Abstract
A genetic screen based on the blue-white β-galactosidase complementation assay designed to detect G→A mutations arising during RNA-dependent DNA synthesis was used to compare the fidelity of mutant human immunodeficiency virus type 1 reverse transcriptases (RTs) with the mutations M230L and M230I with the wild-type enzyme, in the presence of biased deoxynucleoside triphosphate (dNTP) pools. The mutant RTs with the M230L and M230I changes were found to be 20 to 70 times less faithful than the wild-type RT in the presence of low [dCTP]/[dTTP] ratios but showed similar fidelity in assays carried out with equimolar concentrations of each nucleotide. Biased dNTP pools led to short tandem repeat deletions in the target sequence, which were also detectable with the assay. However, deletion frequencies were similar for all of the RTs tested. The reported data suggest that RT pausing due to the low dNTP levels available in the RT reaction mixture facilitates strand transfer, in a process that is not necessarily mediated by nucleotide misinsertion.
Human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) converts the viral genomic RNA to a double-stranded DNA intermediate that is integrated into the host cell DNA. HIV-1 RT is a multifunctional enzyme with RNA- and DNA-dependent DNA polymerase, RNase H, strand transfer, and strand displacement activities (38). The mature enzyme is a heterodimer composed of two subunits of 66 and 51 kDa, with subdomains termed fingers, thumb, palm, and connection in both subunits and an RNase H domain in the large subunit only. The catalytic site resides within the palm subdomain of the 66-kDa subunit, which contains the catalytic residues Asp-110, Asp-185, and Asp-186. Other residues in their vicinity, such as Lys-65, Arg-72, Asp-113, Ala-114, Tyr-115, and Gln-151, are involved in the interaction with the incoming dNTP (8). Mutations affecting those amino acids often result in drastic reductions of the RT's polymerase activity or significant changes in the nucleotide specificity of the enzyme (reviewed in reference 25).
Residues 227 to 235 (FLWMGYELH) of the palm subdomain form the β12-β13 hairpin, which defines the so-called primer grip, a highly conserved motif of retroviral RTs. The primer grip is involved in maintaining the primer terminus in an orientation appropriate for nucleophilic attack on an incoming dNTP. Mutational analysis of primer grip residues has shown their influence on various RT functions, including dNTP binding (44), polypurine tract removal (32), RNase H activity (29), template-primer utilization (5, 14), and fidelity of DNA synthesis (7, 43).
Met-230 is located at the tip of the β12-β13 hairpin. The side chain of Met-230 interacts with the deoxyribose ring at position −2 of the primer and lies in the minor groove of the template-primer complex (4, 8). Substitution of Ala for Met-230 renders an RT with reduced affinity for the incoming deoxynucleoside triphosphate (dNTP) (44) and leads to a noninfectious virus (45). However, substitution of Ile for Met-230 does not impair polymerase function or virus viability. Although Met-230 is highly conserved among HIV-1 isolates, other residues (i.e., Ile, Leu, or Val) have also been found at this position in RTs of viral isolates from HIV-infected individuals (34; http://hivdb.stanford.edu). The mutation M230L has been selected in vitro after passaging of HIV-1IIIB or HIV-2ROD in the presence of delavirdine (13, 28) and has been detected in clinical isolates from patients treated with other RT inhibitors (9). This mutation alone reduced susceptibility to all approved nonnucleoside RT inhibitors (i.e., nevirapine, delavirdine, and efavirenz) by 23- to 58-fold (9). In addition, M230I has been identified in virus from patients treated with the inhibitor HBY097 (18), as well as after passaging of simian immunodeficiency virus in vitro in the presence of delavirdine (12).
Transfection experiments carried out with proviral DNA revealed that mutation M230I compensated for the dNTP binding defect shown by an HIV-1 RT bearing Trp at position 115 (27). In addition, it was recently demonstrated that the amino acid substitution M230I improved the replication capacity of HIV-1 bearing the RT mutation Q151L, thereby facilitating the emergence of multi-dideoxynucleoside-resistant HIV-1 variants (24). Unlike other primer grip mutant RTs (i.e., those with the F227A and W229A changes), that with M230I was relatively error prone in gel-based misincorporation fidelity assays with DNA oligonucleotides as templates. Interestingly, the catalytic efficiency for T · G misinsertions was 3- to 16-fold higher for the mutant enzyme than for the wild-type (WT) RT (7).
G→A transitions have been identified as the most frequent mutations arising during reverse transcription in several retroviruses (30, 39; reviewed in reference 37), and extensive G→A hypermutation has been observed as a particular trait of lentivirus RTs (40, 42). G · T mispairs formed during RNA-dependent DNA synthesis may result in G→A mutations in the viral progeny. The chance for a T to be inserted opposite a G can be increased by lowering the ratio of the concentrations of dCTP and dTTP (20, 21). In this study, we have used a genetic assay that uses a target sequence containing three UGG codons (41) to compare the fidelity of RNA-dependent DNA polymerization displayed by WT and primer grip mutant RTs, in the presence of biased dNTP pools. Higher frequencies of G→A mutations obtained after lowering of the dCTP concentration in the assay were remarkably increased in retrotranscription assays with mutant RTs with M230L and M230I, which were thus characterized as highly mutagenic. Other genetic lesions (i.e., short tandem repeat deletions) were also detected with the assay in the presence of biased dNTP pools. However, their frequency was not influenced by mutations in the primer grip.
RT purification and experimental outline of the genetic assay.
WT HIV-1BH10 RT and mutant RTs were expressed and purified as previously described (7, 22). Mutation M230L was introduced in the RT-coding region with the QuikChange Site-Directed Mutagenesis Kit (Stratagene) with plasmid pRT6 (33) as the template. All RTs were purified as p66-p51 heterodimers, and mutations were introduced into both subunits of the enzymes.
The genetic assay was carried out with a 141-nucleotide (nt) RNA template containing a 66-bp HIV-1 pol fragment that includes the sequence…AAGGAAACAUGGGAAACAUGGUGG… (Trp codons underlined), which encodes RT residues 395 to 402 (Lys-Glu-Thr-Trp-Glu-Thr-Trp-Trp) (41). The RNA was obtained by in vitro transcription with T3 RNA polymerase. Plasmid pBluescript SK (+), containing the pol fragment previously cloned in its unique EcoRI and HindIII restriction sites (kindly provided by M. A. Martínez, Fundació IrsiCaixa, Badalona, Spain), was linearized with HindIII, purified by agarose gel electrophoresis, and used as the template in the transcription reaction. These reactions were carried out at 37°C for 1 h in a 100-μl volume containing 40 mM Tris-HCl (pH 8.0), 6 mM MgCl2, 10 mM dithiothreitol (DTT), 2 mM spermidine, the four ribonucleotides at 0.5 mM each, 1 μg of linearized pBluescript DNA, 0.6 U of RNasin/μl, and 0.5 U of T3 RNA polymerase/μl. Samples were then heated at 65°C for 5 min, and after addition of 10 U of RNase-free DNase, the incubation was continued for another 30 min. The RNA was then phenol extracted and precipitated with ethanol.
The RNA was reverse transcribed in 50 mM HEPES (pH 7.0) containing 15 mM NaCl, 15 mM magnesium acetate, 130 mM potassium acetate, 10 mM DTT, and various dNTP concentrations, depending on the assay. Oligonucleotide 29T3 (5′-GCGGGCAAGCTTGTGGYTTGCCAATAYTYTGT-3′; 8 pmol) was annealed to 10 to 40 pmol of the 141-nt RNA template in 50 μl of the reaction mixture by being first heated to 65°C for 60 s and then incubated at 37°C for another 60 s. Reactions were initiated by addition of 25 U of RNasin and 30 to 300 nmol of the corresponding RT and were then incubated for 3 h at 37°C. In order to recover sufficient material for subsequent cloning, cDNA was amplified by PCR with oligonucleotides 29T3 and 31T (5′-GGCGAATTCTAAATTTAAACTACCCATACAA-3′), producing an 87-bp fragment with EcoRI and HindIII restriction sites at its ends. Optimized PCR conditions were 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 250 μM each dNTP, 360 ng of each primer, 25 μl of the reverse transcription reaction mixture, and 1.25 U of AmpliTaq DNA polymerase in a final volume of 100 μl. PCR amplification was initiated by incubation at 94°C for 2 min, followed by 20 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s and a final incubation at 72°C for 5 min.
The amplified DNA was phenol extracted, precipitated with ethanol, subjected to treatment with EcoRI and HindIII, and then purified from a 1% agarose gel. This fragment encoding three Trp codons was cloned in frame of the lacZα fragment of the replicative form (RF) of bacteriophage M13mp18. For this purpose, a modified M13mp18 RF carrying an exogenous EcoRI-HindIII insert (414 bp) encoding the murine leukemia virus protease (26) was digested first with HindIII and then with EcoRI to obtain the linearized vector, which was purified from a 1% agarose gel and treated with shrimp alkaline phosphatase before ligation to the PCR products. Escherichia coli XL-1 Blue MRF′ was transformed and plated onto minimal agar plates containing 8% X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and 20% IPTG (isopropyl-β-d-thiogalactopyranoside). The plates were incubated at 37°C for approximately 15 h. A few blue plaques (as a control) and all colorless and light blue plaques were analyzed for mutations. Mutant plaques were picked from plates, resuspended in 100 μl of 0.9% NaCl, and stored at 4°C.
Plaques containing the appropriate inserts were identified by PCR amplification with primers 1R (5′-GCAGTGAGCGCAACGCAATTAATG-3′) and 1F (5′-AAAGCGCCATTCGCCATTCAGG-3′). Reaction conditions were 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 100 μM each dNTP, 150 ng of each primer, 3 μl of the solution containing the resuspended plaque, and 1.25 U of AmpliTaq DNA polymerase in a final volume of 50 μl. PCR amplification was initiated by incubation at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s and a final incubation at 72°C for 10 min. Reaction products were analyzed in a 3% agarose gel. Plaques rendering the expected amplification products of approximately 387 bp were easily distinguishable from others derived from vectors lacking the 66-bp EcoRI-HindIII insert (315 bp) or containing the modified M13mp18 RF used as a vector source (723 bp). RF DNA from selected plaques was prepared as previously described (2) and subjected to DNA sequencing.
Analysis of the frequency of G→A transitions arising during reverse transcription in the presence of biased [dCTP]/[dTTP] pools.
The effect of the concentration of dCTP on the frequency of G→A transitions in reactions catalyzed by the WT HIV-1 RT was analyzed with the genetic assay described above. In the presence of saturating concentrations of all dNTPs, the frequency of G→A transitions (fG→A) was 1.07 × 10−4 (Table 1). An inverse relationship was noted between fG→A and the magnitude of the [dCTP]/[dTTP] bias. A bias of around 2 × 10−4 (i.e., 0.1 μM dCTP, 440 μM dTTP) produced a 30-fold increase in fG→A relative to estimates obtained from reactions carried out in the presence of equimolar concentrations of each dNTP. These effects were even more pronounced in assays carried out with mutant RTs. For example, G→A transition frequencies for the mutant RTs with M230I and M230L were very low in reactions carried out with a 50 μM concentration of each dNTP (frequencies of <4 × 10−4) but increased to as much as 0.154 and 0.0634, respectively, in assays done with a [dCTP]/[dTTP] ratio of 1:4,400. These values were also significantly higher than those determined with the WT RT. The mutant RT with M230I was around two to three times more error prone than that with M230L in assays carried out with biased concentrations of dCTP and dTTP.
TABLE 1.
Mutation frequencies accompanying reverse transcription in the presence of biased dNTP pools
| Enzyme and no. of expts | [dNTP] (μM)
|
No. of plaques analyzeda
|
No. of G→A transitions | fG→Ab | fG→A rel.c | ||||
|---|---|---|---|---|---|---|---|---|---|
| dCTP | dTTP | dATP | dGTP | Total | Mutated | ||||
| WT RT | |||||||||
| 12 | 50 | 50 | 50 | 50 | 3,108 | 2 | 2 | 1.07 × 10−4 | |
| 3 | 1 | 440 | 40 | 20 | 585 | 2 | 2 | 5.70 × 10−4 | |
| 4 | 0.5 | 440 | 40 | 20 | 486 | 3 | 3 | 1.03 × 10−3 | |
| 4 | 0.2 | 440 | 40 | 20 | 622 | 5 | 5 | 1.34 × 10−3 | |
| 6 | 0.1 | 440 | 40 | 20 | 1,270 | 23 (4)d | 25 | 3.28 × 10−3 | |
| M230L | |||||||||
| 4 | 50 | 50 | 50 | 50 | 418 | 0 | 0 | <3.99 × 10−4 | <3.7 |
| 3 | 0.5 | 440 | 40 | 20 | 427 | 20 (1) | 20 | 7.81 × 10−3 | 7.6 |
| 2 | 0.2 | 440 | 40 | 20 | 194 | 23 (2) | 27 | 2.32 × 10−2 | 17.3 |
| 3 | 0.1 | 440 | 40 | 20 | 225 | 62 (12) | 86 | 6.34 × 10−2 | 19.3 |
| M230I | |||||||||
| 6 | 50 | 50 | 50 | 50 | 599 | 0 | 0 | <2.78 × 10−4 | <2.6 |
| 3 | 1 | 440 | 40 | 20 | 431 | 17 | 17 | 6.57 × 10−3 | 11.5 |
| 2 | 0.5 | 440 | 40 | 20 | 389 | 136 | 169 | 7.24 × 10−2 | 70.3 |
| 2 | 0.2 | 440 | 40 | 20 | 321 | 152 (2) | 174 | 9.03 × 10−2 | 67.4 |
| 2 | 0.1 | 440 | 40 | 20 | 181 | 115 (22) | 167 | 1.54 × 10−1 | 47.0 |
The number of mutant plaques that carried a G→A substitution in the target sequence and the total number of plaques were scored in 2 to 12 independent experiments.
The frequency of G→A transitions (fG→A) was calculated as follows: (number of G→A substitutions identified at Trp codons)/(number of target nucleotides) × (total number of plaques analyzed).
Relative frequencies (fG→A rel.) are the result of dividing the fG→A value for each mutant enzyme and dNTP concentration by the corresponding value obtained with the WT RT.
Values in parentheses represent the number of clones containing a deletion of 8 to 10 nt in addition to at least one G→A mutation.
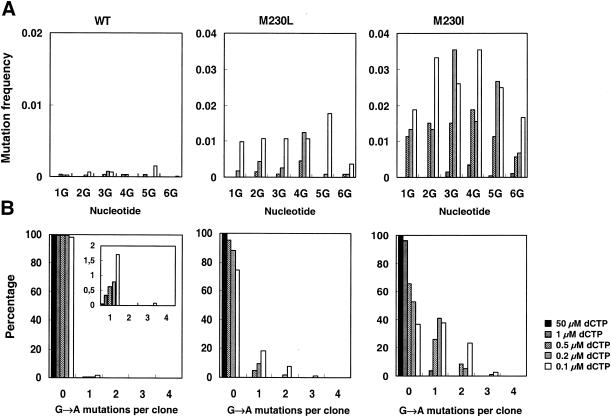
The genetic assay used allows the identification of specific G→A mutations occurring at six positions within the Trp codons present in the amplified insert. We have determined the fG→A values at each position (Fig. 1A). These values were consistently higher for the mutant RT with M230I than for the mutant RT with M230L and very low in the case of the WT RT. Mutations were evenly distributed, and none of the Gs analyzed appeared as a mutational hot spot. Thus, in those cases where the number of identified G→A mutations was higher than 30, the statistical analysis showed that the observed distribution was not significantly different from random (P > 0.2 by the two-tailed chi-square test).
FIG. 1.
Frequency distribution of G→A transitions at specific sites of the tryptophan trap. (A) Frequency distribution for reactions performed with WT RT and the mutant RTs with M230L and M230I. The six G sites indicated correspond to the second and third bases of the Trp codons as they appear in the RNA template (read from the 5′ to the 3′ end). (B) Number of G→A transitions per clone obtained with the three RTs. With decreasing dCTP concentrations, the number of highly mutated clones (containing two or three G→A transitions) increased, particularly in the case of the mutant RT with M230I. Mutated clones were rarely found with the WT RT and represented less than 2% of the total number of clones analyzed (data shown in the insert).
Plaques containing two or more G→A transitions per clone were rarely found in assays done with the WT RT but were relatively frequent with the mutant RT with M230I (Fig. 1B), indicating that this variant enzyme is strongly hypermutagenic in the presence of highly biased dNTP pools. The frequencies of plaques containing two or more G→A substitutions obtained with the mutant RT with M230I fit well to binomial distributions whose corresponding P values were the frequencies reported in Table 1 and were not higher than those expected by random chance (P > 0.5 by the two-tailed chi-square test).
Fidelity assessments based on primer extension assays.
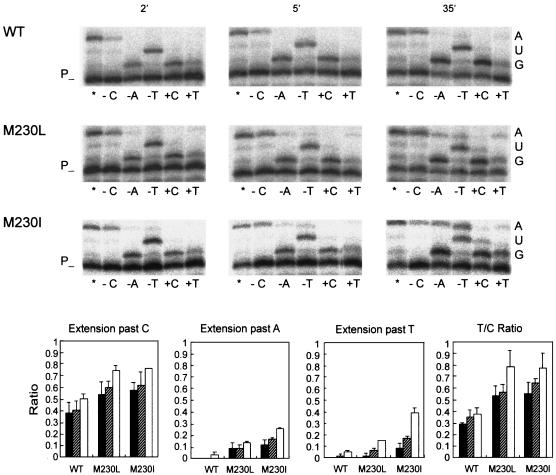
The higher efficiency of misincorporation of T opposite G during RNA-dependent DNA synthesis shown by the mutant RTs with M230I and M230L in comparison with the WT RT was further confirmed in a gel-based fidelity assay with a 25-mer oligonucleotide primer (2TRP [5′-TGTGGCTTGCCAATACTCTGTCCAC-3′]) and the 141-nt RNA template used in the genetic assay. Nucleotide incorporations at positions +1, +2, and +3 were determined in the presence of different combinations of dNTPs. As shown in Fig. 2, in the presence of dCTP, dATP, and dTTP (lane *), all three enzymes showed similar kinetics, with a final product of 28 nt. When dCTP, dATP, or dTTP was absent from the reaction mixture (lane −C, −A, or −T, respectively), the percentage of extension past the barrier site was highest for the mutant RT with M230I and lowest for the WT RT. With all of the enzymes, the largest percentages were observed in lane −C, which would involve misincorporation of A or T opposite G and further extension of mispaired template-primers. Misincorporation of T opposite G was about twofold more efficient with the mutant RTs with M230L and M230I than with the WT RT in comparison with the incorporation of C opposite G (compare lane +T with lane +C).
FIG. 2.
Extension of primer 2TRP by WT and mutant RTs in assays containing an RNA template. The nucleotides used in these assays were dATP-dCTP-dTTP (lane *), dATP-dTTP (lane −C), dCTP-dTTP (lane −A), dATP-dCTP (lane −T), dCTP alone (lane +C), and dTTP alone (lane +T). Template RNA nucleotides at positions +1, +2 and +3 are shown on the right and include the first 2 nt of the second Trp codon in the insert analyzed. The labeled template-primer at 30 nM was incubated with 20 to 40 nM RT at 37°C for up to 35 min in a total volume of 24 μl containing 50 mM HEPES (pH 7.0), 15 mM NaCl, 15 mM magnesium acetate, 130 mM KCH3COO, 1 mM DTT, and 5% polyethylene glycol 6000 (23). The concentration of all nucleotides in the extension reaction mixtures was 5 μM. Histograms below show the relative fidelities of the WT and mutant RTs. The amount of extension products was quantitated by phosphorimaging. Maximum extensions were determined for each enzyme and time point, as the sum of the intensities of bands at positions +1, +2 and +3, relative to the intensity of the band corresponding to the unextended primer, in reactions carried out with 3 nt (lane *). In the absence of dCTP (lane −C), dATP (lane −A), or dTTP (lane −T), misinsertion and mispair extension errors render bands at position +3 (i.e., in reactions shown in lanes −A and −T) or bands at positions +2 and +3 (lane −C). Reported ratios were obtained from the intensities of these bands referred to those obtained for unextended or shorter primers and then divided by the maximum intensity as obtained from results in lane *. The T/C ratio was determined as the relative amount of extended primer in the presence of dTTP (lane +T) compared with that in the presence of dCTP (lane +C) for each enzyme and time point. All results represent the average ± the standard deviation from three independent experiments.
Analysis of deletions arising during reverse transcription.
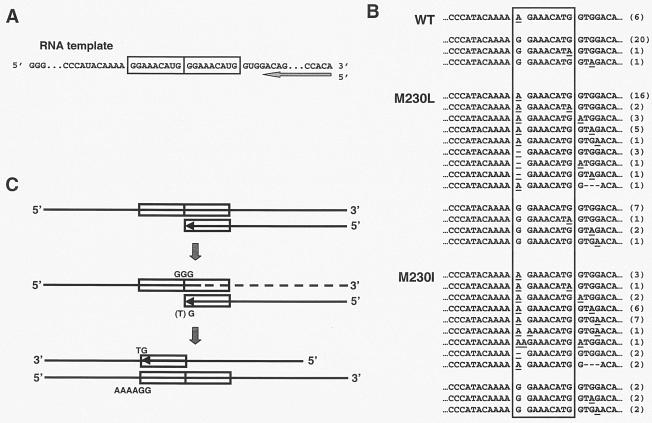
A number of colorless plaques rendering DNA fragments of around 380 bp upon PCR amplification with primers 1R and 1F were found to contain deletions of 8 to 10 nt that involved a tandem repeat sequence found in the reverse-transcribed 66-nt pol fragment (Fig. 3A). These deletions appeared more frequently in DNAs from plaques obtained in the assays carried out with the lowest concentrations of dCTP (Table 1), thereby indicating that they arise during reverse transcription. Since that type of error may not necessarily be associated with G→A mutations in these assays, we screened all plaques (blue and white) for tandem repeat deletions. The results are shown in Table 2. The deletion frequencies were at least 18-fold higher with biased [dCTP]/[dTTP] pools than with equimolar dNTP concentrations, for all three of the RTs tested. However, differences between the enzymes were not statistically significant.
FIG. 3.
Analysis of deletions generated in the genetic assay. (A) Nucleotide sequence of the RNA template containing the 9-nt tandem repeat sequence (boxed). (B) DNA sequences of deletion-containing clones identified with the genetic screening assay. Underlined nucleotides represent mutations found in each clone. The total number of occurrences of each sequence is shown on the right. (C) Proposed model for the mechanism leading to the observed deletions. The low concentration of dCTP in the biased dNTP assays leads to RT pausing and degradation of the RNA template by the RT's RNase H activity while copying the first repeat sequence. Strand transfer through annealing of the synthesized cDNA to the second repeat sequence in the RNA template would then allow fixation of the deletion. The relatively high proportion of G→A mutations at the strand transfer site found in clones obtained with the mutant RTs with M230L and M230I results from the higher frequency of misincorporation of T opposite G displayed by those enzymes.
TABLE 2.
Effects of RT mutations on the frequency of tandem repeat deletions
| Enzyme and [dCTP] (μM)a | No. of clones containing deletions of 8-10 nt/total no. of plaques analyzedb | fΔ[8-10nt]c |
|---|---|---|
| WT RT | ||
| 50 | 0/182 | <5.5 × 10−3 |
| 0.1 | 58/146 | 0.397 |
| M230L | ||
| 50 | 0/141 | <7.1 × 10−3 |
| 0.1 | 36/205 | 0.176 |
| M230I | ||
| 50 | 1/224 | 4.5 × 10−3 |
| 0.1 | 18/222 | 0.081 |
Nucleotide concentrations in the assays were either 50 μM each dNTP or 0.1 μM dCTP, 440 μM dTTP, 40 μM dATP, and 20 μM dGTP.
The number of clones harboring deletions of 8 to 10 nt in the target sequence and the total number of plaques were determined from two or three independent experiments. Blue and white plaques were both screened for deletions. The proportion of blue and white plaques analyzed was consistent with the data shown in Table 1. For example, in reactions carried out with the RT with M230I in the presence of a biased [dCTP]/[dTTP] ratio, we analyzed 80 blue plaques and 142 white plaques.
Deletion frequencies (fΔ[8-10nt]) were calculated as the ratio of the number of plaques containing a deletion of 8 to 10 nt to the total number of plaques analyzed. Deletions were identified by nucleotide sequencing. Frequencies obtained in different experiments were within a threefold range.
Mutations at the first nucleotide of the repeat sequence (i.e., G→A transitions, 1-nt deletions, and 1-nt insertions) were often detected after sequencing of mutant clones containing the deletion (Fig. 3B). The percentage of clones having these mutations was three- to fourfold higher in reactions carried out with primer grip mutant RTs than in reactions performed with the WT RT. Thus, mutations were found at the relevant site in 33 out of 44 clones with M230L and in 25 out of 31 clones with M230I. In contrast, only 6 out of 28 clones obtained with the WT RT were found to contain a G→A mutation at the first nucleotide of the repeat sequence.
Our results are consistent with a model in which deletion mutant RTs would result from strand transfer mediated by HIV-1 RT (Fig. 3C). RT pausing at the GGG stretch located between the two tandem repeat sequences could facilitate RNase H-mediated degradation of the template, thus favoring a subsequent transfer event involving annealing of the synthesized cDNA to the second repeat sequence within the same RNA template or to a different acceptor RNA molecule (11, 35, 36). Our data indicate that reduced reverse transcription rates and RT pausing result from the low dCTP concentrations available in reactions carried out with strongly biased dNTP pools.
Discussion.
Understanding the basis of HIV hypermutability is crucial for AIDS management. Mutational analyses of RTs have been extensively used to elucidate the role of different residues in DNA synthesis fidelity (25, 37). Methods to detect differences in nucleotide specificity among mutant RTs include in vitro assays based on the determination of kinetic constants for the incorporation of dNTPs on specific template-primers (gel-based assays) (6) and assays in which the single-stranded region of a gapped-duplex DNA, which includes a reporter gene (typically lacZα), is copied by the RT in the presence of dNTP substrates (genetic assays). Errors arising during this process are estimated after analyzing the phenotype of plaques generated by infection of host bacteria (2). Although this method is useful for the analysis of a wide range of sequence contexts and has been widely used to estimate the fidelity of RT variants, its adaptation to the study of RNA-dependent DNA synthesis fidelity is relatively complex (3, 10, 15, 16). In addition, these assays have to be done in the presence of high concentrations of dNTPs to ensure complete synthesis of the gapped sequence, and effects on fidelity resulting from alterations in nucleotide affinity may be difficult to detect.
In this study, we used an alternative assay that allows easy determination of G→A transition frequencies during RNA-dependent DNA synthesis and their sensitivity to biased dNTP pools, i.e., low [dCTP]/[dTTP] ratios. In agreement with previous reports, our results obtained with WT RT show an inverse relationship between fidelity and the magnitude of the [dCTP]/[dTTP] bias (20, 21). This correlation was also observed with the mutant RTs with M230L and M230I, which were much more mutagenic than the WT enzyme in the presence of strongly biased dNTP pools. For WT RT, G→A transition frequencies were up to 30-fold higher when the most unbalanced dNTP pools were used than when equimolar nucleotide concentrations were used, in contrast to the >150- and >550-fold increases observed with the mutant RTs with M230L and M230I, respectively. These data reveal that primer grip mutant RTs exhibit a strong tendency to form rG · dT mispairs.
These results are also consistent with our previous reports showing that substitution of Ile for Met-230 confers reduced fidelity of DNA synthesis in gel-based misincorporation fidelity assays (7). Interestingly, our results show that the mutant RT with M230L has similar properties but its influence on fidelity is relatively smaller. Crystallographic studies of HIV-1 RT have demonstrated that Met-230 interacts with the primer along the minor groove of DNA-DNA complexes (4). Residues lining the minor groove include Gly-262 and Trp-266, whose effect on fidelity has been previously shown (1). In addition, one residue that interacts with the primer at the RNase H domain of murine leukemia virus RT (Tyr-586) has been shown to increase the mutation rate in the vicinity of adenine-thymidine tracts (46). Taken together, all of these data indicate that the template-primer is an important structural determinant of fidelity in retroviral RTs.
Besides G→A mutations, other types of errors (i.e., short deletions) in the reporter sequence were also detected in the presence of the low [dCTP]/[dTTP] ratios. The mutational pattern observed in these reactions was consistent with a model in which deletions result from primer dissociation after copying of the first tandem repeat sequence, followed by annealing to the second repeat, probably on a different RNA template molecule (Fig. 3C). This event would be favored by RNA template degradation mediated by the RT's RNase H activity (11, 35, 36). Deletion frequencies in the presence of biased dNTP pools were similar but relatively high for all of the RTs tested, suggesting that RT pausing due to the low dCTP concentration was not influenced by primer grip mutations. On the other hand, the frequency of G→A mutations at the strand transfer site was higher for the mutant RTs with M230L and M230I than for the WT RT. These data indicate that, in the sequence context analyzed, misinsertion of T opposite G does not produce a significant increase in the deletion frequency, although the mutator phenotype of primer grip mutant RTs explains the occurrence of G→A mutations at the strand transfer site.
Estimates of dNTP concentrations in different cell types revealed a 3- to 15-fold excess of dTTP over dCTP (41). These ratios are rather small in comparison with those used in our assays, but there are no data on the variance or the distribution of dNTP concentrations for individual cells or subcellular compartments. In any case, intracellular dNTP pools are known to affect the rate and spectrum of retroviral mutations (17), as well as the frequency of tandem repeat deletions during reverse transcription (31), and are likely to have a large influence on retroviral mutation rates. Since M230L and M230I arise during treatment with RT inhibitors (9, 12, 18), we could envision a situation similar to that recently described for mutant RTs arising during treatment with zidovudine (i.e., M41L/T215Y, M41L/D67N/K70R/T215Y), which were shown to increase HIV mutation frequencies, particularly in the presence of nucleoside analogue inhibitors (19). Nonnucleoside RT inhibitors per se are not expected to modify viral mutation rates. However, primer grip mutations arising during treatment with those drugs may have a significant impact on HIV-1 population dynamics, a possibility that needs to be investigated further.
Acknowledgments
We thank E. Domingo, M. A. Martínez, and E. Viguera for valuable discussions and critical reading of the manuscript.
This work was supported by grant 01/0067-01 from the Fondo de Investigación Sanitaria (FIS)-FEDER (Spain) and by an institutional grant from the Fundación Ramón Areces. Support from the Red Temática Cooperativa de Investigación en SIDA (FIS G03/173) is also acknowledged.
REFERENCES
- 1.Beard, W. A., S. J. Stahl, H.-R. Kim, K. Bebenek, A. Kumar, M.-P. Strub, S. P. Becerra, T. A. Kunkel, and S. H. Wilson. 1994. Structure/function studies of human immunodeficiency virus type 1 reverse transcriptase—alanine scanning mutagenesis of an α-helix in the thumb subdomain. J. Biol. Chem. 269:28091-28097. [PubMed] [Google Scholar]
- 2.Bebenek, K., and T. A. Kunkel. 1995. Analyzing fidelity of DNA polymerases. Methods Enzymol. 262:217-232. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, J. C., K. Bebenek, and T. A. Kunkel. 1992. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc. Natl. Acad. Sci. USA 89:6919-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, Jr., A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structural and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 Å resolution. J. Mol. Biol. 284:1095-1111. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh, M., P. S. Jacques, D. W. Rodgers, M. Ottmann, J.-L. Darlix, and S. F. J. Le Grice. 1996. Alterations to the primer grip of p66 HIV-1 reverse transcriptase and their consequences for template-primer utilization. Biochemistry 35:8553-8562. [DOI] [PubMed] [Google Scholar]
- 6.Goodman, M. F., S. Creighton, L. B. Bloom, and J. Petruska. 1993. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28:83-126. [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez-Rivas, M., and L. Menéndez-Arias. 2001. A mutation in the primer grip region of HIV-1 reverse transcriptase that confers reduced fidelity of DNA synthesis. Nucleic Acids Res. 29:4963-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 9.Huang, W., N. T. Parkin, Y. S. Lie, T. Wrin, R. Haubrich, S. Deeks, N. Hellman, C. J. Petropoulos, and J. M. Whitcomb. 2000. A novel HIV-1 RT mutation (M230L) confers NNRTI resistance and dose-dependent stimulation of replication. Antivir. Ther. 5(Suppl. 3):24-25. [Google Scholar]
- 10.Hübner, A., M. Kruhoffer, F. Grosse, and G. Krauss. 1992. Fidelity of human immunodeficiency virus type 1 reverse transcriptase in copying natural RNA. J. Mol. Biol. 223:595-600. [DOI] [PubMed] [Google Scholar]
- 11.Hwang, C. K., E. S. Svarovskaia, and V. K. Pathak. 2001. Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. USA 98:12209-12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isaka, Y., S. Miki, S. Kawauchi, A. Suyama, H. Sugimoto, A. Adachi, M. Hayami, O. Yoshie, T. Fujiwara, and A. Sato. 2000. Isolation and characterization of simian immunodeficiency virus variants that are resistant to nonnucleoside reverse transcriptase inhibitors. Arch. Virol. 145:2481-2492. [DOI] [PubMed] [Google Scholar]
- 13.Isaka, Y., S. Miki, S. Kawauchi, A. Suyama, H. Sugimoto, A. Adachi, T. Miura, M. Hayami, O. Yoshie, T. Fujiwara, and A. Sato. 2001. A single amino acid change at Leu-188 in the reverse transcriptase of HIV-1 and SIV renders them sensitive to non-nucleoside reverse transcriptase inhibitors. Arch. Virol. 146:743-755. [DOI] [PubMed] [Google Scholar]
- 14.Jacques, P. S., B. M. Wöhrl, M. Ottmann, J.-L. Darlix, and S. F. J. Le Grice. 1994. Mutating the “primer grip” of p66 HIV-1 reverse transcriptase implicates tryptophan-229 in template-primer utilization. J. Biol. Chem. 269:26472-26478. [PubMed] [Google Scholar]
- 15.Ji, J., and L. A. Loeb. 1992. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry 31:954-958. [DOI] [PubMed] [Google Scholar]
- 16.Ji, J., and L. A. Loeb. 1994. Fidelity of HIV-1 reverse transcriptase copying an hypervariable region of the HIV-1 env gene. Virology 199:323-330. [DOI] [PubMed] [Google Scholar]
- 17.Julias, J. G., and V. K. Pathak. 1998. Deoxyribonucleoside triphosphate pool imbalances in vivo are associated with an increased retroviral mutation rate. J. Virol. 72:7941-7949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleim, J.-P., M. Winters, A. Dunkler, J.-R. Suarez, G. Riess, I. Winkler, J. Balzarini, D. Oette, T. C. Merigan, and The HBY 097/2001 Study Group. 1999. Antiviral activity of the human immunodeficiency virus type 1-specific nonnucleoside reverse transcriptase inhibitor HBY 097 alone and in combination with zidovudine in a phase II study. J. Infect. Dis. 179:709-713. [DOI] [PubMed] [Google Scholar]
- 19.Mansky, L. M., D. K. Pearl, and L. C. Gajary. 2002. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J. Virol. 76:9253-9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez, M. A., M. Sala, J.-P. Vartanian, and S. Wain-Hobson. 1995. Reverse transcriptase and substrate dependence of the RNA hypermutagenesis reaction. Nucleic Acids Res. 23:2573-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez, M. A., J.-P. Vartanian, and S. Wain-Hobson. 1994. Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc. Natl. Acad. Sci. USA 91:11787-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martín-Hernández, A. M., E. Domingo, and L. Menéndez-Arias. 1996. Human immunodeficiency virus type 1 reverse transcriptase: role of Tyr115 in deoxynucleotide binding and misinsertion fidelity of DNA synthesis. EMBO J. 15:4434-4442. [PMC free article] [PubMed] [Google Scholar]
- 23.Martín-Hernández, A. M., M. Gutiérrez-Rivas, E. Domingo, and L. Menéndez-Arias. 1997. Mispair extension fidelity of human immunodeficiency virus type 1 reverse transcriptases with amino acid substitutions affecting Tyr115. Nucleic Acids Res. 25:1383-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumi, S., P. Kosalaraksa, H. Tsang, M. F. Kavlick, S. Harada, and H. Mitsuya. 2003. Pathways for the emergence of multi-dideoxynucleoside-resistant HIV-1 variants. AIDS 17:1127-1137. [DOI] [PubMed] [Google Scholar]
- 25.Menéndez-Arias, L. 2002. Molecular basis of fidelity of DNA synthesis and nucleotide specificity of retroviral reverse transcriptases. Prog. Nucleic Acid Res. Mol. Biol. 71:91-147. [DOI] [PubMed] [Google Scholar]
- 26.Menéndez-Arias, L., I. T. Weber, and S. Oroszlan. 1995. Mutational analysis of the substrate binding pocket of murine leukemia virus protease and comparison with human immunodeficiency virus proteases. J. Biol. Chem. 270:29162-29168. [DOI] [PubMed] [Google Scholar]
- 27.Olivares, I., V. Sánchez-Merino, M. A. Martínez, E. Domingo, C. López-Galíndez, and L. Menéndez-Arias. 1999. Second-site reversion of a human immunodeficiency virus type 1 reverse transcriptase mutant that restores enzyme function and replication capacity. J. Virol. 73:6293-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olmstead, R. A., D. E. Slade, L. A. Kopta, S. M. Poppe, T. J. Poel, S. W. Newport, K. B. Rank, C. Biles, R. A. Morge, T. J. Dueweke, Y. Yagi, D. L. Romero, R. C. Thomas, S. K. Sharma, and W. G. Tarpley. 1996. (Alkylamino)piperidine bis(heteroaryl)piperizine analogs are potent, broad-spectrum nonnucleoside reverse transcriptase inhibitors of drug-resistant isolates of human immunodeficiency virus type 1 (HIV-1) and select for drug-resistant variants of HIV-1IIIB with reduced replication phenotypes. J. Virol. 70:3698-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palaniappan, C., M. Wisniewski, P. S. Jacques, S. F. J. Le Grice, P. J. Fay, and R. A. Bambara. 1997. Mutations within the primer grip region of HIV-1 reverse transcriptase result in loss of RNase H function. J. Biol. Chem. 272:11157-111564. [DOI] [PubMed] [Google Scholar]
- 30.Pathak, V., and H. M. Temin. 1990. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc. Natl. Acad. Sci. USA 87:6019-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer, J. K., R. S. Topping, N.-H. Shin, and A. Telesnitsky. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J. Virol. 73:8441-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Powell, M. D., M. Ghosh, P. S. Jacques, K. J. Howard, S. F. J. Le Grice, and J. G. Levin. 1997. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J. Biol. Chem. 272:13262-13269. [DOI] [PubMed] [Google Scholar]
- 33.Quiñones-Mateu, M. E., V. Soriano, E. Domingo, and L. Menéndez-Arias. 1997. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology 236:364-373. [DOI] [PubMed] [Google Scholar]
- 34.Rhee, S.-Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, C. M., J. S. Smith, and M. J. Roth. 1999. RNase H requirements for the second strand transfer reaction of human immunodeficiency virus type 1 reverse transcription. J. Virol. 73:6573-6581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suo, Z., and K. A. Johnson. 1997. Effect of RNA secondary structure on RNA cleavage catalyzed by HIV-1 reverse transcriptase. Biochemistry 36:12468-12476. [DOI] [PubMed] [Google Scholar]
- 37.Svaroskaia, E. S., S. R. Cheslock, W.-H. Zhang, W.-S. Hu, and V. K. Pathak. 2003. Retroviral mutation rates and reverse transcriptase fidelity. Front. Biosci. 8:D117-D134. [DOI] [PubMed] [Google Scholar]
- 38.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. W. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 39.Vartanian, J.-P., A. Meyerhans, B. Åsjö, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vartanian, J.-P., A. Meyerhans, M. Sala, and S. Wain-Hobson. 1994. G→A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc. Natl. Acad. Sci. USA 91:3092-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vartanian, J.-P., U. Plikat, M. Henry, R. Mahieux, L. Guillemot, A. Meyerhans, and S. Wain-Hobson. 1997. HIV genetic variation is directed and restricted by DNA precursor availability. J. Mol. Biol. 270:139-151. [DOI] [PubMed] [Google Scholar]
- 42.Wain-Hobson, S., P. Sonigo, M. Guyader, A. Gazit, and M. Henry. 1995. Erratic G→A hypermutation within a complete caprine arthritis-encephalitis virus (CAEV) provirus. Virology 209:297-303. [DOI] [PubMed] [Google Scholar]
- 43.Wisniewski, M., C. Palaniappan, Z. Fu, S. F. J. Le Grice, P. Fay, and R. A. Bambara. 1999. Mutations in the primer grip region of HIV reverse transcriptase can increase replication fidelity. J. Biol. Chem. 274:28175-28184. [DOI] [PubMed] [Google Scholar]
- 44.Wöhrl, B. M., R. Krebs, S. H. Thrall, S. F. J. Le Grice, A. J. Scheidig, and R. S. Goody. 1997. Kinetic analysis of four HIV-1 reverse transcriptase enzymes mutated in the primer grip region of p66—implications for DNA synthesis and dimerization. J. Biol. Chem. 272:17581-17587. [DOI] [PubMed] [Google Scholar]
- 45.Yu, Q., M. Ottmann, C. Pechoux, S. Le Grice, and J.-L. Darlix. 1998. Mutations in the primer grip of human immunodeficiency virus type 1 reverse transcriptase impair proviral DNA synthesis and virion maturation. J. Virol. 72:7676-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, W.-H., E. S. Svarovskaia, R. Barr, and V. K. Pathak. 2002. Y586F mutation in murine leukemia virus reverse transcriptase decreases fidelity of DNA synthesis in regions associated with adenine-thymine tracts. Proc. Natl. Acad. Sci. USA 99:10090-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]