach (Deng et al., 2009). Chromosome fusions likely arose in cells in which endogenous Trf2 was nearly completely depleted and functionally replaced by Trf2 L286R. It is likely that Cre-mediated hit-and-run deletion of Trf2F/F cannot achieve the same level of efficiency, since even in the best scenario Cre-mediated deletion of targeted alleles in MEFs only approaches 90% (S.C., personal observation; Wu et al., 2006).
Structural basis of the fission yeast S. pombe Taz1-SpRap1 interaction
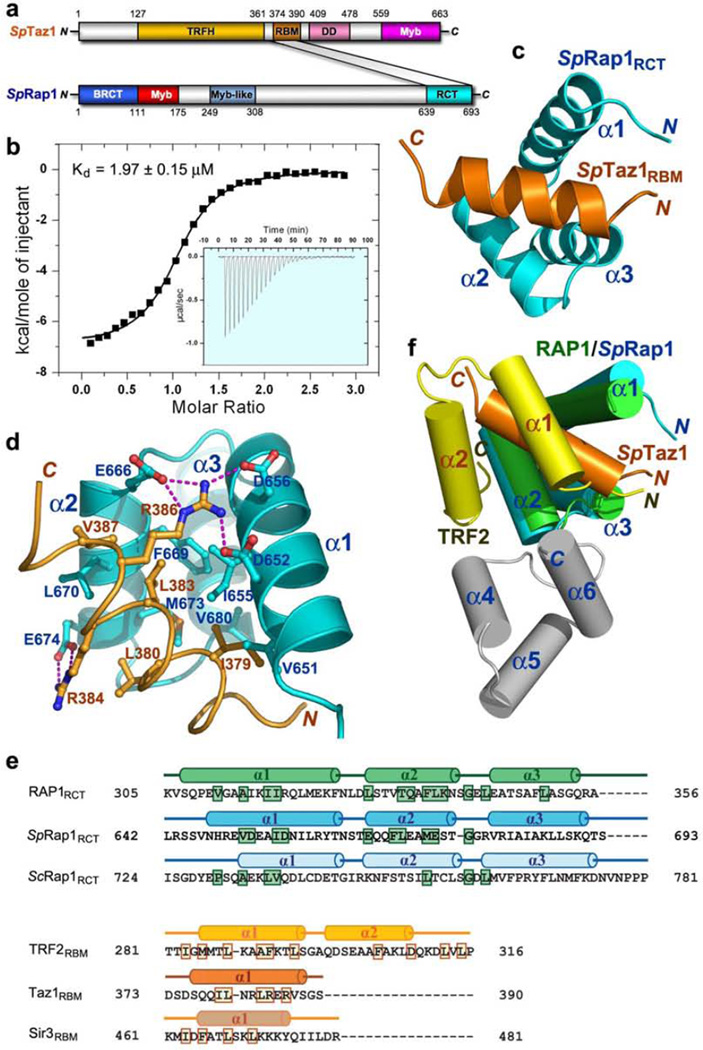
Fission yeast SpRap1 protein plays an important role in telomere length homeostasis and telomere protection19,20,39. Similar to mammalian RAP1, SpRap1 also associates with the double-stranded telomeric DNA-binding protein Taz1, an ortholog of human TRF proteins19,20. However, unlike its budding yeast and mammalian counterparts, SpRap1 lacks a recognizable RCT domain19,20. Furthermore, bioinformatic approaches failed to identify a TRF2RBM-like motif in Taz1 (YC, FW, and ML, unpublished result). To determine the mechanism how SpRap1 interacts with Taz1, we characterized the Taz1-SpRap1 interaction by gel filtration chromatography (data not shown). Various fragments of SpRap1 were evaluated for their ability to interact with Taz1. Our data revealed that, similar to the mammalian TRF2-RAP1 interaction, the C-terminus of SpRap1 (resides 639–693) is sufficient for interaction with Taz1 (Fig. 5a). Using a similar strategy, a short 32-residue fragment of Taz1 (residues 365–396) was found to be the minimal region that is necessary and sufficient for binding to Rap1 (Fig. 5a). Hereafter, Taz1365–396 and SpRap1639–693 will be referred to as Taz1RBM and SpRap1RCT, respectively (Fig. 5a). Taz1RBM binds to SpRap1RCT with an equilibrium dissociation constant (Kd) of 2.0 µM (Fig. 5b), ∼100-fold weaker that the interaction between human TRF2RBM and RAP1RCT (Fig. 1b).
Figure 5.
Structure of the fission yeast Taz1RBMSpRap1RCT complex. (a) Domain organization of the Taz1 and SpRap1 polypeptide chains. In Taz1, the putative TRFH domain is colored in brown, the C-terminal Myb domain in magenta, the SpRap1-binding motif (RBM) in orange, the dimerization domain (DD) in pink, and the C-terminal Myb domain in magenta. This domain organization is based on the crystal structures of Taz1 TRFH and dimerization domains (FW, YY, and ML unpublished results). In SpRap1, the N-terminal BRCT domain is in blue, the Myb domain in red, the Myb-like domain in light-blue, and the C-terminal RCT domain in cyan. The shaded area between Taz1 and SpRap1 indicates that the Taz1-SpRap1 interaction is mediated by Taz1RBM and SpRap1RCT. (b) In vitro ITC measurement of the interaction between Taz1RBM with SpRap1RCT. Insert is the ITC titration data. (c) Overall structure of the Taz1RBMSpRap1RCT complex. Taz1RBM and SpRap1RCT are colored in orange and cyan, respectively. (d) Hydrophobic and electrostatic interactions between the Taz1RBM helix and helices α1 and α2 of SpRap1RCT. Taz1RBM and SpRap1RCT are shown in ribbon model and colored as in panel c. The interacting residues of Taz1RBM and SpRap1RCT are presented as ball-and-stick models. Intermolecular electrostatic interactions are shown as dashed magenta lines. (e) Upper panel: structure-based sequence alignment of the RAP1 RCT domains from humans, S. pombe and S. cerevisiae. Lower panel: structure-based sequence alignment of the RBM regions of human TRF2, S. pombe Taz1 and S. cerevisiae Sir3. Secondary structure assignments from the human TRF2RBM-RAP1RCT crystal structure, the fission yeast Taz1RBMSpRap1RCT solution structure, and the budding yeast Sir3RBMScRap1RCT crystal structure (see Figure 7 below) are shown as colored cylinders (α helices) above the aligned sequences. Residues that are important or predicted to be important for the interactions based on the structures are highlighted in colored boxes. (f) Superposition of the solution structure of the fission yeast Taz1RBMSpRap1RCT complex on the crystal structure of the human TRF2RBM-RAP1RCT complex. Helices are shown as colored cylinders; human TRF2RBM is in yellow and human RAP1RCTin green (helices α1, α2, and α3) and in gray (helices α4, α5, and α6) whereas fission yeast Taz1RBM and SpRap1RCT in orange and cyan, respectively.
To reveal the structural basis of SpRap1 recognition by Taz1, we reconstituted the Taz1RBM-SpRap1RCT complex and determined its solution structure by nuclear magnetic resonance (NMR) (Supplementary Figs. 5a and 5b, Supplementary Table 2). To simplify the 15N- and 13C-labeled NMR sample preparation, we linked SpRap1RCT to Taz1RBM with a 14-residue linker. The linker is flexible and long enough so that it does not influence the proper interaction between Rap1RCT and Taz1RBM (Supplementary Figs. 5c and 5d). The structure of the Taz1RBM-SpRap1RCT complex reveals a compact globular fold (Fig. 5c). Taz1RBM contains a single α helix, while SpRap1RCT consists of three helices (Fig. 5c). Together, these helices are arranged into an intermolecular four-helix bundle. The Taz1RBM-SpRap1RCT interface buries a total of ∼1,680 Å2 solvent accessible surface area (Fig. 5d), which is substantially less than the interface area between human TRF2RBM and RAP1RCT. This is consistent with the much weaker binding affinity between Taz1RBM and SpRap1RCT (Fig. 5b).
Surprisingly, the structure of SpRap1RCT closely resembles the N-terminal three-helix bundle of the RCT domain of human RAP1 (Fig. 5d). Based on amino acid sequence alignment alone, the presence and extent of the RCT domain of SpRap1 could not have been correctly predicted (Fig. 5e). Notably, the structural similarity is not only limited to the Rap1RCT moiety of the complex; the helix of Taz1RBM interacts with SpRap1RCT in a fashion remarkably similar to the α1 helix of TRF2RBM in the TRF2RBM-Rap1RCT complex (Fig. 5d). The hydrophobic portion of the Taz1RBM helix packs into a hydrophobic groove formed by helices α1 and α2 of SpRap1RCT. Similar to Leu288 in the human TRF2CBM, the side chains of Ile379 and Leu383 of Taz1 point into the hydrophobic groove of SpRap1RCT with complementary surface (Fig. 5d). Although the Taz1RBM-SpRap1RCT interface is predominantly hydrophobic, electrostatic interactions provide additional specificity and stability to the complex. At both side of the Taz1RBM helix, the side chains of two arginine residues (Arg384 and Arg386) mediate a total of six electrostatic interactions with four acidic amino acids in SpRap1 (Glu674, Asp652, Asp656, and Glu666), helping anchor the Taz1RBM helix into the hydrophobic groove of SpRap1RCT (Fig. 5d). Despite the fact that Taz1RBM lacks the second helix α2 in TRF2RBM (Fig. 5f), the striking structural similarity between Taz1RBM-SpRap1RCT and TRF2RBM-RAP1RCT strongly support the notion that the interaction between Rap1 and the double-stranded telomeric DNA-binding protein is evolutionarily conserved from fission yeast to higher eukaryotes.
Mutational and functional analyses of the Taz1-SpRap1 interface
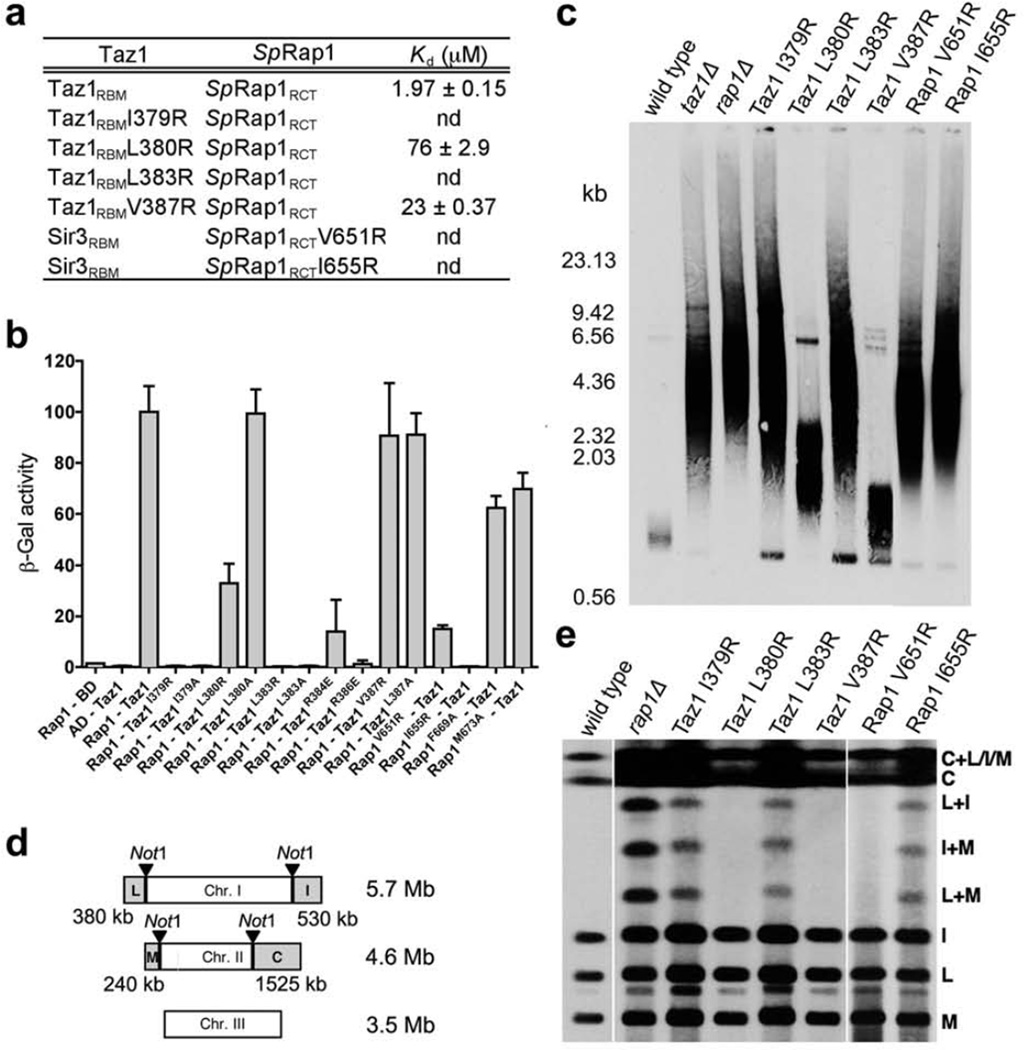
To investigate the significance of the Taz1-SpRap1 interaction, we first used ITC to measure the binding of a panel of missense mutations targeting interacting residues in both Taz1RBM and SpRap1RCT. An arginine substitution of Ile379 or Leu383 of Taz1 or Ile655 of SpRap1 at the center of the hydrophobic interface completely abolished the Taz1RBM-SpRap1RCT interaction (Fig. 6a). By contrast, three point mutations (Taz1 L380R, Taz1 V387R, and Rap1 V651R) weakened but did not disrupt the interface (Fig. 6a). These results are consistent with the solution structure: the side-chain of Taz1 Leu380, Taz1 Val387, and Rap1 Ile651 are all located at the periphery region of the interface, and thus make less contributions to the interaction (Fig. 5b). The effects of these mutants were also confirmed by yeast two-hybrid assays (Fig. 6b). Furthermore, mutants Taz1 R384E and R386E, designed to eliminate the electrostatic contacts between Taz1 and SpRap1, either completely abrogates or greatly weakened the interaction (Fig. 6b). Taken together, these results demonstrated that both hydrophobic and electrostatic interactions are crucial for the Taz1-SpRap1 complex formation.
Figure 6.
Mutational analysis of the Taz1-SpRap1 interaction. (a) Yeast two-hybrid assays to ascertain the effects of the Taz1 and SpRap1 mutations on the Taz1-SpRap1 interaction. Interaction of LexA-Taz1 with GAD-SpRap1 was measured as β-galactosidase activity. Data are averages of three independent β-galactosidase measurements normalized to the wild-type Taz1-SpRap1 interaction, arbitrarily set to 100. (b) In vitro ITC binding data of the wild-type and mutant Sir3RBM-ScRap1RCT interactions. (c) Analyses of telomere length in the various taz1 or rap1 mutants. EcoRI-digested genomic DNAs from indicated strains were subjected to Southern hybridization using the telomere repeats as the probe. (d) Schematic representation of NotI restriction sites on fission yeast genome. (e) Analyses of the various taz1 or rap1 mutants for telomere protection. Chromosomal DNAs were prepared in agarose plugs and separated by PFGE after NotI digestion. The gel was transferred to a nylon membrane and hybridized with a probe specific for telomere repeats. (f) Analyses of the effects on telomere protection when exo1, lig4 or rad3 are deleted in the presence (wild-type) or absence of taz1 or rap1. Experiments were performed as in panel e.
To address the in vivo consequence of the Taz1-SpRap1 interaction in telomere maintenance and protection, we first analyzed the telomere length phenotypes of the Taz1 and SpRap1 mutants that disrupted to varying degrees the Taz1-SpRap1 interaction in ITC assays (Supplementary Table 3). All the mutant proteins were expressed at near wild-type levels in yeast cells (Supplementary Figs. 6a and 6b), suggesting that residues at the Taz1-SpRap1 interface are not required for protein stability. Consistent with the published results, deletion of taz1+ or rap1+ from yeast cells resulted in a dramatic increase in telomere length and length heterogeneity compared to wild-type cells (Fig. 6c). Notably, all of the mutants exhibited partial or complete loss of telomere length regulation, in a manner that is consistent with the severity of the Taz1-SpRap1 interaction defect (Figs. 6b and 6c). Three point mutants (Taz1 I379R, Taz1 L383R, and Rap1 I655R) that completely abolished the Taz1-SpRap1 interaction in the ITC assay displayed a rap1Δ– and taz1Δ-like telomere length defect (Figs. 6b and 6c). In contrast, the Taz1 L387R mutant that retained the most similar-to-wide-type Rap1-binding activity exhibited the least defect in suppressing telomere length elongation (Figs. 6b and 6c). To analyze how the Taz1-SpRap1 interaction contributes to telomere end protection, we next examined the frequency of NHEJ-dependent telomere fusions at the G1 phase exhibited by these mutants by pulsed field gel electrophoresis (PFGE) of NotI-digested chromosomal DNA (Fig. 6d). Three mutants (Taz1 I379R, Taz1 L383R, and Rap1 I655R) with no detectable Taz1-SpRap1 interaction clearly exhibited altered mobility bands representing intra-chromosome fusions (Fig. 6e). In comparison, the mutants (Taz1 L380R, Taz1 V387R, and Rap1 V651R) that maintained partial Taz1-SpRap1 interaction activity completely protected telomeres from fusions (Fig. 6d), suggesting that these weakened Taz1-SpRap1 interactions are still able to mediate end protection. Taken together, both in vitro and in vivo studies indicated that the interactions between Taz1RBM and SpRap1RCT observed in the solution structure are essential for telomere end protection and maintenance.
Next, to examine whether these telomere defects were caused by the failure of telomere targeting of SpRap1 by Taz1, we analyzed the cellular localization of SpRap1 in yeast cells expressing mutant Taz1-mCherry and SpRap1-GFP proteins. Surprisingly, IF data showed that all the SpRap1 and Taz1 mutations, including those that completely disrupted the Taz1-SpRap1 interaction, only partially weakened the telomere localization of SpRap1 (Supplementary Figs. 6c and 6d). This result indicated that the binding of SpRap1 to Taz1 is not the only mechanism for targeting SpRap1 to telomeres. Unlike mammalian RAP1 that only binds to TRF2 at telomeres, SpRap1 interacts with two telomeric proteins, Taz1 and Poz1, simultaneously. Therefore, it is likely that both Taz1 and Poz1 can recruit SpRap1 to telomeres. To test this idea, we examined the SpRap1 localization in poz1Δ cells. Strikingly, deletion of poz1+ completely abolished the telomere localization of SpRap1 (Supplementary Figs. 6c and 6d), indicating that Poz1 is the key in targeting SpRap1 to telomeres. Collectively, these date suggested that, instead of serving as the telomere recruitment mechanism for SpRap1, the Taz1-SpRap1 interaction plays a more direct role in telomere regulation and protection than previously thought.
Notwithstanding the remarkable structural similarity between the Taz1RBM-SpRap1RCT and TRF2RBM-RAP1RCT complexes, fission yeast SpRap1 seems to have different functions at telomeres compared to its mammalian counterpart. First, deletion of SpRap1 did not result in telomere attrition as seen in Trf2 L286R expressing MEFs. Instead, telomere length in rap1Δ cells became ∼10 times longer with increased heterogeneity (Fig. 6b). Second, the interaction between Taz1 and SpRap1, unlike mammalian TRF2-RAP1 interactions, only plays a minor role in the telomere localization of SpRap1. Third, as reported previously39, telomere fusions in rap1Δ cells were completely rescued by lig4Δrap1Δ double mutation, indicating that these fusions at the G1 phase were mediated by NHEJ that is suppressed by SpRap1 in normal yeast cells (Fig. 6f). Furthermore, telomere fusions in rap1Δ cells require neither Exo1 nor checkpoint kinase Rad3 (the ATR ortholog) (Fig. 6f). This is in contrast to the Trf2 L286R-induced end-to-end chromosome fusions observed in mouse cells, in which chromosome fusions occurred independent of the NHEJ pathway and HDR mediated telomere attrition requiring both ExoI and Rad51 preceded these fusions (Figs. 4a–4f). Taken together, we propose that fission yeast and mammals employ an evolutionary conserved interaction mode to mediate the interactions of the RAP1 proteins, which protect chromosome ends through different mechanisms.
Structural basis of the budding yeast S. ceravesiae Sir3-ScRap1 interaction and its importance in telomeric silencing
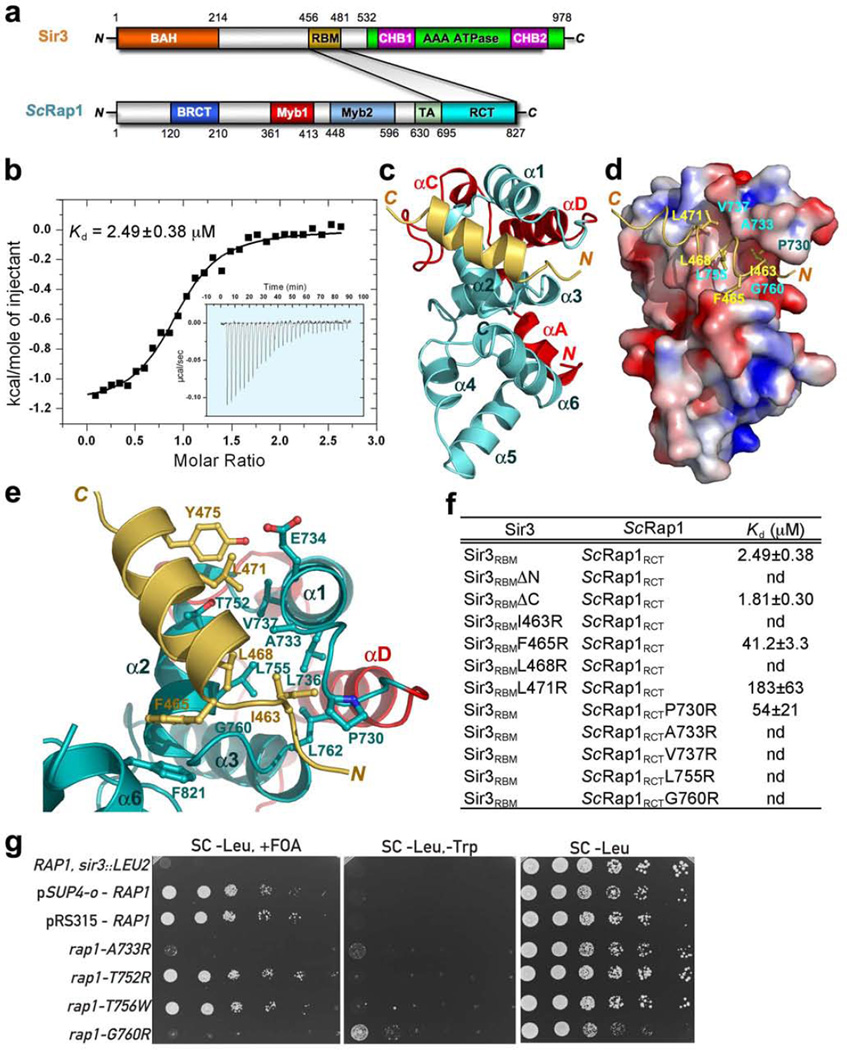
The budding yeast ScRap1 protein plays multiple roles in telomere maintenance and transcription regulation14. Unlike mammalian and fission yeast RAP1 proteins, ScRap1 localizes to telomeres by direct DNA binding to TG1–3 repeats through its two Myb domains15. Accordingly, budding yeast does not use a TRF2/Taz1-like protein as its major telomere-binding protein. ScRap1 mediates silencing at telomeres and at the silent HM mating-type loci by recruiting the Sir3 and Sir4 proteins via its C-terminal RCT domain (Fig. 7a)16. Previous studies revealed that a fragment of 26 amino acids of Sir3 (residues 456 – 481; referred to as Sir3RBM) is sufficient to mediate a yeast two-hybrid interaction with ScRap1RCT (Fig. 7a)16. We verified the direct binding of Sir3RBM to ScRap1RCT by ITC. The equilibrium dissociation constant (Kd) between Sir3RBM and ScRap1RCT is ∼ 1.8 µM (Fig. 7b), comparable to the fission yeast Taz1RBM-SpRap1RCT interaction (Fig. 5b).
Figure 7.
Structure of the budding yeast Sir3RBMScRap1RCT complex and its role in telomeric silencing. (a) Domain organization of the Sir3 and ScRap1 polypeptide chains. In Sir3, the N-terminal BAH domain is colored in orange, the C-terminal AAA ATPase domain in green with the two embedded CHB motifs in magenta, the ScRap1-binding motif (RBM) in light-orange. In ScRap1, the N-terminal BRCT domain is in blue, the two Myb domains in red and light-blue, the transcription activation (TA) domain in light-green, and the C-terminal RCT domain in cyan. (b) In vitro ITC measurement of the interaction between Sir3RBM and ScRap1RCT. Insert is the ITC titration data. (c) Overall structure of the Sir3RBMScRap1RCT complex. Sir3RBM is colored in light-orange. The core of the RCT domain contains helices α1-α6 (in cyan). The non-conserved N-terminal four-helix extension of ScRap1RCT (in red) folds onto the other side of ScRap1RCT and thus makes no contribution to the Sir3RBMScRap1RBM interaction. (d) The Sir3RBM helix (in light-orange) binds in a hydrophobic groove formed by helices α1 and α2 of ScRap1RCT. The Sir3RBM binding site of ScRap1RCT is shown in surface representation and colored according to its electrostatic surface potential (positive potential: blue; negative potential: red). (e) Hydrophobic interactions between the Sir3RBM helix and helices α1 and α2 of ScRap1RCT. Sir3RBM and ScRap1RCT are shown in ribbon model and colored as in panel c. The interacting residues of Sir3RBM and ScRap1RCT are presented as ball-and-stick models. (f) In vitro ITC binding data of the wild-type and mutant Sir3RBMScRap1RCT interactions. (g) Silencing phenotypes of ScRap1 mutants. Left panel: silencing of telomeres was tested in a telomeric silencing assay. Serial dilutions of the indicated mutant strains were plated on medium containing 5-FOA, which is lethal to cells expressing URA3, or medium lacking tryptophan (SC-Trp). Inability to grow on 5-FOA indicates a loss of telomeric silencing. Middle panel: silencing of the mating-type loci was tested in an HMR silencing assay. Serial dilutions of each mutant strain were plated on medium lacking tryptophan (SC-Trp) or histidine (SC-Leu). Growth on SC-Trp indicates a loss of mating-type locus silencing. Right panel: SC-Leu was used as a growth control. The genotype of the RAP1, sir3::LEU2 strain is MATα adh4::URA3 (tel VII-L) HMR::ADE2 sir3::LEU2. It was a control to show that derepression of the URA3 telomeric reporter gene (adh4::URA3) by mutation of SIR3 would cause a loss of growth on the FOA plates.
Despite the lack of apparent sequence similarity between Sir3RBM with either TRF2RBM or Taz1RBM, our findings that both mammalian and fission yeast RAP1 proteins use their RCT domains to recognize a short helical region of their interacting partners (Figs. 1c and 5c) prompted us to ask whether ScRap1RCT utilizes the same mechanism to bind to Sir3RBM. To test this hypothesis, we crystallized the Sir3RBM-ScRap1RCT complex and solved its structure by molecular replacement at a resolution of 2.0 Å (Supplementary Table 4). Surprisingly, the complex structure revealed a 2:1 stoichiometry between ScRap1RCT and Sir3RBM in the asymmetric unit (Supplementary Fig. 6a). Both ScRap1RCT molecules exhibit essentially the same conformation as the previously reported unliganded ScRap1RCT structure (Fig. 7c and Supplementary Fig. 6a)21. The central region of Sir3RBM adopts a helical conformation, which together with the N-terminal extension interacts with one ScRap1RCT molecule in the asymmetric unit, whereas the C-terminus of Sir3RBM (residues 461–464) contacts with the other ScRap1RCT (Supplementary Fig. 6a). ITC measurements using Sir3RBM peptides lacking either the N- or C- terminal tails demonstrated that the Sir3RBM C-terminus is dispensable for ScRap1RCT interaction (Fig. 7f and Supplementary Fig. 6b). Thus, the second interaction mode between ScRap1RCT and Sir3RBM observed in the crystals is likely due to lattice packing effects. We will focus our subsequent analysis on the first interaction mode here.
As we predicted, the binding mode of Sir3RBM to ScRap1RBM closely resembles the interactions between TRF2RBM and RAP1RCT and between Taz1RBM and SpRap1RCT (compare Figs. 1c, 5c, and 7c). The Sir3CBM helix packs against a hydrophobic groove formed by helices α1 and α2 of ScRap1RCT (Figs. 7d and 7e). The formation of the binary complex causes the burial of ∼ 1,700 Å2 of surface area at the interface. The core of this hydrophobic interface consists of the side chains of eight residues (Ala733, Val737, Leu755, and Gly760 in ScRap1RCT, and Ile463, Phe465, Leu468, and Leu471 in Sir3RBM) (Figs. 7d and 7e). Similar to the TRF2RBM-RAP1RCT interface, the N-terminal tail of Sir3RBM binds into a deep hydrophobic cleft formed by loops LD1 (between helices αD and α1) and L23 (between α2 and α3) (Figs. 7d and 7e). The side chain of Sir3 Ile463 (equivalent to TRF2 Ile283) is surrounded by a group of hydrophobic resides of ScRap1 (Figs. 7d and 7e), whose equivalent residues in mammalian and fission yeast RAP1 proteins also play important roles in the TRF2RBM-RAP1RCT and the Taz1RBM-SpRap1RCT interactions (Fig. 5e). Notably, the unique feature of ScRap1 is that it employs its RCT domain to recruit Sir3 to telomeres, whereas both mammalian RAP1 and fission yeast SpRap1 are recruited to telomeres through their RCT domains by interacting with other telomeric DNA-binding proteins.
The crystal structure of the Sir3RBM-ScRap1RCT complex is corroborated by mutagenesis. Mutations of the hydrophobic residues at the interface either completely abolished or greatly weakened the binding of the Sir3RBM helix to ScRap1RCT (Fig. 7f). To further examine the functional significance of the Sir3RBM-ScRap1RCT interaction, we tested the effects of two ScRap1 mutants (A733R and G760R) that disrupt the hydrophobic Sir3-binding groove of ScRap1 (Figs. 7e and 7f) on telomeric silencing. We employed a standard assay in which the URA3 gene is placed immediately adjacent to telomere VII-L created at the ADH4 locus40. Both mutants exhibited a strong loss of silencing, as manifested by a >1,000-fold decrease in the ability to form colonies on medium containing 5-fluoroorotic acid (5-FOA), a drug that kills cells expressing the URA3 gene (Fig. 7g)41. Next, the effects on mating-type silencing by the same two ScRap1 mutants were assayed in a strain in which the TRP1 gene replaced the mating-type genes adjacent to a mutated HMR-E silencer element (HMRΔA::TRP1)42. Both RAP1 mutations caused minor derepression of this silent locus, as indicated by a slight increase in growth on medium lacking tryptophan (Fig. 7g). This striking difference in the effect of the RAP1 mutations in telomeric versus HM silencing most likely reflects the well established redundancy of the HMR silencers, even in the absence of the A element at the HMR-E silencer16, and more specifically, the ability of Abf1, which still binds to this silencer, to recruit Sir3 independently (P. Moretti and D. Shore, unpublished data). Taken together, we conclude that the Sir3RBM-ScRap1RCT interaction plays an important role in telomeric silencing but is less important in mating-type silencing, where other pathways for Sir protein recruitment are likely to compensate for the loss of this interaction.
CONCLUSIONS
More than a decade of structural-function studies have revealed that the oligonucleotide/oligosaccharide binding (OB) folds and the Myb domains function as evolutionarily conserved protein motifs utilized by telomere proteins to bind to single-stranded or double-stranded telomeric DNAs15,43–48. However, it is not known whether evolutionarily conserved protein-protein interaction motifs also exist among telomere proteins. The TRFH domain in mammalian TRF proteins was proposed to be just such a motif, since a putative TRFH domain was identified in S. pome Taz15. However, our recent structural studies of S. pombe Taz1 dispelled this notion, because this putative TRFH domain has a completely different 3D structure and does not contain a peptide-binding pocket found in mammalian TRFH containing proteins (FW, YY, and ML, unpublished results)25.
In this study, by utilizing comparative structural analysis of RAP1 proteins from diverse organisms, we uncovered an evolutionarily conserved protein-protein interaction module, the RCT domain of RAP1. Our structure-based functional studies revealed that the RAP1 RCT domains of both mammalian and fission yeast mediate interactions with another telomere-binding protein (TRF2 in mammals, Taz1 in fission yeast) for chromosome end protection. In contrast, the RCT domain of budding yeast Rap1 recruits Sir3 to telomeres for transcriptional silencing. These results thus highlight the remarkable functional plasticity of this structurally conserved motif. Given the almost undetectable sequence similarity among some of the RCT domains and their binding partners in different organisms, it is unlikely that bioinformatics approaches will reveal additional RCT domains. Thus, our structural-functional studies provide the foundation for the study of the RCT domain of additional RAP1 proteins. For example, an RAP1-like protein (TbRap1) was recently identified in the parasite Trypanosoma brucei49. TbRap1 is an intrinsic component of the T. brucei telomere complex and a major regulator for silencing variant surface glycoprotein (VSG) expression sites49. Interestingly, our secondary structural analysis revealed a clear helix-rich pattern at the C-terminus of TbRap1 (data not shown). It is thus of obvious interest to understand whether this helical region is the equivalent RCT motif of TbRap1, whether it mediates the interaction with TbTRF (a mammalian TRF1/2-like protein50) for telomere localization, and whether it recruits a Sir3-like protein for VSG silencing. Answers to these questions will provide further insight into the structure and function of the RCT domains.
METHODS
Protein Expression and Purification
The human TRF2RBM-RAP1RCT complex
Human TRF2RBM (residues 275 – 316) was cloned into a GST fusion protein expression vector, pGEX6p-1 (GE healthcare) and RAP1RCT (residues 303 – 399) into a modified pET28b vector with a Sumo protein fused at the N-terminus after the His6 tag 48. The TRF2RBM-RAP1RCT complex was coexpressed in E. coli BL21(DE3). After induction for 16 hours with 0.1 mM IPTG at 25°C, the cells were harvested by centrifugation and the pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 50 mM NaH2PO4, 400 mM NaCl, 3 mM imidazole, 10% glycerol, 1 mM PMSF, 0.1 mg ml−1 lysozyme, 2 mM 2-mercaptoethanol, and home-made protease inhibitor cocktail). The cells were then lysed by sonication and the cell debris was removed by ultracentrifugation. The supernatant was mixed with Ni-NTA agarose beads (Qiagen) and rocked for 6 hours a t˚ C4 before elution with 250 mM imidazole. Then Ulp1 protease was added to remove the His6-Sumo tag. The complex was then mixed with glutathione sepharose beads (GE Healthcare) and rocked for 8 hours at 4˚ C before elution with 15 mM glutathione. Protease 3C was added to remove the GST-tag. Finally, the TRF2RBM-RAP1RCT complex was further purified by passage through Mono-Q ion-exchange column and by gel-filtration chromatography on Hiload Superdex200 equilibrated with 25 mM Tris-HCl pH 8.0, 150 mM NaCl and 5 mM dithiothreitol (DTT). The purified TRF2RBM-RAP1RCT complex was concentrated to 25 mg ml−1 and stored at 80˚C. The Se -Met substituted complex was similarly purified. For the ITC assay, wt and mutant proteins of TRF2RBM and RAP1RCT were individually expressed in E. coli and purified following the same procedure as described above except for only one affinity chromatography step was used according to the tags of the proteins.
The fission yeast Sp Rap1RCT-Taz1RBM Fusion Protein
To facilitate protein purification and structure determination, we made a fusion protein construct that contains SpRap1RCT (residues 639–693), a 14-residue linker (NH2- GGSGGSKLGGSGGS-COOH), and Taz1RBM (residues 362–395). The SpRap1RCT-Taz1RBM fusion protein was cloned into the modified pET28b vector with a His6–Sumo tag 48. The protein was expressed in E. coli BL21(DE3). 10 mL of overnight culture was transferred to 1 L of M9 minimal media supplemented with 15NH4Cl or 15NH4Cl/[13C]-glucose for the preparation of 15N-labeled or 15N/13C-labeled proteins, respectively. When the culture reached an OD600 of 0.6, expression of the fusion protein was induced by adding 0.2 mM IPTG. The proteins were purified by Ni-NTA affinity chromatography. The Sumo-tag was removed by on-column cleavage with ULP1. The proteins were further purified by gel filtration on a Superdex-75 column (GE healthcare).
The budding yeast Sir3RBM-Sc Rap1RCT complex
Sir3RBM (residues 456 – 481) was cloned into a GST fusion protein expression vector, pGEX6p-1 (GE healthcare) and ScRap1RCT (residues 679 – 827) into the modified pET28b vector with a His6-Sumo tag 48. The Sir3RBM-ScRap1RCT complex was coexpressed in E. coli BL21(DE3). The purification procedure is the same for the human TRF2RBM-RAP1RCT complex as described above except for the last step. Due to the relatively weak interaction between Sir3RBM and ScRap1RCT, some of the Sir3RBM peptides were lost during the purification process. Thus, in the final purification step (gel filtration chromatography) we mixed additional individually purified Sir3RBM peptides with the complex to make sure that there were enough Sir3RBM peptides in the final sample. Therefore, the material used for crystallization was not a stable 1:1 complex.
Crystallization, Data Collection and Structure Determination
The human TRF2RBM-RAP1RCT complex
The TRF2RBM-RAP1RCT complex was crystallized by hanging-drop-vapor-diffusion at 4°C. The precipitant/well solution contained 100 mM sodium citrate pH 5.2, 17% PEG 2000, 16% isopropanol and 10 mM DTT. Crystals were gradually transferred to a harvesting solution containing 100 mM sodium citrate pH 5.2, 19% PEG2000, 20% Glycerol, 16% Isopropanol, and 10 mM DTT before being flash-frozen in liquid nitrogen for storage and data collection under cryogenic conditions (100K). Se-Met-SAD (at Se peak wavelength) dataset with a resolution of 1.95 Å was collected at beam line 21ID-D at APS and processed using HKL2000 51. Crystals belong to space group P212121 and contain three TRF2RBM-RAP1RCT complexes per asymmetric unit. Nine selenium sites were located and refined, and SAD phases calculated using SHARP 52. The initial SAD map was significantly improved by solvent flattening. A model was automatically built into the modified experimental electron density using ARP/WARP 53; the model was then further refined using simulated-annealing and positional refinement in CNS54 with manual rebuilding using program O55.
The budding yeast Sir3RBM-Sc Rap1RCT complex
The Sir3RBM-ScRap1RCT complex was crystallized by hanging-drop-vapor-diffusion at 4°C. The precipitant/well solution contained 100 mM sodium sitrate pH 4.8, 30% PEG4K, 200 mM ammonium acetate, and 10 mM DTT. Crystals were gradually transferred to a harvesting solution containing 100 mM sodium citrate pH 4.8, 30% PEG4K, 200 mM ammonium acetate, 20% glycerol, and 10 mM DTT before flash-frozen in liquid nitrogen for storage and data collection under cryogenic conditions (100K). A native dataset with a resolution of 2.0 Å was collected at beam line 21ID-D at APS and processed using HKL2000 51. Crystals belong to space group R32 with a = b = 89.831 Å, c = 211.791 Å. The structure was determined by molecular replacement method using Phaser in the CCP4i suite 56 and the crystal structure of ScRap1RCT (PDB code: 3CZ6) as the initial model. The Sir3RBM fragment was manually built into the electron density using O 55 and then further refined in CNS 54.
NMR Spectroscopy and Structure Determination of the Fission Yeast Taz1RBM-Sp Rap1RCT Complex
The NMR experiments were carried out at 25 °C on Bruker 600- and 800-MHz spectrometers equipped with four RF channels and triple resonance pulsed-field gradient cryoprobes. The chemical shifts were referenced to internal 2, 2-dimethyl-2-silapentanesulfonic acid (DSS). The samples were prepared with 1.5 mM Taz1RBM-SpRap1RCT Complex dissolved in a buffer of 90% H2O/10% D2O containing 20 mM sodium phosphate (pH 6.5) and 50 mM NaCl. All NMR spectra were processed with NMRPipe 57 and analyzed with Sparky. Two-dimensional 15N- and 13C-edited HSQC, (HB)CB(CGCD)HD, (HB)CB(CGCDCE)HE, and three-dimensional HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, C(CCO)NH, H(CCCO)NH, HCCH-TOCSY and CCH-COSY spectra were recorded to obtain the chemical shift assignments of backbone and side-chain atoms. The three-dimensional 15N- and 13C-edited NOESY-HSQC spectra (mixing time 100 ms) were collected to generate the distance restraints for structure calculations.
Initial structures were calculated using the program ARIA 2.2 58, NOE peaks were assigned with SANE 59 and CYANA 2.1 60, and the final structures were refined with Amber 9.0 61. Distance restraints were derived from interproton NOEs. Backbone dihedral angle (φ and ψ) restraints were generated from chemical shift data using TALOS 62. Hydrogen bond restraints were determined using the secondary structure information from CSI 63 and confirmed by intermediate range NOEs. The 20 lowest energy structures from ARIA were selected as models for SANE to extend the NOE assignments. The final set of distance restraints were obtained after several rounds of SANE/CYANA calculations. Two hundred structures from CYANA were refined by restrained molecular dynamics calculations with Amber using generalized Born salvation model to account for solvent effects. The 20 refined structures with the lowest energy were analyzed using PROCHECK-NMR64
Isothermal Titration Calorimetry (ITC)
The equilibrium dissociation constants of the TRF2rbm-RAP1rct, the Taz1RBM-SpRap1RCT, and the Sir3RBM-ScRap1RCT interactions were determined by using a VP-ITC calorimeter (MicroCal). The enthalpies of interactions were measured at 20°C in 25 mM Tris (pH 8.0) and 150 mM NaCl. Two independent experiments were performed for every interaction described here. ITC data were subsequently analyzed and fit using Origin 7 software (OriginLab) with blank injections of peptides into buffer subtracted from the experimental titrations prior to data analysis.
Yeast Two-hybrid Assay
The yeast two-hybrid assays were performed using L40 strains harboring pBTM116 and PACT2 (Clontech) fusion plasmids and selected on–Leu–Trp plates. β-galactosidase activities were measured according to Clontech MATCHMAKER library protocol and the averages from three individual transformants were reported.
Co-immunoprecipitation (Co-IP)
293T cells were transfected by the calcium-phosphate coprecipitation method using 4 µg of total plasmid DNA per well in 6-wells dishes. For immunoprecipitations, cells were lysed in ice-cold buffer (50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 400 mM NaCl, 0.2% NP40, 0.1% SDS, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 g ml−1 of aprotinin, 10 g ml−1 of pepstatin and 1 g ml−1 of leupeptin). Supernatants collected were used for immunoprecipitation with anti-Myc antibody conjugated agarose beads (Sigma). We washed the beads four times with lysis buffer, eluted proteins with Laemmli loading buffer and analyzed them by SDS-PAGE.
shRNA Sequence
The murine Trf2 shRNA targeting sequence GAACAGCTGTGATGATAA was cloned into pRetro-Super vector (Stratagene) and used as described in the text. shRNA against Rad51 (pRetroSuper encoding RAD51 (GGGAAUUAGUGAAGCCAAA)) was obtained from Magdalena Tarsounas (University of Oxford).
Generation of MEFs and Retroviral Infection of Cell Lines
MEFs were isolated from embryonic day (E) 13.5 embryos grown in standard culture condition. Primary MEFs isolated from Ku70−/−, Lig4−/−, ATM−/−, Apollo−/− and ExoI−/−embryos were immortalized at passage 2 by transfection with pBabeSV40LT. mTrf2 shRNA was generated in pSuper as described 26. 293T cells were transiently transfected with shRNA resistant DNA constructs for viral particle packaging using Lipofectamine Plus (Invitrogen). Viral supernatant were collected 48–72 hours post-transfection, filtered through 0.45 µm membrane, and directly used to infect the SV40LT immortalized MEFs. MEFs infected with shRNA resistant cDNAs were further infected by two consecutive rounds of mTrf2 shRNA at 12 h intervals. After four days of infection, cells selected in puromycin were harvested for chromosome analysis.
Immunoblotting
Cell extracts were isolated and western blot was performed as described26. Antibodies were: anti-Chk2 from BD Biosciences; anti-Flag, anti-γ-tubulin and anti-haemagglutinin from Sigma; anti-γH2AX from Upstate; anti-myc from Santacruz, anti-RAP1 from Abcam; anti-53BP1 antibody was obtained from Dr. Philip Carpenter at the UT Medical School; and anti-TRF2 antibody was obtained from Dr. J. Karlseder at the Salk Institute.
Immunofluorescence and fluorescent in situ hybridization (IF-FISH)
Cells grown on coverslips were fixed for 10 min in 2% sucrose and 2% paraformaldehyde at RT followed by PBS washes. Coverslips were blocked for one hour in blocking solution (0.2% fish gelatin and 0.5% BSA in 1XPBS). The cells were incubated with primary antibodies (ant-HA and anti-Myc for HeLa cells; anti-γ-H2AX, anti-53BP1, anti-Trf2, and anti-Rap1 for MEFs) for 2 hour at RT. After PBS washes, coverslips were incubated with the appropriate Alexa fluor secondary antibody for one hour followed by washes in PBS. Next, the coverslips were fixed with 4% paraformaldehyde for 10 min at RT, washed extensively in PBS. Hybridizing mix (70 % formamide, 2% BSA, 100µg/ml tRNA) containing peptide-nucleic acid (PNA) 5’-Tam-OO-(CCCTAA)4-3’ probe (Applied Biosystem)26 was added to each cover slip and the cells were denatured by heating for 3 min at 80°C on a heat block. After 2 hour incubation at RT in the dark, cells were washed twice with 70 % formamide, 0.1% Tween 20, 0.1% BSA, 10 mM Tris-HCl and pH 7.5 followed by 3 washes in 50mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% BSA and 0.1% Tween-20. DNA was counterstained with 4.6-diamidino-2-phenylindole (DAPI) and slides were mounted in 90% glycerol/10% PBS containing 1 µg/ml p-phenylene diamine (Sigma). Digital images were captured with a Nikon Eclipse 800 microscope.
PNA FISH and CO-FISH
Metaphase chromosomes from MEFs were prepared 4 to 7h after colcemid treatment. Chromosomes were fixed and telomere FISH with peptide-nucleotide acid (PNA) Tam-OO-(CCCTAA)4 probe (Applied Biosystem) was performed as described previously 26. For CO-FISH, metaphase spreads were incubated sequentially with Tam-OO-(CCCTAA)4-3’ and 5’-FITC-CO-(TTAGGG)4 probes as described previously 8,26,65. Images were captured on a Nikon Eclipse 800 microscope and processed with MetaMorph Premier (Molecular Devices). A minimum of 30 metaphases from each sample were analyzed.
Strains and General Techniques for Fission Yeast
The S. pombe strains used in this study are listed in Table S3. Yeast extract media YES and EMM (with or without nitrogen) were used to grow cells. Growth media, basic genetics, and biochemical techniques for fission yeast were described previously66. Gene disruption was performed by the replacement of most of each ORF with the ura4+, kanMX6 (kanr), or hphMX6 (hygr) cassettes67,68. C-terminal tagging of each gene was performed by the insertion of a tag with kanMX6 (kanr) or hphMX6 (hygr) cassettes at each chromosomal gene locus67,68. Mutations in the taz1+ or rap1+ genes were created using QuikChange (Stratagene), and each mutated DNA fragment was used for transformation of the taz1::ura4+ or rap1::ura4+ strains to replace the ura4+ cassette with mutated DNA. Mutagenesis of the chromosomal gene was confirmed by genome sequencing.
Telomere Southern Analysis for Fission Yeast
Southern analyses for detection of telomere repeats were performed as previously described 20. Genomic DNAs were digested by EcoRI. Telomere repeats (∼ 300 bp) were used as the probe.
Pulse Field Gel Electrophoresis (PFGE) for Fission Yeast
PFGE for detection of telomere end fusion was performed as previously described69. Cells were grown in EMM with nitrogen, and then incubated in EMM without nitrogen for 24 hours to arrest cells in G0 phase. Genomic DNAs were digested by Not1. Telomere repeats (∼ 300 bp) were used as the probe.
Microscopic Analysis of Sp Rap1 Localization in Fission Yeast
Cells were grown in EMM medium, and fluorescence microscope images for live cells were taken using a fluorescence microscope system (DeltaVision; Applied Precision). A 3D stack of images spanning 13 focal planes at 0.3 µm increments was recorded. Projection images were generated using a maximum intensity method.
S. cerevisiae Silencing Assays
Point mutations in RAP1 were generated by site-directed mutagenesis and cloned in a LEU2-CEN-ARS plasmid (pRS315;70). Strains for the silencing assays were generated by sporulation of a W303 MATa/MATα diploid 71 heterozygous for rap1::KanMX (a complete ORF replacement allele), the telomeric silencing reporter adh4::URA3-Tel VII-L 40, and the HMR silencing reporter HMRAA::TRP142. This diploid strain also contained a SUP4-o centromeric plasmid carrying the wild-type RAP1 gene. A MATα haploid segregant carrying the two silencing reporter genes, the rap1::KanMX allele and the RAP1-SUP4-o plasmid was identified. This strain was then transformed with pRS315, pRS315-RAP1 (wt), or the pRS315-rap1 mutant plasmids. In strains transformed with either wild type RAP1 or the mutants, the RAP1-SUP4-o plasmid was counter-selected on plates containing canavanine and its loss was confirmed by showing that the resulting strains were Ade-. Silencing assays were performed as described 40,42, by spotting 5 µl aliquots of successive 5-fold dilutions of overnight cultures on the indicated selective media. Plates were incubated at 30 °C for 3 days before being photographed.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. Jan Karlseder, Lenhard Rudolph, Madalena Tarsounas and Philip Carpenter for providing valuable reagents. We thank Fuyuki Ishikawa for strains, Yuki Ogiyama and Kojiro Ishii for technical support on PFGE. M.L. acknowledges generous financial support from NIH (RO1 GM083015), the American Cancer Society and the Sidney Kimmel Foundation. M.L. is a Howard Hughes Medical Institute Early Career Scientist. S.C. acknowledges generous financial support from the NIA (RO1 AG028888), the NCI (RO1 CA129037), the Welch Foundation, the Susan G. Koman Race for the Cure Foundation, the Abraham and Phyllis Katz Foundation and the Michael Kadoorie Cancer Genetic Research Program. J.K. was supported by grants from OSAKA UNIVERSITY Life Science Young Independent Researcher Support Program (Japan Science and Technology Agency), from the Japanese Ministry of Education, Culture, Sports, Science and Technology, from Inamori Foundation, and from Astellas Foundation for Research on Metabolic Disorders, and from Takeda Science Foundation. Work in the laboratory of D.S. was supported by a grant from the Swiss National Fund (grant 31003A116716), by the Swiss National Fund “Frontiers in Genetics” NCCR program, and by the Canton of Geneva. Use of Life Sciences Collaborative Access Team Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. DE-AC02-06CH11357.
REFERENCES
- 1.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 2.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.Linger BR, Price CM. Conservation of telomere protein complexes: shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101:471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 6.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 7.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 8.Guo X, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. Embo J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 10.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 11.Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 12.Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. Embo J. 2009 doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huet J, et al. A general upstream binding factor for genes of the yeast translational apparatus. Embo J. 1985;4:3539–3547. doi: 10.1002/j.1460-2075.1985.tb04114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 15.Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- 16.Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 18.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 19.Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- 20.Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 21.Feeser EA, Wolberger C. Structural and functional studies of the Rap1 C-terminus reveal novel separation-of-function mutants. J Mol Biol. 2008;380:520–531. doi: 10.1016/j.jmb.2008.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J Mol Biol. 1991;218:183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- 23.Lenain C, et al. The Apollo 5' exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 24.van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- 26.Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996;10:1310–1326. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 28.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Bailey SM, Goodwin EH, Meyne J, Cornforth MN. CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis. 1996;11:139–144. doi: 10.1093/mutage/11.2.139. [DOI] [PubMed] [Google Scholar]
- 30.Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 32.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 33.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonetti D, Martina M, Clerici M, Lucchini G, Longhese MP. Multiple pathways regulate 3' overhang generation at Scerevisiae telomeres. Mol Cell. 2009;35:70–81. doi: 10.1016/j.molcel.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Shen B, Nolan JP, Sklar LA, Park MS. Functional analysis of point mutations in human flap endonuclease-1 active site. Nucleic Acids Res. 1997;25:3332–3338. doi: 10.1093/nar/25.16.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 37.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 38.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 39.Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. Embo J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at Scerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 41.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 42.Sussel L, Shore D. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc Natl Acad Sci U S A. 1991;88:7749–7753. doi: 10.1073/pnas.88.17.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 45.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 46.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 47.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 48.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 49.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Espinal A, Cross GA. Trypanosome telomeres are protected by a homologue of mammalian TRF2. Mol Cell Biol. 2005;25:5011–5021. doi: 10.1128/MCB.25.12.5011-5021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otwinowski Z, Minor W. in Method in Enzymology. Vol. 276. Academic Press; 1997. Processing of X-ray Diffraction Data Collected in Oscillation Mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 52.La Fortelle Ed, Bricogne G. in Method in Enzymology. Vol. 276. Academic Press; 1997. Maximum-likelihood Heavy-Atom Parameter Refinement for Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods; pp. 472–494. [DOI] [PubMed] [Google Scholar]
- 53.Lamzin VS, Perrakis A, Wilson KS. The ARP/WARP suite for automated construction and refinement of protein models. In: Rossmann MG, Arnold E, editors. In Int. Tables for Crystallography Vol. F: Crystallography of biological macromolecules. The Netherlands: Dordrecht, Kluwer Academic Publishers; 2001. pp. 720–722. [Google Scholar]
- 54.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 55.Jones TA, Zou JY, Cowan SW, Kjeldgaaed M. Improved methods for building protein models in electron density maps and the location of errors in these methods. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 56.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 58.Rieping W, et al. ARIA2: automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- 59.Duggan BM, Legge GB, Dyson HJ, Wright PE. SANE (Structure Assisted NOE Evaluation): an automated model-based approach for NOE assignment. J Biomol NMR. 2001;19:321–329. doi: 10.1023/a:1011227824104. [DOI] [PubMed] [Google Scholar]
- 60.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 61.Case DA, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 63.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 64.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 65.Else T, et al. Genetic p53 deficiency partially rescues the adrenocortical dysplasia phenotype at the expense of increased tumorigenesis. Cancer Cell. 2009;15:465–476. doi: 10.1016/j.ccr.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 67.Bahler J, et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 68.Hentges P, Van Driessche B, Tafforeau L, Vandenhaute J, Carr AM. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast. 2005;22:1013–1019. doi: 10.1002/yea.1291. [DOI] [PubMed] [Google Scholar]
- 69.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 70.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.