Abstract
The Gag proteins of a number of different retroviruses contain late or L domains that promote the release of virions from the plasma membrane. Three types of L domains have been identified to date: Pro-Thr-Ala-Pro (PTAP), Pro-Pro-X-Tyr, and Tyr-Pro-Asp-Leu. It has previously been demonstrated that overexpression of the N-terminal, E2-like domain of the endosomal sorting factor TSG101 (TSG-5′) inhibits human immunodeficiency virus type 1 (HIV-1) release but does not affect the release of the PPPY-containing retrovirus murine leukemia virus (MLV), whereas overexpression of the C-terminal portion of TSG101 (TSG-3′) potently disrupts both HIV-1 and MLV budding. In addition, it has been reported that, while the release of a number of retroviruses is disrupted by proteasome inhibitors, equine infectious anemia virus (EIAV) budding is not affected by these agents. In this study, we tested the ability of TSG-5′, TSG-3′, and full-length TSG101 (TSG-F) overexpression, a dominant negative form of the AAA ATPase Vps4, and proteasome inhibitors to disrupt the budding of EIAV particles bearing each of the three types of L domain. The results indicate that (i) inhibition by TSG-5′ correlates with dependence on PTAP; (ii) the release of wild-type EIAV (EIAV/WT) is insensitive to TSG-3′, whereas this C-terminal TSG101 fragment potently impairs the budding of EIAV when it is rendered PTAP or PPPY dependent; (iii) budding of all EIAV clones is blocked by dominant negative Vps4; and (iv) EIAV/WT release is not impaired by proteasome inhibitors, while EIAV/PTAP and EIAV/PPPY release is strongly disrupted by these compounds. These findings highlight intriguing similarities and differences in host factor utilization by retroviral L domains and suggest that the insensitivity of EIAV to proteasome inhibitors is conferred by the L domain itself and not by determinants in Gag outside the L domain.
Retroviral Gag precursor proteins are necessary and sufficient for the formation of virus-like particles (VLPs) (70, 82). During or shortly after release from the cell, the Gag precursor is proteolytically cleaved by the viral protease to generate the mature Gag proteins, which generally include matrix (MA), capsid, and nucleocapsid, and a variety of spacer peptides and domains unique to individual retrovirus genera (14, 70). Distinct domains within the Gag polyprotein provide specific functions that are essential for virus assembly and release. The membrane-binding (M) domain, located in MA, is required for Gag binding to the plasma membrane; the interaction (I) domain, which maps to capsid and basic residues within nucleocapsid, promotes Gag-Gag interaction; and the late (L) domain, located at a variety of positions in the retroviral Gag precursor, stimulates particle release from the plasma membrane.
L domains have been described for the majority of retroviruses and for the rhabdoviruses and filoviruses (for reviews, see references 15 and 59). Genetic disruption of these motifs results in severe defects in virion release. It is well established that a PTAP motif near the N terminus of p6 acts as the L domain of human immunodeficiency virus type 1 (HIV-1) (19, 26). PPPY motifs within the p2b domain of Rous sarcoma virus (RSV) (81), p12 of murine leukemia virus (MLV) (85), the MA of bovine leukosis virus (79) and human T-cell leukemia virus type 1 (33), pp16 of Mason-Pfizer monkey virus (M-PMV) (83), and the MA of the rhabdovirus vesicular stomatitis virus (10, 23, 27) are critical for the release of these viruses. Both PTAP and PPPY motifs display L-domain activity in the context of Ebola virus VP40 (22, 38) and M-PMV (20). The L domain of the nonprimate lentivirus equine infectious anemia virus (EIAV) maps to a YPDL motif in the p9 domain of Gag (60).
It is now believed that L domains act in concert with components of the cellular ubiquitination and endosomal sorting machinery to promote virus particle budding and release (for reviews, see references 7, 15, 59, and 77). The connection between ubiquitination, endosomal sorting, and virus release derives support from a number of lines of evidence. (i) Several L-domain-containing retroviral Gag proteins, including p6 of HIV-1 and simian immunodeficiency virus, p9 of EIAV, and p12 of MLV, are ubiquitinated (46-48). (ii) The PPPY L-domain motifs of RSV, M-PMV, vesicular stomatitis virus, and Ebola virus reportedly interact with E3 ubiquitin ligases related to Nedd4 (21, 31, 84). (iii) EIAV Gag, in a YPDL-dependent fashion, interacts with the AP-50 subunit of AP-2, a protein complex involved in endocytosis (61). (iv) TSG101, which contains at its N terminus a domain highly related to E2 ubiquitin (Ub)-conjugating enzymes (32, 37), interacts with the PTAP L-domain motif of HIV-1 (11, 17, 41, 74), HIV-2 (44), and Ebola virus (38, 41, 72) and functions in the release of these viruses. Finally, (v) proteasome inhibitors disrupt the release of HIV-1, simian immunodeficiency virus, MLV, RSV, and M-PMV (48, 51, 52, 64). Proteasome inhibitors induce the accumulation of polyubiquitinated proteins, thereby depleting intracellular pools of free Ub (43, 49) and inhibiting Gag ubiquitination (48, 49, 64). However, proteasome inhibitors affect a variety of pathways in the cell, including the sorting and lysosomal degradation of certain cell surface receptors, such as the epidermal growth factor receptor (EGF-R) (35, 39). It is therefore not clear whether the activity of proteasome inhibitors is due to a direct impact on Gag ubiquitination or whether it is mediated indirectly through the effect of these inhibitors on the ubiquitination, trafficking, or degradation of cellular protein(s). Interestingly, although EIAV p9 is ubiquitinated, the release of this lentivirus is insensitive to proteasome inhibitors (48, 51). The intriguing possibility has been suggested that a Ub-like motif in p9 might confer upon EIAV insensitivity to proteasome inhibitors (51).
The endosomal sorting pathway functions in the downregulation of cell surface receptors (e.g., EGF-R) and in the delivery of proteolytic enzymes (e.g., carboxypeptidease S and cathepsin D) to the vacuole in yeast and the lysosome in mammalian cells (for reviews, see references 29, 34, 54, and 66). A key aspect of the endosomal sorting pathway is the delivery of cargo proteins to vesicles that bud into late endosomes to form multivesicular bodies (MVBs). The contents of these vesicles are ultimately delivered to the lysosome or vacuole, where they are degraded or serve a proteolytic function (40, 45). Monoubiquitination of cargo proteins often plays an essential role in their recognition by the endosomal sorting machinery (13, 25, 55). It has long been recognized that the so-called class E vacuolar protein sorting (Vps) proteins function in endosomal sorting in yeast (63), and homologues of these factors exist in mammalian cells (for a review, see reference 29). Disruption of the genes encoding the class E Vps proteins leads to the formation of an enlarged, aberrant endosomal compartment referred to as the class E compartment (63) (for a review, see reference 54). The topological equivalence of vesicle budding into the MVB and enveloped virus budding from the plasma membrane is noteworthy (52).
A number of class E Vps proteins interact to form three multiprotein complexes that appear to play a sequential role in the sorting of proteins into the MVB pathway (9). These complexes have been termed ESCRT-I, II, and III (for endosomal sorting complex required for transport) (2, 3, 28). ESCRT-I is composed of TSG101 (Vps23 in yeast) and Vps28; in yeast it contains a third component, Vps37, for which no mammalian homologue has thus far been identified (6). The N-terminal, E2-like domain of TSG101 interacts directly with Ub on cargo proteins and also contains the binding determinant for the PTAP motif of HIV-1 p6 (4, 17, 28, 56, 57). It has been suggested (59) and recently demonstrated (58) that the physiological ligand for the PTAP-binding surface of TSG101 is Hrs (Vps27 in yeast), which contains a PSAP motif and plays a key role in the sorting of ubiquitinated proteins into the MVB pathway (62, 65). The C terminus of TSG101 interacts with Vps28, and, in yeast, with Vps37 (6, 42). TSG101 therefore serves to link cargo proteins, or HIV-1 Gag, to ESCRT-I. Yeast ESCRT-II consists of Vps22, 25, and 36, and the ESCRT-III complex is composed of two subcomplexes, Vps2/Vps24 and Snf7/Vps20 (2, 3). The mammalian cell homologues of the ESCRT-II proteins are Eap25, Eap30, and Eap45, and the so-called Chmp proteins constitute the ESCRT-III homologues (29). The catalytic activity of the AAA ATPase Vps4 is apparently essential for the disassociation of ESCRT components from the endosomal membrane and is required for the inward budding of vesicles into the MVB (29). Catalytically inactive, dominant negative forms of Vps4 potently inhibit not only HIV-1 budding but also the release of MLV (17) and an HIV-1 chimera in which p6 is replaced by EIAV p9 (42).
It is believed that cargo proteins are sorted into the MVB pathway by interacting sequentially with the ESCRT-I, II, and III complexes before being packaged into vesicles that bud into the MVB. Proteins may also be able to access this pathway downstream of ESCRT-II and III by being recruited directly to ESCRT-III. The class E Vps protein AIP1/Alix (PalA in Aspergillus nidulans and Vps31, Bro1, or Npi3 in yeast) (1, 76) interacts with the ESCRT-III component Vps32/Snf7 (53), also known as Chmp4 in mammalian cells. Interestingly, AIP1/Alix recognizes proteins bearing a YPXL sequence (75). Because EIAV L-domain activity is provided by a YPDL motif in p9 (60), it has been suggested that AIP1/Alix could recruit EIAV Gag to ESCRT-III (75).
It was recently demonstrated that overexpressing the N-terminal, E2-like domain of TSG101 (TSG-5′) inhibits HIV-1 budding. A mutation that disrupts the ability of TSG-5′ to bind Gag suppresses its ability to inhibit HIV-1 release (11, 18), suggesting that TSG-5′ blocks budding by directly binding the PTAP L domain of p6. We also observed that overexpression of full-length TSG101 (TSG-F) inhibits HIV-1 budding (18). Confocal microscopy demonstrated that TSG101 overexpression leads to the formation of swollen, aberrant endosomes, suggesting that TSG-F disrupts budding via perturbation of the endosomal sorting machinery. Finally, we observed that overexpression of the C-terminal domain of TSG101 (TSG-3′) potently inhibits both HIV-1 and MLV budding and induces the formation of large intracellular vesicles that are morphologically distinct from the swollen endosomes induced by TSG-F or a dominant negative form of Vps4, Vps4EQ (18).
The aim of the present study was to gain insight into the cellular machinery used for retrovirus budding by examining the impact of a wide variety of budding inhibitors on particle release mediated by the three known classes of L domains. To this end, we examined the impact of the proteasome inhibitors TSG-5′, TSG-F, TSG-3′, and Vps4EQ on the release of EIAV particles bearing either the native YPDL L-domain sequence or replaced with the heterologous PTAP and PPPY motifs (EIAV/PTAP and EIAV/PPPY, respectively). We report that inhibition mediated by TSG-5′ is PTAP dependent, confirming earlier work suggesting that this N-terminal TSG101 fragment inhibits budding by directly blocking PTAP activity. Intriguingly, TSG-F and TSG-3′ suppress the release of both EIAV/PTAP and EIAV/PPPY but are largely ineffective against wild-type EIAV (EIAV/WT). As reported previously (48, 51), EIAV/WT is resistant to proteasome inhibitors, but interestingly, both EIAV/PTAP and EIAV/PPPY are sensitive. The release of VLPs produced by all three classes of EIAV Gags analyzed here is blocked by Vps4EQ. These results support the hypothesis that retroviral Gags bearing PTAP, PPPY, and YPDL L domains all interact with class E Vps machinery to promote VLP release but suggest that WT EIAV enters this pathway downstream from the block imposed by proteasome inhibitors and by overexpression of TSG-F and TSG-3′.
MATERIALS AND METHODS
Plasmids, DNA cloning, and mutagenesis.
The TSG-5′ expression vector pcGNM2/TSG-5′ (11, 69) and the TSG-3′ expression vector pcGNM2/TSG-3′ (18, 69) were kindly provided by Z. Sun (Stanford University). The full-length TSG101 expression vector (pcGNM2/TSG-F) was constructed as described previously (18). All TSG101-derived expression vectors used in this study encode proteins with N-terminal influenza hemagglutinin (HA) epitope tags. Plasmid eGFP-hVPS4(EQ), a catalytically inactive mutant of hVps4 bearing an E→Q substitution in position 223, fused to the green fluorescent protein (eGFP) (5), was kindly provided by P. Woodman (University of Manchester, Manchester, United Kingdom). Construction of the EIAV Gag expression plasmid pPRE/Gag was described previously (51). The pPRE/Gag derivatives expressing chimeric EIAV Gag with the parental YPDL L-domain replaced by heterologous L-domains were generated using a two-step PCR strategy (8). The critical amino acids of each L domain (underlined) plus adjacent residues were exchanged; the QNLYPDLS sequence of EIAV Gag was changed to RPEPTAPP in the EIAV-PTAP construct, and QNLYPDLSEI was changed to ASAPPPPYVG in EIAV-PPPY. The ΔYPDL mutant was constructed by deleting residues YPDL and replacing them with SRSA.
Cell culture, transfections, radioimmunoprecipitation, and Western blotting.
293T cells were maintained in culture and were transfected either with the calcium phosphate method as previously described (16) or with Lipofectamin 2000 (Invitrogen). Transfections were performed in 6-well dishes plated at 4 × 105 cells/well. The methods used for metabolic labeling of transfected cells, preparation of cell and viral lysates, immunoprecipitation analysis, and Western blotting have been described previously (16, 30). EIAV Gag proteins were immunoprecipitated with polyclonal anti-EIAV horse antiserum (36). HIV-1 proteins were immunoprecipitated with anti-HIV immunoglobulin (HIV-Ig) (obtained from the National Institutes of Health AIDS Research and Reference Reagent Program). EIAV Gag Western blotting was performed with anti-EIAV horse antiserum and secondary anti-horse immunoglobulin (36). Detection of TSG-F, TSG-5′, and TSG-3′ was performed by Western blotting using anti-HA mouse monoclonal antibody (Sigma, St. Louis, Mo.). Anti-eGFP antibody was also obtained from Sigma.
Protease inhibitor treatment.
Treatment of virus-producing cells with the peptide aldehyde n-carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (zLLL or MG132) (Calbiochem) has previously been described (51, 64). Briefly, 12-h posttransfection, cells were treated for 90 min with either dimethyl sulfoxide (DMSO) or 10 μM MG132 (Calbiochem) dissolved in DMSO. The treated cells were metabolically labeled for 2.5 h in either DMSO or 10 μM MG132 dissolved in DMSO.
RESULTS
The virus budding defect induced by deletion of the YPDL motif in EIAV p9 is reversed by introducing PPPY or PTAP motifs.
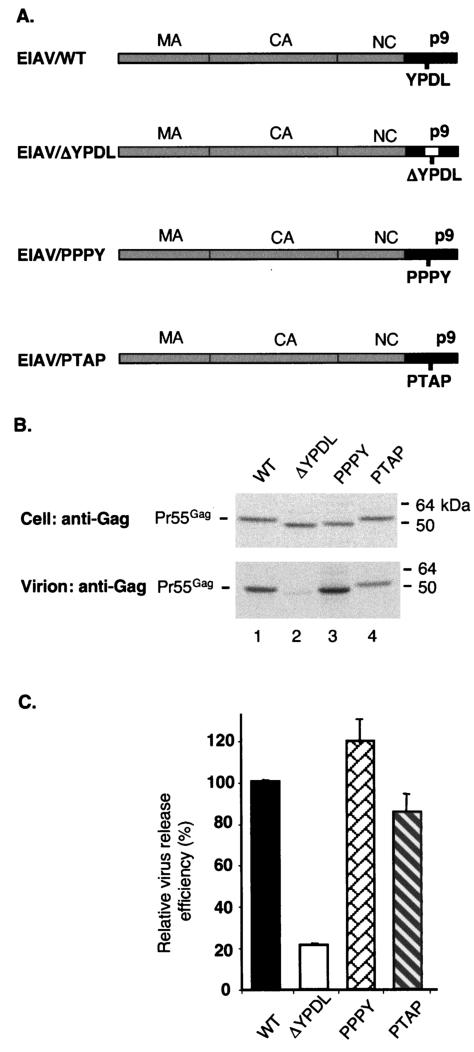
It was previously demonstrated in the context of a full-length EIAV molecular clone that PTAP and PPPY motifs could functionally replace the YPDL L domain present in the p9 domain of Gag (36). To confirm this observation in the context of an EIAV Gag expression vector, we introduced the YPDL deletion (ΔYPDL) and the PTAP- and PPPY-containing substitutions into the Gag expression vector PRE/Gag (51) (Fig. 1A and Materials and Methods). The resulting WT, ΔYPDL, PPPY, and PTAP Gag expression vectors were tested by radioimmunoprecipitation analysis for their ability to direct VLP production in transfected 293T cells (Fig. 1B). As previously demonstrated (36, 60), deletion of the YPDL motif markedly impaired VLP production, resulting in a fivefold reduction in the release of VLP-associated Gag (Fig. 1B, lane 2, and C). As expected, replacement of the YPDL motif with sequences encoding PPPY or PTAP restored VLP production to near-WT levels (Fig. 1B, lanes 3 and 4, and C). These data indicate that in the context of Gag-only expression, PPPY and PTAP can functionally replace the L-domain activity of the YPDL motif.
FIG. 1.
The virus budding defect induced by deletion of the YPDL motif in EIAV p9 is reversed by introducing the PPPY or PTAP motif. (A) The schematic organization of the WT and mutant EIAV Gag chimeras used in this study. The location of the L domain of EIAV (YPDL) is shown. The open box within p9 represents the YPDL deletion (ΔYPDL). The EIAV/WT p9 sequence YPDL is replaced by SRSA (EIAV/ΔYPDL), ASAPPPPYVG (EAIV/PPPY), or RPEPTAPP (EIAV/PTAP). CA, capsid protein; NC, nucleocapsid protein. (B) 293T cells were transfected with vectors expressing EIAV/WT Gag, the ΔYPDL deletion mutant, or the PPPY and PTAP chimeras. Transfected cells were metabolically labeled overnight with [35S]Met-[35S]Cys; cell and viral lysates were radioimmunoprecipitated with horse anti-EIAV antiserum (Materials and Methods). The position of the EIAV Gag precursor (Pr55Gag) is indicated on the left, and the position of molecular mass markers is shown on the right. (C) Virus release efficiency, determined by phosphorimager analysis and calculated as the amount of VLP-associated Gag divided by the total (cell plus VLP) Gag. The data are averages from at least three independent experiments plus or minus the standard error.
TSG-5′ potently inhibits EIAV/PTAP but not EIAV/WT or EIAV/PPPY particle release.
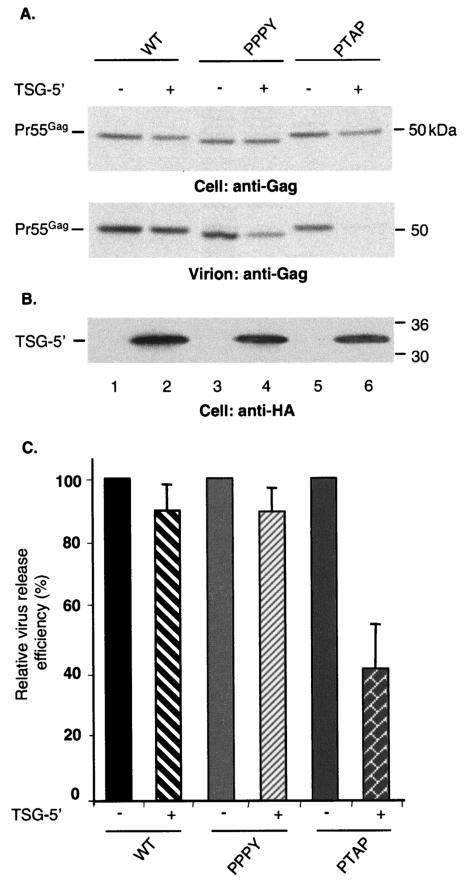
It was previously reported that overexpression of TSG-5′ potently inhibits HIV-1, but not MLV, budding (11). It was subsequently observed (18) that a mutation in TSG-5′ that blocks the interaction between Gag and TSG101 (the TYN− mutation) disrupts the inhibitory activity of TSG-5′. These results suggest that TSG-5′ suppresses virus release by binding and inactivating the PTAP L domain. To determine the effect of TSG-5′ overexpression on the production of EIAV/WT, EIAV/PTAP, and EIAV/PPPY particles, we cotransfected the plasmids encoding these Gag derivatives with the TSG-5′ expression vector at a 1:1 DNA ratio. A marked defect in virus release was observed for EIAV/PTAP (Fig. 2A, lanes 5 and 6, and C), whereas the production of EIAV/WT particles was unaffected (Fig. 2A, lanes 1 and 2, and C). Even at 1:2 and 1:4 ratios of EIAV/WT DNA to TSG-5′ expression vector, no significant inhibition of VLP release was observed (data not shown). Only a slight impairment was detected with EIAV/PPPY (Fig. 2A, lanes 3 and 4, and C). Similar levels of TSG-5′ were expressed in all transfected cultures (Fig. 2B). These results demonstrate that the inhibition of virus budding by TSG-5′ is largely PTAP dependent and support the hypothesis (18) that TSG-5′ inhibition is a result of a direct binding between this N-terminal TSG101 fragment and the PTAP motif. To confirm this hypothesis, we tested the ability of EIAV/PTAP versus EIAV/WT VLPs to incorporate TSG-5′. Consistent with the model, we observed that TSG-5′ is incorporated into EIAV/PTAP, but not EIAV/WT, VLPs (Fig. 3).
FIG. 2.
TSG-5′ inhibits EIAV/PTAP, but not EIAV/WT or EIAV/PPPY, VLP release. 293T cells were transfected with vectors expressing EIAV/WT, EIAV/PPPY, or EIAV/PTAP Gag alone (lanes 1, 3, and 5) or were cotransfected with Gag vectors and the TSG-5′ expression vector at a 1:1 DNA ratio (lanes 2, 4, and 6). (A) Cell and viral lysates were radioimmunoprecipitated with horse anti-EIAV antiserum. (B) The cell lysates were immunoblotted with anti-HA antiserum to detect TSG-5′ expression. For panels A and B, the positions of the EIAV Gag precursor (Pr55Gag) and TSG-5′ are indicated on the left, and the positions of molecular mass markers are shown on the right. (C) Virus release efficiency, calculated as described in the legend to Fig. 1. The data are averages from at least three independent experiments plus or minus the standard error.
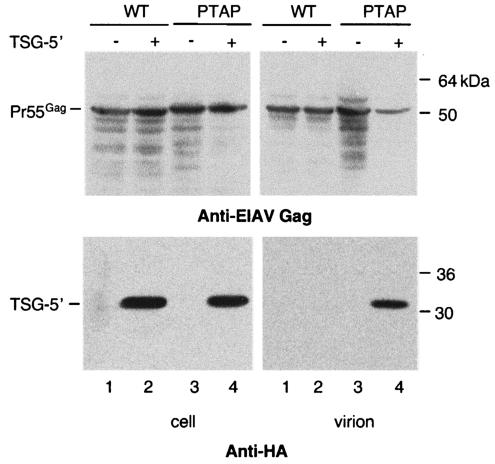
FIG. 3.
EIAV/PTAP, but not EIAV/WT, VLPs incorporate TSG-5′. 293T cells were transfected with vectors expressing EIAV/WT or EIAV/PTAP Gag alone (lanes 1 and 3) or were cotransfected with the Gag plasmids and the TSG-5′ expression vector at a 1:1 DNA ratio (lanes 2 and 4). Cell and viral lysates were immunoblotted with horse anti-EIAV antiserum (top panel) or anti-HA antiserum (lower panel). The positions of Pr55Gag and TSG-5′ are indicated at the left; the positions of molecular mass markers are shown on the right.
EIAV/WT and EIAV/PTAP display significant differences in their sensitivities to TSG101-based inhibitors of the endosomal sorting machinery.
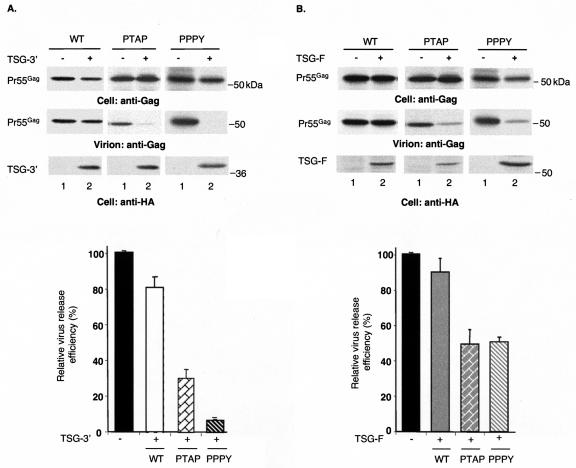
Previous data (11, 18) and the results presented above indicate that TSG-5′ suppresses virus release specifically by binding the PTAP motif and blocking L-domain function. In contrast, overexpression of TSG-3′ or TSG-F inhibits particle budding by disrupting the host MVB machinery that promotes the pinching off of virions from the plasma membrane (18). TSG-3′ is a particularly potent inhibitor of virus budding and is not specific for HIV-1; both HIV-1 and MLV release are reduced 5- to 10-fold by this C-terminal TSG101 fragment (18). The EIAV Gag mutants in the present study allow us to examine the host machinery required for the budding activity mediated by each of the three known L domains. To determine the manner in which EIAV/WT uses the endosomal sorting machinery for its release, we examined the effect of TSG-3′ and TSG-F overexpression on the budding of EIAV/WT. Surprisingly, given its potent impact on HIV-1 and MLV budding and its dramatic effect on the morphology of intracellular membranes (18), TSG-3′ had little or no effect on EIAV/WT release, even at a 1:2 or 1:4 ratio of EIAV/WT DNA to TSG-3′ expression vector (Fig. 4; data not shown). At a 1:1 ratio of EIAV/WT to TSG-F-expressing DNA, TSG-F also had no significant effect on VLP production (Fig. 4). At a 1:2 ratio, TSG-F caused a relatively modest but statistically significant reduction (∼40%) in virus release (data not shown). In striking contrast, both TSG-3′ and TSG-F strongly inhibited the release of EIAV/PTAP and EIAV/PPPY virions at a 1:1 DNA ratio (Fig. 4). These results indicate that the YPDL L-domain motif present in EIAV/WT confers a high degree of resistance to TSG101-based inhibitors of the endosomal sorting pathway.
FIG. 4.
EIAV/PTAP and EIAV/PPPY, but not EIAV/WT, VLP release is inhibited by TSG-F and TSG-3′. 293T cells were transfected with vectors expressing EIAV/WT, EIAV/PPPY, or EIAV/PTAP Gag alone (lanes 1) or were cotransfected with Gag plasmids and vectors encoding TSG-3′ (A) or TSG-F (B) at 1:1 DNA ratios (lanes 2). Cell and viral lysates were radioimmunoprecipitated with horse anti-EIAV antiserum, and the cell lysates were subjected to immunoblotting with anti-HA antiserum for the detection of TSG-3′ and TSG-F. Virus release efficiency was calculated as described in the legend to Fig. 1. The data are averages from at least three independent experiments plus or minus the standard error. Molecular mass markers are indicated on the right.
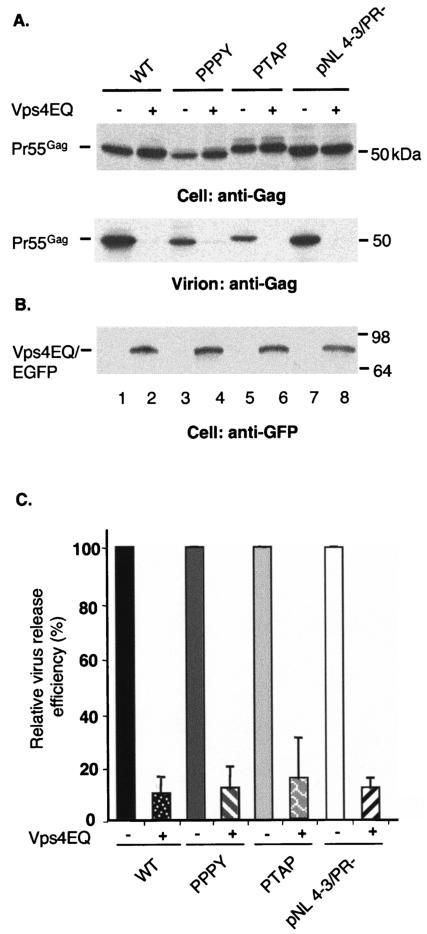
EIAV/WT release is suppressed by Vps4EQ, a dominant negative inhibitor of the class E Vps machinery.
As mentioned above, we observed that TSG101-based inhibitors had little or no effect on WT EIAV particle release, whereas EIAV/PTAP and EIAV/PPPY were strongly impaired by TSG-3′ and TSG-F overexpression. To determine whether WT EIAV interacts with other elements of the endosomal sorting pathway downstream of ESCRT-I, we determined the effect of a dominant negative mutant of Vps4, Vps4EQ (5), on particle release. We cotransfected EIAV/WT Gag and Vps4EQ expression vectors at a 1:1 DNA ratio and analyzed the efficiency of VLP production (Fig. 5). As a positive control, we assessed the impact of Vps4EQ on HIV-1 virion release. As previously reported (17), Vps4EQ strongly inhibited HIV-1 release. Particle production mediated by EIAV/PPPY and EIAV/PTAP was also markedly reduced. Interestingly, EIAV/WT particle production was also potently inhibited; a 10-fold reduction in the efficiency of VLP-associated Gag release into the medium was observed (Fig. 5). These data indicate that EIAV/WT Gag budding is sensitive to perturbation of the class E Vps machinery.
FIG. 5.
EIAV/WT, EIAV/PPPY, and EIAV/PTAP VLP release is suppressed by Vps4EQ. 293T cells were transfected with vectors expressing EIAV/WT, EIAV/PPPY, EIAV/PTAP Gag, or pNL4-3/PR− alone (lanes 1, 3, 5, and 7) or were cotransfected with these DNAs and a vector encoding an EGFP-Vps4EQ fusion protein (lanes 2, 4, 6, and 8). Cell and viral lysates were radioimmunoprecipitated with horse anti-EIAV antiserum (lanes 1 through 6) or anti-HIV Ig (lanes 7 and 8) (A) or were immunoblotted with an anti-eGFP antibody to detect eGFP-Vps4EQ (B). Molecular mass markers are indicated on the right. Virus release efficiency was calculated as described in the legend to Fig. 1 C. The data are averages from at least three independent experiments plus or minus the standard error.
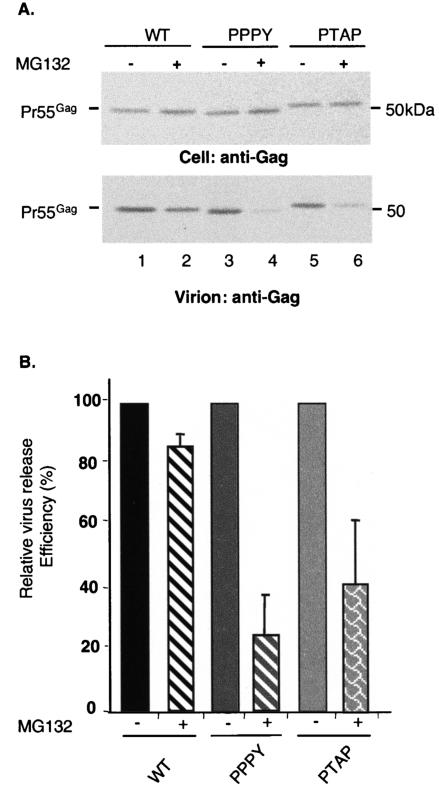
The resistance of EIAV Gag to proteasome inhibitors is L domain dependent.
Several studies have observed that the budding of a number of divergent retroviruses is disrupted by proteasome inhibitors (48, 51, 52, 64, 67). In contrast, the release of EIAV is not affected by these agents (48, 51). It has been proposed that a Ub-like motif located in the p9 C-terminus might confer resistance to proteasome inhibitors (51). To examine the basis for this unusual property of EIAV, we examined the effect of MG132 and epoxomycin on the release of EIAV/WT, EIAV/PTAP, and EIAV/PPPY particles. As reported previously (48, 51), EIAV/WT particle release was not significantly reduced by either inhibitor (Fig. 6; data not shown). In contrast, the release of EIAV/PPPY and EIAV/PTAP virion-associated Gag was markedly impaired (by approximately three- to fivefold) (Fig. 6). These data suggest that the insensitivity of EIAV to proteasome inhibitors is conferred by the L domain itself and not by downstream sequence(s) in p9.
FIG. 6.
The resistance of EIAV Gag to proteasome inhibitors is L domain dependent. (A) 293T cells were transfected in duplicate with EIAV/WT, EIAV/PTAP, or EIAV/PPPY. Twelve hours posttransfection, the cells were treated for 90 min with DMSO (lanes 1, 3, and 5) or with 10 μM MG132 (lanes 2, 4, and 6). The treated cells were metabolically labeled for 2.5 h in the presence or absence of MG132. Cell and viral lysates were radioimmunoprecipitated with horse anti-EIAV antiserum. Molecular mass markers are indicated on the right. (B) Virus release efficiency was calculated as described in the legend to Fig. 1. The data are averages from at least three independent experiments plus or minus the standard error.
DISCUSSION
HIV-1 budding and release are facilitated by an interaction between the L domain in p6 and the ESCRT-I component TSG101. Other studies have reported the use of TSG101-based inhibitors to disrupt the budding of HIV-1 and MLV. In the present study, we extended these findings by using a panel of EIAV clones to examine the effect of budding inhibitors on the release of VLPs promoted by all three known classes of viral L domains: PTAP, PPPY, and YPDL. We also examined the role of class E Vps machinery in the context of these divergent L domains. We show that inhibition of VLP release by TSG-5′ correlates with dependence on PTAP, that host factor requirements can be shifted by exchanging L-domain usage, and that TSG-3′ impairs budding of PPPY- and PTAP-containing viruses but not that of YPDL-dependent (WT) EIAV. The release of VLPs containing all three classes of L domains is strongly suppressed by Vps4EQ, suggesting that all retroviruses bearing these motifs utilize class E Vps machinery for budding from the plasma membrane. Finally, we report data suggesting that sensitivity to proteasome inhibitors is conferred by the L domain itself and not by other determinants in Gag distinct from the L domain.
It was initially determined by Parent et al. (50) that L domains of two unrelated Gag proteins (HIV-1 and RSV) are exchangeable and can function in a positionally independent manner. L-domain interchangeability has also been reported in the context of MLV (85) and EIAV (36). In the present study, we have taken advantage of these observations by constructing vectors that express chimeric EIAV Gag proteins containing PPPY and PTAP in place of YPDL. As previously reported (36, 60), we confirm that deletion of the YPDL motif results in severe inhibition of VLP release and that this defect can be rescued by substituting PPPY or PTAP for YPDL.
We report here that overexpression of TSG-5′ results in a marked reduction in EIAV/PTAP VLP production but has no effect on the release of EIAV/WT and EIAV/PPPY (Fig. 2). In contrast, TSG-F overexpression inhibits both EIAV/PTAP and EIAV/PPPY release (Fig. 4); significant effects on EIAV/WT budding are seen only at higher TSG-F expression levels (unpublished data). The data extend previous analyses (11, 18) and support the hypothesis that inhibition by TSG-5′ is PTAP dependent, whereas the inhibition induced by TSG-F is the result of a broad perturbation of the endosomal sorting pathway.
Recent reports have demonstrated that the C terminus of TSG101 is required for the recruitment of Vps28 to the ESCRT-I complex (6, 42). We observed that overexpression of TSG-3′ potently inhibited the release of both HIV-1 and MLV virions (18). Consistent with the latter report, we show here that the release of EIAV/PTAP and EIAV/PPPY is strongly suppressed by TSG-3′. Remarkably, in light of the drastic effect of TSG-3′ on HIV-1 and MLV release and on the morphology of intracellular vesicles (18), EIAV/WT is resistant to the effects of TSG-3′ (Fig. 4). The absence of strong inhibition of EIAV/WT release by both TSG-F and TSG-3′ prompted us to examine whether EIAV utilizes class E Vps machinery for its release. We observed that overexpression of a dominant negative mutant of Vps4, Vps4EQ (5), broadly inhibits the release of all the VLPs tested in this study, including EIAV/WT (Fig. 5). These data are in agreement with a recent report by Martin-Serrano et al. (41) demonstrating that the budding of an HIV-1 chimera in which p6 was replaced by p9 is sensitive to inhibition by a dominant negative Vps4. Also consistent with this conclusion, while this article was in preparation, Tanzi et al. (71) reported a block to EIAV release by dominant negative Vps4. Taken together, these data suggest that while host factor utilization involved in YPDL-mediated budding diverges in significant respects from that mediated by PTAP or PPPY, all three L domains share the need for a functional Vps4.
It is intriguing that overexpression of TSG-F, TSG-3′, and Vps4EQ all induce defects in the endosomal sorting pathway, yet only Vps4EQ potently inhibits EIAV/WT release. These results indicate that fundamental differences exist in either the magnitude or nature of the endosomal sorting defects induced by these inhibitors, consistent with our previous analysis of the effects of these proteins on EGF-R sorting and on endosomal morphology (18). More work will be required to understand fully the precise nature of the sorting defects induced by these proteins and the consequences of these defects for retrovirus budding.
The mechanism by which proteasome inhibitors block retrovirus budding and the explanation for the resistance to these compounds exhibited by EIAV remain undefined. It has been widely suggested that depletion of free Ub induced by proteasome inhibitors prevents Gag ubiquitination, thereby in some manner disrupting particle release. Indeed, the recognition of cargo proteins by the cellular endosomal sorting machinery is often dependent upon cargo ubiquitination, as certain key components of the sorting machinery (e.g., TSG101/Vps23 and Hrs) contain Ub-interacting motifs (see above). Furthermore, directly linking Ub to RSV Gag renders RSV budding partially resistant to proteasome inhibitors. However, the role played by Gag ubiquitination in retroviral budding is uncertain. (i) It has been reported that brief treatment of Gag-expressing cells with proteasome inhibitors can disrupt budding, even though these treatments do not substantially deplete intracellular pools of free Ub (49). (ii) Mutation of the sites of p6 ubiquitination does not impair HIV-1 budding (12, 47), indicating that p6 ubiquitination is not required for HIV-1 release. (iii) TSG101 can bind p6 in the absence of the Gag-Ub modification, though ubiquitination may enhance binding (17, 57, 74). (iv) Mouse mammary tumor virus Gag and EIAV p9 are ubiquitinated (49), yet the budding of both of these viruses is insensitive to proteasome inhibitors (49, 51). The latter observation led to the proposal that EIAV p9 harbors a Ub-like motif that would make modification of p9 by ubiquitination unnecessary and would confer resistance to proteasome inhibitors (51). However, the finding that EIAV mutants containing PTAP or PPPY motifs in place of the native YPDL sequence are sensitive to proteasome inhibitors suggests that resistance to these inhibitors is conferred by the YPDL motif itself and not by downstream sequences in p9. It remains possible, however, that the putative Ub-like motif in p9 is functional in the native context but not in the context of PTAP or PPPY. Additional support for the hypothesis that sensitivity to proteasome inhibitors is determined solely by the L domain itself might be provided by testing the impact of proteasome inhibitors on a YPDL-dependent HIV-1 clone. We attempted this analysis; however, we observed that HIV-1 Gag chimeras in which PTAP was replaced by sequences from EIAV spanning the YPDL motif exhibited a marked release defect (M. Shehu-Xhilaga et al., unpublished data), making any meaningful interpretation of the impact of proteasome inhibitors on the release of these clones difficult. We are presently modifying these mutants in an attempt to obtain release-proficient, YPDL-dependent HIV-1 clones. It is important to emphasize that ubiquitination affects the trafficking and degradation of a large number of cellular proteins, including components of the sorting machinery itself (for reviews see references 24, 29, 73, and 80). The impact of proteasome inhibitors on virus release could therefore be indirect and not specifically attributable to effects on Gag ubiquitination. We propose that YPDL-containing Gag enters the MVB sorting pathway at a point downstream from the block imposed by proteasome inhibitors, most likely due to recruitment of EIAV Gag to ESCRT-III via a direct binding between the YPDL motif and AIP1/ALIX (68, 75, 78). It is interesting to note the parallel spectrum of inhibition towards retrovirus budding exhibited by proteasome inhibitors and TSG-3′.
In conclusion, the data reported here provide significant insights into the utilization of class E Vps machinery in retrovirus budding. A more in-depth understanding of the interplay between host factors and virus release may suggest novel strategies for therapeutic interventions that target this step of the virus replication cycle.
Acknowledgments
We thank A. Ono and R. Goila-Gaur for critical review of the manuscript and helpful discussions, A. Buckler-White for DNA sequencing, and P. Woodman and Z. Sun for plasmids. The reagent AIDS patient Ig was obtained from the NIH AIDS Research and Reference Reagent Program.
This work was supported in part by NIH grant 2RO1 CA49296 (to R.C.M.) and NIH postdoctoral training grant T32 AI49820 (to C.C.). M.S.-X. was supported in part by a C. J. Martin fellowship.
REFERENCES
- 1.Amberg, D. C., E. Basart, and D. Botstein. 1995. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2:28-35. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, N., A. Horman, and P. Woodman. 2002. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157:91-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, N., and P. Woodman. 2001. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 7.Carter, C. A. 2002. Tsg101: HIV-1's ticket to ride. Trends Microbiol. 10:203-205. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C., F. Li, and R. C. Montelaro. 2001. Functional roles of equine infectious anemia virus Gag p9 in viral budding and infection. J. Virol. 75:9762-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conibear, E. 2002. An ESCRT into the endosome. Mol. Cell 10:215-216. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., R. N. Harty, J. Paragas, P. Palese, and J. W. Wills. 1999. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J. Virol. 73:3359-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupre, S., C. Volland, and R. Haguenauer-Tsapis. 2001. Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11:R932-R934. [DOI] [PubMed] [Google Scholar]
- 14.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freed, E. O., and M. A. Martin. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for hiv-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 18.Goila-Gaur, R., D. G. Demirov, J. M. Orenstein, A. Ono, and E. O. Freed. 2003. Defects in human immunodeficiency virus budding and endosomal sorting induced by TSG101 overexpression. J. Virol. 77:6507-6519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottwein, E., J. Bodem, B. Muller, A. Schmechel, H. Zentgraf, and H. G. Krausslich. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474-9485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harty, R. N., M. E. Brown, J. P. McGettigan, G. Wang, H. R. Jayakar, J. M. Huibregtse, M. A. Whitt, and M. J. Schnell. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J. Virol. 75:10623-10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harty, R. N., J. Paragas, M. Sudol, and P. Palese. 1999. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J. Virol. 73:2921-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 25.Hicke, L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell 106:527-530. [DOI] [PubMed] [Google Scholar]
- 26.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jayakar, H. R., K. G. Murti, and M. A. Whitt. 2000. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J. Virol. 74:9818-98127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 29.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 30.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koonin, E. V., and R. A. Abagyan. 1997. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat. Genet. 16:330-331. [DOI] [PubMed] [Google Scholar]
- 33.Le Blanc, I., M. C. Prevost, M. C. Dokhelar, and A. R. Rosenberg. 2002. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J. Virol. 76:10024-10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemmon, S. K., and L. M. Traub. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457-466. [DOI] [PubMed] [Google Scholar]
- 35.Levkowitz, G., H. Waterman, E. Zamir, Z. Kam, S. Oved, W. Y. Langdon, L. Beguinot, B. Geiger, and Y. Yarden. 1998. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12:3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, F., C. Chen, B. A. Puffer, and R. C. Montelaro. 2002. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J. Virol. 76:1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, L., and S. N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319-329. [DOI] [PubMed] [Google Scholar]
- 38.Licata, J. M., M. Simpson-Holley, N. T. Wright, Z. Han, J. Paragas, and R. N. Harty. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J. Virol. 77:1812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Longva, K. E., F. D. Blystad, E. Stang, A. M. Larsen, L. E. Johannessen, and I. H. Madshus. 2002. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luzio, J. P., B. A. Rous, N. A. Bright, P. R. Pryor, B. M. Mullock, and R. C. Piper. 2000. Lysosome-endosome fusion and lysosome biogenesis. J. Cell Sci. 113:1515-1524. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2003. Role of ESCRT-I in retroviral budding. J. Virol. 77:4794-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mimnaugh, E. G., H. Y. Chen, J. R. Davie, J. E. Celis, and L. Neckers. 1997. Rapid deubiquitination of nucleosomal histones in human tumor cells caused by proteasome inhibitors and stress response inducers: effects on replication, transcription, translation, and the cellular stress response. Biochemistry 36:14418-14429. [DOI] [PubMed] [Google Scholar]
- 44.Myers, E. L., and J. F. Allen. 2002. Tsg101, an inactive homologue of ubiquitin ligase e2, interacts specifically with human immunodeficiency virus type 2 gag polyprotein and results in increased levels of ubiquitinated gag. J. Virol. 76:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odorizzi, G., M. Babst, and S. D. Emr. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95:847-858. [DOI] [PubMed] [Google Scholar]
- 46.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 47.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Penalva, M. A., and H. N. Arst, Jr. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Piper, R. C., and J. P. Luzio. 2001. Late endosomes: sorting and partitioning in multivesicular bodies. Traffic 2:612-621. [DOI] [PubMed] [Google Scholar]
- 55.Polo, S., S. Sigismund, M. Faretta, M. Guidi, M. R. Capua, G. Bossi, H. Chen, P. De Camilli, and P. P. Di Fiore. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451-455. [DOI] [PubMed] [Google Scholar]
- 56.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 57.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pornillos, O., D. S. Higginson, K. M. Stray, R. D. Fisher, J. E. Garrus, M. Payne, G.-P. He, H. E. Wang, S. G. Morham, and W. I. Sundquist. 2003. HIV Gag mimics the TSG101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pornillos, O. W., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:469-579. [DOI] [PubMed] [Google Scholar]
- 60.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raiborg, C., K. G. Bache, D. J. Gillooly, I. H. Madshus, E. Stang, and H. Stenmark. 2002. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4:394-398. [DOI] [PubMed] [Google Scholar]
- 63.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shih, S. C., D. J. Katzmann, J. D. Schnell, M. Sutanto, S. D. Emr, and L. Hicke. 2002. Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4:389-393. [DOI] [PubMed] [Google Scholar]
- 66.Stahl, P. D., and M. A. Barbieri. 2002. Multivesicular bodies and multivesicular endosomes: the “ins and outs” of endosomal traffic. Sci. STKE 2002:PE32. [DOI] [PubMed] [Google Scholar]
- 67.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:1-20. [DOI] [PubMed] [Google Scholar]
- 69.Sun, Z., J. Pan, W. X. Hope, S. N. Cohen, and S. P. Balk. 1999. Tumor susceptibility gene 101 protein represses androgen receptor transactivation and interacts with p300. Cancer 86:689-696. [DOI] [PubMed] [Google Scholar]
- 70.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 71.Tanzi, G. O., A. J. Piefer, and P. Bates. 2003. Equine infectious anemia virus utilizes host vesicular protein sorting machinery during particle release. J. Virol. 77:8440-8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Timmins, J., G. Schoehn, S. Ricard-Blum, S. Scianimanico, T. Vernet, R. W. Ruigrok, and W. Weissenhorn. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J. Mol. Biol. 326:493-502. [DOI] [PubMed] [Google Scholar]
- 73.Varshavsky, A. 1997. The ubiquitin system. Trends Biochem. Sci. 22:383-387. [DOI] [PubMed] [Google Scholar]
- 74.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vincent, O., L. Rainbow, J. Tilburn, H. N. Arst, Jr., and M. A. Penalva. 2003. YPXL/I is a protein interaction motif recognized by aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 23:1647-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vito, P., L. Pellegrini, C. Guiet, and L. D'Adamio. 1999. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J. Biol. Chem. 274:1533-1540. [DOI] [PubMed] [Google Scholar]
- 77.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H.-Y. Chung, E. Morita, H. E. Wang, T. Davis, G.-P. He, D.-M. Cimbora, A. Scott, H.-G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 79.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus Gag membrane targeting and late domain function. J. Virol. 76:8485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 81.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 83.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]