Abstract
Transgenic photo-thermo sensitive genic male sterility Oryza sativa L. cv. “261S” plants with the anti-Waxy gene were successfully obtained using an Agrobacterium tumefaciens-mediated co-transformation method. Marker-free homozygous transgenic lines with the anti-Waxy gene were obtained. The setting seed rates of the transgenic plants via self-pollination or via crossing with the restorer line WX99075 rice and the 1000-grain weight of the transgenic plants and the F2 hybrid seeds obtained by crossing the transgenic or non-transgenic plants with the restorer line WX99075 rice, and the number of panicles of the transgenic plants and yields of the F2 hybrid rice, were analysed. Quality indexes of the transgenic plants and of the F2 hybrid seeds were analysed. Our researches results indicate that hybrid female and hybrid descendant edibility could be improved via the introduction of the anti-Waxy gene, but the grain yields of the reserve seeds via self-pollination of the transgenic photo-thermo sensitive genic sterile lines and of the hybrid rice were not affected.
Keywords: photo-thermo sensitive genic male sterility rice, anti-Waxy gene, rate of setting seeds, yield, quality indexes of the grains
Introduction
Amylose content (AC) is one of the most important indexes that affect rice edibility. Many investigations have attempted to improve grain edibility by introducing the anti-Waxy gene into rice. In 1993, Shimada et al.’s research was the first to demonstrate that an anti-Waxy gene could reduce amylose content in the grains (Shimada et al. 1993). In 2000, Terada et al.’s report demonstrated that an integrated anti-Waxy gene caused a reduction in amylose synthesis, not only in the endosperms but also in the pollen grains of transgenic rice (Terada et al. 2000).
Recently, research to improve rice edibility has been of great interest in China. An expression vector p13W4 plasmid with the anti-Waxy-gus fusion gene under the Waxy gene promoter has been constructed by the National Laboratory of Plant Molecular Genetics at the Institute of Plant Physiology & Ecology of the Chinese Academy of Science (Liu et al. 2003) and have been used to transform the leading conventional rice cultivars and the major parents to produce hybrid rice in China for improving the grain edibility (Chen et al. 2002, Li et al. 2007, 2008, 2009, Liu et al. 2003, Liu et al. 2006, Shen et al. 2004a, Wang et al. 2003, Yin et al. 2001). Because the T-DNA region of the p13W4 vector contains not only the anti-Waxy-gus fusion gene but also the hygromycin phosphotransferase gene (hpt), which is driven by the CaMV35S promoter, these DNA sequences exist in transgenic rice.
The public’s concerns about the biosafety of transgenic crops have been raised in recent years as more commercial products of transgenic crops become available. Another expression vector p13W8 plasmid without hpt gene coding sequence and the gus reporter gene in the T-DNA has also been constructed by the National Laboratory of Plant Molecular Genetics at the Institute of Plant Physiology & Ecology of the Chinese Academy of Science (Shen et al. 2003) and been used in improving rice edibility (Cheng et al. 2010, Ding et al. 2009, Li et al. 2005, Shen et al. 2004b, Wu et al. 2008); however, the 35S promoter driving expression of the hpt gene has been not eliminated from p13W8, and therefore, the progeny contain the p35S promoter sequence.
The most important aspect of the safety of transgenic plants is that the expression vectors are safe. The 35S promoter is widely used in transgenic crop plants. Although it is has not been determined whether transgenic plants with the 35S promoter are a risk to human health, there are now two different views on 35S promoter hazards (Ho et al. 1999, Josef et al. 2003). In order to establish a complete risk assessment for the environmental release of transgenic plants and to rapidly apply transgenic plants to food production, it is preferable to remove any superfluous DNA sequences that are not the target genes or their essential promoters. In order to demonstrate the safety of these sequences from the source species, it is especially important that there are not any disputed DNA sequences, such as in the T-DNA of the expression vectors. Thus, our lab has constructed the new expression vector p13AWY-1, with the anti-Waxy gene, which is driven by the Waxy gene promoter (Yang et al. 2009). This vector does not contain any drug resistance selection marker genes or the 35S promoter.
Hybrid rice “Minyou Xiangjing”, whose female and male parents are photo-thermo sensitive genic male sterile rice cv. 261S and recovery line WX99075 respectively, has been applied widely in agriculture of the Yangtse Delta, China. “Minyou Xiangjing” is a high-yield rice variety and its grains have fragrance, but it contains a high level of amylose content in the grains (Cao et al. 2004). In this research, the expression vector p13AWY-1 was used to transform the PTGMS 261S rice plants by an Agrobacterium tumefaciens-mediated co-transformation method for improving PTGMS 261S rice and “Minyou Xiangjing” hybrid rice edibility and characteristics of the transgenic rice progeny were analysed.
Materials and Methods
Plasmid vectors
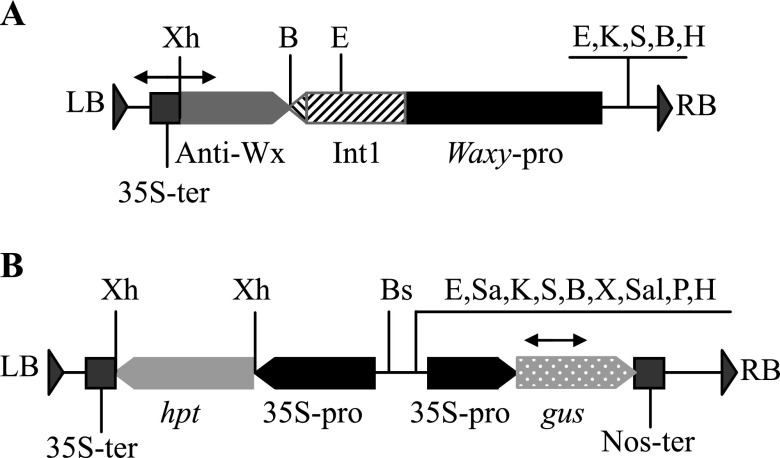
Two kinds of plasmids, p13AWY-1 (Fig. 1A) and pCAMBIA1301 (Fig. 1B), were also used in this study. Between the left and right border of the T-DNA of p13AWY-1, there is a 1288 bp Waxy gene promoter, the 1116 bp first intron of the Waxy gene, the 756 bp anti-Waxy gene and the 35S terminator. pCAMBIA1301 contains the hpt gene and the gus reporter gene, which are guided by the 35S promoter in the T-DNA region. The two vectors were introduced into the Agrobacterium tumefaciens strain EHA105, respectively, for use in the rice co-transformation.
Fig. 1.
The structure of the T-DNA region in the binary vectors of p13AWY-1 (A) and pCAMBIA1301 (B). Waxy-pro and Int1: the promoter (1288 bp) and first intron (1116 bp) of the Waxy gene from rice cv. 232; Anti-Wx: an antisense DNA fragment (756 bp) of the Waxy gene from rice cv. 232; hpt: the hygromycin phosphotransferase gene; gus: the β-glucuronidase (GUS) gene; 35S-pro and 35S-ter: the 35S promoter and terminator of CaMV; Nos-ter: the terminator of the nos synthase gene; LB and RB: the left and right border of the T-DNA; Xh: XhoI; B: BamHI; E: EcoRI; Bs: BstXI; Sa: SacI; K: KpnI; S: SmaI; X: XbaI; Sal: SacI; P: PstI and H: HindIII. The arrows show the sizes and locations of the PCR products.
Obtaining the transgenic rice
The male-sterile rice 261S (Oryza sativa L. japonica cv. 261S) was used as the acceptor in this study. The calli derived from the mature embryos of the rice seeds were used as the explant in this study. The co-transformation method followed that described by Li et al. (2005). Two separate stains of A. tumefaciens, which contained p13AWY-1 and pCAMBIA1301 mixed at a ratio of 9 : 1, were used to transform the calli. The media and protocols of the induction of the calli, the transforming, the selection of the hygromycin and the regeneration of the transgenic rice followed that described by Li et al. (2008).
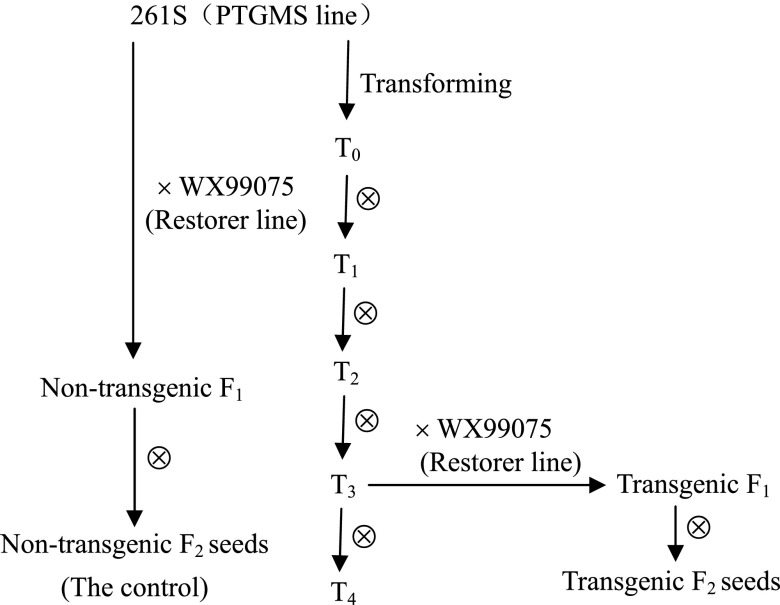
Fig. 2 is the scheme for the development of plant materials used in this study.
Fig. 2.
Scheme for the development of plant materials used in this study.
PCR testing of transgenic plants
According to the DNA sequence of pCAMBIA1301, gus gene-specific primers were designed, wherein the primer sequences were 5′GGAATCCATCGCAGCGTAATG3′ (GUS1) and 5′GCCGACAGCAGCAGTTTCATC3′ (GUS2). According to the DNA sequence of pCAMBIA1301, hpt gene-specific primers were designed, wherein the primer sequences were 5′TGACATTGGGGAGTTTAGCG 3′ (Z16) and 5′TGACATTGGGGAGTTTAGCG 3′ (Z17). According to the DNA sequence of p13AWY-1, specific primers that were homologous to the DNA sequence of the anti-Waxy gene of rice, between the 35S terminator and the left border of the T-DNA were designed. The primer sequences were 5′CTCTCGGAGAGGTTCAGCTC3′ (YW5) and 5′CTTAATAACACATTGCGGACG 3′ (YWf). The PCR parameters consisted of 30 cycles of 94°C for 40 s, 53.5°C for 40 s and 72°C for 45 s after 5 min at 94°C for an initial denaturalization and then an extension step at 72°C for 10 min.
Southern blot testing of the co-transformed plants
According to the DNA sequence of p13AWY-1, specific primers that were homologous to the DNA sequences of the promoter of the Waxy gene were designed. The primer sequences were 5′AGCTTATTACAGCCGTGGGAGA3′ (P13W4c) and 5′TAGCGGCTGCTTGTACGTGCT3′ (P13W4i). Using p13AWY-1 as a template, a 540 bp fragment was amplified. This PCR product was used as the probe in a Southern blot. The total DNA of the co-transformed plants was digested using KpnI. The procedures for the Southern blot followed the protocol of the Amersham ECL labelling kit.
GUS staining of transgenic plants and the obtaining of marker-free progeny and homozygous transgenic plants
The expression of GUS was assayed according to the method of Jefferson (1987). The GUS-negative seeds that contained embryos were germinated on 1/2 MS medium and were then transplanted to experimental fields. The T1 plants, which contained the target gene, were selected by the primers YW5 and YWf. The seeds of the T1 plants were separately harvested from each plant. The T2 and T3 plants were also tested for the target gene by PCR and homozygous T4 seeds from the T3 plants containing the anti-Waxy gene were obtained.
Analyses of agronomic traits, the rate of setting seeds by self-pollination and the 1000-grain weight
Heading date of the transgenic and non-transgenic rice plants were recorded. When the seeds of the T4 plants were harvested in the experimental fields, five plants from the sections that contained homozygous transgenic rice and non-transgenic 261S separately were randomly selected. The number of filled grains per panicle, the number of unfilled grains per panicle, the rate of setting seeds by the self-pollination of the five plants were calculated, and number of panicles of the homozygous transgenic and non-transgenic 261S were measured. The 1000-grain weights were also analysed. The resulting data were then statistically analysed (Tong 1986).
Analysis of the rate of setting seeds by cross pollination
The seedlings of the transgenic plants, the non-transgenic 261S and the recovery line WX99075 were planted in the experimental fields following the protocol of seed setting by conventional cross-pollination. In the flowering period, we simulated natural conditions, wherein we used bamboo to beat the male parents so that pollen was spread every day at noon so as to set the seeds of the transgenic rice and control plants by cross-pollination. When the seeds were ripe, five plants from the sections of the homozygous transgenic rice and non-transgenic 261S separately were randomly selected. The number of filled grains per panicle, the number of unfilled grains per panicle and the rate of setting seeds via the cross-pollination of each plant were calculated. The resulting data were then statistically analysed (Tong 1986).
Obtaining the F2 hybrid seeds and analysis of the 1000-grain weight of rice
F1 hybrid seeds, which were obtained by separately crossing the transgenic plants and the non-transgenic 261S with the recovery line WX99075, were planted. During the growing period, the anti-Waxy genes of the F1 plants were assayed by PCR. The F2 seeds were harvested when the seeds were ripe and the 1000-grain weights were also analysed. The resulting data were statistically analysed (Tong 1986).
Determination of the amylose content of rice
Approximately 30–50 mature seeds at the top of each predetermined plant’s panicles were dehulled and floured. The brown rice powder was sifted through a sieve with 100 tiny holes and dried overnight at 60°C. The amylose contents of the brown rice were measured by the colourisation method of Juliano (1971). The resulting data were statistically analysed (Tong 1986).
Results and Analyses
Cultivating the transgenic rice plants and PCR analysis of the transgenic plants
Seedlings were obtained from calli that had been infected by Agrobacterium after the process of co-culture, two rounds of hygromycin selection, pre-differentiation, differentiation and rooting. In this study, the seedlings were obtained from a total of 58 resistant calli and 45 transgenic plants that regenerated from the resistant calli survived. There was data recorded during the co-transformation experiments in the Table 1.
Table 1.
Data statistic of transgenic rice plants
| Mature embryo data inoculated | Calli data infected | Survival calli data after two rounds of hygromycin selection | No of calli regenerated | Differentiation rate (%) | No. of survival regenerated plants |
|---|---|---|---|---|---|
| 1409 | 1284 | 324 | 84 | 25.93 | 45 |
A PCR analysis was performed by a pair of specific primers, GUS1 and GUS2, for the gus gene in the T0 generation of the transgenic plants and the pCAMBIA1301 vector and 261S genomic DNA were used as the templates for the positive control and negative control, respectively. The results indicate that products of about 560 bp were amplified from the genomic DNA of all of the transgenic plants and the positive control, whereas no band was detected in the negative control (Fig. 3).
Fig. 3.
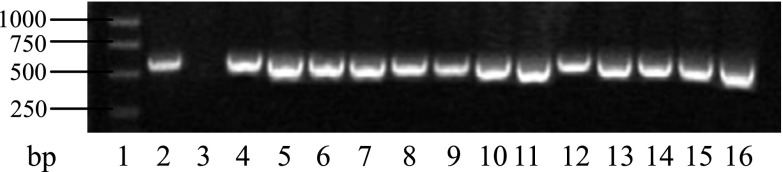
PCR testing of some of the T0 transgenic plants with the T-DNA of pCAMBIA1301. 1: Marker DNA; 2: pCAMBIA1301; 3: Wild type; 4–16: Transgenic plants.
The second pair of primers, YW5 and YWf, was used to identify whether the lines carried the anti-Waxy gene in the T0 generation of the 45 transgenic plants. The results indicate that a set of products of about 650 bp was amplified from 12 of 45 transgenic plants, whereas the negative control produced no products (Fig. 4). PCR analysis indicated that 12 co-transformed plants were obtained, giving a co-transformation rate of 26.67%.
Fig. 4.
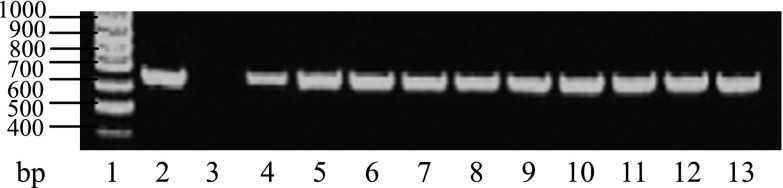
PCR testing of the T0 co-transformed plants with the T-DNA of p13AWY-1. 1: Marker DNA; 2: p13AWY-1; 3: Wild type; 4–13: Co-transformed plants.
Southern blot analysis of transgenic plants
The T0 generation of eight co-transformed transgenic plants were identified by Southern blot, wherein a fragment in the promoter of the Waxy gene served as the probe, whereas the p13AWY-1 vector and the non-transgenic rice served as the positive and negative controls, respectively. The results of the Southern blot indicate that there was one same hybridization band produced by the probe with the internal Waxy gene promoter in the rice genomic DNA and that more than one copy of the exogenetic gene may have integrated into the genome of the transgenic individuals. Transgenic plants JJ8, JJ12, JJ14, JJ16 and D4 contained a single copy of the anti-Waxy gene, whereas JJ19 contained two copies. Furthermore, both JJ10 and D2 had three copies (Fig. 5).
Fig. 5.

Southern blot testing of some of the transgenic plants with the T-DNA of p13AWY-1. 1: p13AWY-1; 2: Wild type; 3–10: Co-transformed plants (3: JJ10; 4: JJ8; 5: JJ12; 6: JJ14; 7: JJ16; 8: JJ19; 9: D2; and 10: D4). The arrow shows the hybridization bands produced by the probe with the internal Waxy gene promoter in the rice genomic DNA.
GUS staining assay and the selection of marker-free, homozygous transgenic lines
The seeds were harvested from the seven T0 co-transformation plants (JJ8, JJ10, JJ12, JJ14, JJ16, JJ19 and JJ36) and analysed by GUS staining of the brown rice in the T1 seeds. Those that contained the resistance gene and the reporter gene were quickly removed from the T1 seeds by the GUS assay. The negative seeds of the seven transformants were plated on 1/2 MS medium and then developed into T1 plants.
PCR amplification of the total DNA from the T1 generation of seven transgenic plants via the use of YW5 and YWf primers in combination with p13AWY-1 and the genomic DNA of non-transgenic rice 261S, which were used as templates for the positive and negative controls, respectively, was carried out. The result indicated that the T1 generation of these seven transgenic plants yielded a product of about 650 bp, which was amplified from JJ10, JJ12, JJ14, JJ16 and JJ36 and agreed with the product that was found in the positive control. For the progeny of the T1 plants in these lines, the ratio of those with exogenous genes to those without exogenous genes followed the Mendelian Law of 3 : 1 (Table 2). According to the results, it was speculated that a single copy of the anti-Waxy gene was integrated into the genome of each transgenic plant, including JJ10, JJ12, JJ14, JJ16 and JJ36. No PCR product was amplified from JJ8 or JJ19, most likely because the T-DNAs of both vectors, p13AWY-1 and pCAMBIA1301, were integrated into the same chromosome.
Table 2.
The segregation of plants with the anti-Waxy gene in the GUS-negative T1 grains
| Transgenic lines | Positive/Negative | Expected ratio | χ2 value | P value |
|---|---|---|---|---|
| JJ10 | 28/10 | 3 : 1 | 0.035 | 0.70–0.95 |
| JJ12 | 26/6 | 3 : 1 | 0.667 | 0.70–0.95 |
| JJ14 | 20/4 | 3 : 1 | 0.889 | 0.70–0.95 |
| JJ16 | 28/6 | 3 : 1 | 0.980 | 0.30–0.50 |
| JJ36 | 24/7 | 3 : 1 | 0.097 | 0.70–0.95 |
A PCR analysis was carried out to determine whether the hpt and gus genes existed in the genomes of the T1 generation using two pairs of primers, specifically Z16 and Z17 and GUS1 and GUS2, which could be used to amplify the hpt gene and the gus gene, respectively. In the five plants that contained anti-Waxy genes, the pCAMBIA1301 vector and the non-transgenic plants were used as positive and negative controls, respectively. The results of the PCR analysis indicate that products could only be amplified from pCAMBIA1301 and not from the five transgenic plants and non-transgenic plants (data not shown). This indicates that the genomes of the transgenic plants did not contain the hpt or gus genes.
Seeds were selected from five plants of each line of the T1 generation that contained the anti-Waxy gene, which were conformed by PCR analysis and then put into a group. A PCR analysis was performed to identify the genotypes of the T2 plants via the use of YW5 and YWf primers. The T3 seeds were selected from the T2 plants that contained the anti-Waxy gene and were planted in the field. Likewise, PCR analyses were carried out to identify the steady and hereditary transgenic homozygous progeny.
Analyses of agronomic traits, the rate of setting seed by self-pollinating and the 1000-grain weight
The heading dates of the transgenic rice and non-transgenic plants were also from September 1 to 8 when they were transplanted on June 20. There were few differences in traits, such as plant height and plant type, between the genetically modified rice and the non-transgenic plants. Several characteristics, including the number of filled and unfilled seeds per plant, the self-pollinating seed setting rate, the 1000-grain weights and number of panicles, were analysed by separately selecting five individuals from the three homozygous lines (JJ10, JJ12, JJ16) and the non-transgenic 261S. The results of the statistical analyses indicate that there was no significant difference between the transgenic plants and non-transgenic 261S for the self-pollinating seed setting rate or the 1000-grain weights or number of panicles (P > 0.05) (Table 3). This finding demonstrates that the anti-Waxy gene that integrated into 261S had no adverse effect on the characteristics of the self-pollinating seed setting rate, the grain weight and number of panicles of the acceptor.
Table 3.
Rate of setting seeds by self-pollination, 1000-grain and panicles number of the transgenic rice and wild type rice
| Transgenic lines and wild type | Rate of setting seeds by self-pollination ± standard error (%) | 1000-grain weight ± standard error (g) | Number of panicles ± standard error |
|---|---|---|---|
| JJ10 | 29.03 ± 1.29 a | 25.22 ± 0.44 a | 13.0 ± 0.99 a |
| JJ12 | 29.28 ± 1.37 a | 24.90 ± 0.19 a | 12.5 ± 0.73 a |
| JJ16 | 28.84 ± 1.57 a | 25.01 ± 0.31 a | 11.5 ± 1.01 a |
| Wild type | 28.86 ± 1.32 a | 25.17 ± 0.24 a | 11.5 ± 0.54 a |
Note: Values that are followed by different small letters (or capital letters) in the same column are significantly different at the 0.05 (or 0.01) probability level, respectively.
Analyses of setting seeds rate and 1000-grain weight of seeds by cross fertilization
The three homozygous transgenic lines (JJ10, JJ12, JJ16) and the 261S control were crossed with the restorer line WX99075, respectively, to produce hybrid seeds in the field. The results of the statistical analysis indicate that there was no significant difference (P > 0.05) in the outcrossing rate among the transgenic plants and the 261S that had been crossed with the WX99075 restorer line (Table 4), which indicates that the integration of the anti-Waxy gene into 261S had no adverse effect on the outcrossing rate of the acceptor.
Table 4.
The rate of setting seeds and 1000-grain weight of transgenic rice and wild type rice by cross fertilization
| Transgenic lines and wild type | Rate of setting seeds by cross fertilization ± standard error (%) | 1000-grain weight of the four kinds of hybrid rice F2 seeds ± standard error (g) |
|---|---|---|
| JJ10 | 12.42 ± 0.57 a | 27.91 ± 0.34 a |
| JJ12 | 12.55 ± 0.50 a | 27.73 ± 0.56 a |
| JJ16 | 12.84 ± 0.45 a | 28.21 ± 0.52 a |
| Wild type | 12.51 ± 0.41 a | 28.24 ± 0.33 a |
Note: Values that are followed by different small letters (or capital letters) in the same column are significantly different at the 0.05 (or 0.01) probability level, respectively.
The hybrid F1 seeds were harvested and were planted in the second year. The 1000-grain weights of the 4 kinds of F2 seeds harvested from the 4 kinds of F1 hybrid plants were analysed. The results of the statistical analysis indicate that there was no significant difference in the grain weight of the F2 seeds among the 4 kinds of hybrid rice (Table 4), which indicates that the use of the male-sterile line that contained the anti-Waxy gene as the female parent did not impact the grain weight of the hybrid rice.
Analysis of the amylose content in brown rice in the T1 and T4 generations of transgenic rice
The analysis of the amylose content was carried out in the T1 generation of five high yield co-transformants. Fifty mature seeds were selected from the top of the panicle of the transgenic rice. Each transgenic line was analysed three times per plant. The results suggested that there was no obvious difference in terms of amylose content between transgenic plant JJ8 and the control. The amylose contents in JJ12 and JJ16 slightly decreased by 13.17 and 5.95%, respectively, which reached the significance level (P < 0.05) in comparison to the control, whereas in JJ10 and JJ19, it significantly decreased (P < 0.01) by 80.65 and 33.48%, respectively (Table 5).
Table 5.
Amylose contents in T1 and T4 transgenic brown rice
| Transgenic lines and wild type | Mean of AC in T1 brown rice ± standard error (%) | Mean of AC in T4 brown rice ± standard error (%) |
|---|---|---|
| JJ8 | 13.12 ± 0.27 aA | |
| JJ10 | 2.61 ± 0.30 dD | 11.15 ± 0.11 dC |
| JJ12 | 11.67 ± 0.27 bB | 12.69 ± 0.15 bB |
| JJ14 | 11.90 ± 0.15 cC | |
| JJ16 | 12.64 ± 0.28 bB | 12.92 ± 0.06 bB |
| JJ19 | 8.94 ± 0.37 c | |
| JJ36 | 14.17 ± 0.20 aA | |
| Wild type | 13.44 ± 0.05 aA | 14.37 ± 0.08 aA |
Note: Values that are followed by different small letters (or capital letters) in the same column are significantly different at the 0.05 (or 0.01) probability level, respectively.
The T4 seeds were selected from the individual T3 plants of the JJ10, JJ12 and JJ16 homozygous lines and also from T3 plants of the JJ14 and JJ36 lines, which contained anti-Waxy genes according to PCR analysis. Thirty mature seeds were selected from the top of the panicle from the transgenic rice to assay the amylose content. Each transgenic line underwent five plants replicates. The results suggest that the amylose content of the T4 seeds of transgenic lines JJ10, JJ12, JJ14 and JJ16 decreased by different amounts; however, each reached the significance level (P < 0.05) or extremely significance level (P < 0.01) when compared to the control, except for the seeds of plant JJ36 T4. Moreover, the amylose content of the JJ10 T4 seeds was the most severely decreased of the five investigated transgenic lines, wherein the content was 11.15%, which is a decrease of approximately 22.42% in comparison to the control (Table 5).
Analysis of the yield and grain quality index of F2 hybrid rice that contain the anti-Waxy gene
The actual yield of the hybrid rice JJ10 × WX99075 and JJ12 × WX99075 and the control rice 261S × WX99075 was assayed and the actual yield of the three combinations were 8779.5, 8994 and 8566.5 kg/ha, respectively. The combinations of the control 261S × WX99075 and JJ12 × WX99075, for which the yields were much higher, were sent to The Quality Supervision, Inspection and Testing Centre For Rice and Rice Products, Ministry of Agriculture of China in order to test the grain quality indexes. Each combination was analysed two times. The results indicate that the alkali spreading value (ASV) of the two grains were almost the same, the amylose content of JJ12 × WX99075 was reduced by up to 9.60% and the gel consistency (GC) was increased by 14 mm in comparison to the control 261S × WX99075 (Table 6).
Table 6.
ASV, GC and AC in the F2 grains of JJ12 × WX99075 and 261S × WX99075
| Different combinations | Mean ASV ± standard error (degree) | Mean GC ± standard error (mm) | Mean AC ± standard error (%) |
|---|---|---|---|
| JJ12 × WX99075 | 7.0 ± 0 | 75 ± 0 A | 15.96 ± 0.19 b |
| 261S × WX99075 | 7.0 ± 0 | 61 ± 1 B | 17.68 ± 0 a |
Note: Values that are followed by different small letters (or capital letters) in the same column are significantly different at the 0.05 (or 0.01) probability level, respectively.
Discussion
The amylose contents of hybrid rice grains are related to the amylose contents of the male and female parent grains (Chen et al. 2002, Li et al. 1994, Liao et al. 2003). Huang et al. have suggested that one of two parents for producing hybrid rice ought to have lower amylose contents when breeding elite soft rice (1990). The new photo-thermo sensitive genic male sterile lines of rice with lower amylose contents have successfully bred in this research, which will not only improve hybrid rice edibility of “Minyou Xiangjing” and also enlarge the selection scope of male parents for crossing with the sterile lines and provide new resources of sterile lines for breeding elite soft rice. Our previous report has discussed that the GC in the grains would distinctly increase with reductions in AC if the anti-Waxy gene were introduced into conventional rice (Li et al. 2009), however, the present research result further shows that it is also effective for the anti-Waxy gene to increase the GC in the hybrid F2 grains that are produced by cross-breeding WX99075 rice with a parent that contains the anti-Waxy gene.
Although there have been some reports about improving the edibility of hybrid rice parents via the use of the anti-Waxy gene, these reports have primarily described the function of the anti-Waxy gene as a means to reduce the amylose contents of the hybrid rice parent grains (Chen et al. 2002, Li et al. 2008, Liu et al. 2003, Liu et al. 2006, Shen et al. 2004a) or the hybrid rice progeny grains (Li et al. 2008). These reports did not research the other characteristics of the transgenic hybrid rice parents.
The Waxy gene is responsible for the synthesis of amylase in the endosperm and pollen (Okagaki et al. 1988). Terada et al. (2000) suggest that the integrated anti-Waxy gene causes a reduction in amylase synthesis in the pollen grains of transgenic rice (Terada et al. 2000). Whether the pollen grain of rice is normal might affect the rate of setting seeds, and the rate of setting seeds is of great concern to the grain yield. The setting of seeds via self-pollination and cross fertilization is also critically important to the photo-thermo sensitive genic male sterile line when it is applied to two-line hybrid rice production. Our research suggests that the integrated anti-Waxy gene does not influence the rates of setting rice seeds. The grain weight and number of panicles are also ones of the major indices that affect rice yield. Our research on the 1000-grain weight and number of panicles suggests that the grain yields of the transgenic sterile line via self-pollination and the hybrid rice F2 seeds that are produced by crossing between the transgenic sterile line and the restorer line are not reduced by the integration of the anti-Waxy gene.
The degree of amylose content reduction in transgenic grains with two anti-Waxy gene copies has been shown to be more distinct than that of those with one anti-Waxy gene copy (Li et al. 2009, Yin et al. 2007). These prior results were similar to the results of our research. Furthermore, the degree of amylose content reduction in transgenic JJ10 T1 grains with three anti-Waxy gene copies was even more distinct. Although the transgenic line JJ10 contains three copies of the anti-Waxy gene as seen in the Southern blot analysis of the T0 plant, it is estimated that there is only one anti-Waxy gene copy in the JJ10 GUS-negative T1 plants according to an analysis of the segregation ratio of the anti-Waxy gene by PCR. It is inferred that the other two anti-Waxy gene copies are likely integrated into the same chromosome that contains the T-DNA of the plasmid pCAMBIA1301.
Acknowledgements
This work is funded by Developing Agriculture Key Project through science and technology of Shanghai Agricultural Commission (Project Number: Agricultural attack word 2004-2-17) and Leading Academic Discipline Project of Shanghai Municipal Education Commission (Project Number: J50401), respectively.
Literature Cited
- Cao, Y.Q., Zhu, X.M., Li, S.Z., Lu, Y.X., Wang, Y.X., and Lin, F. (2004) Minyou Xiangjing, a new quasi-aromatic two-line japonica hybrid rice combination. Hybrid Rice 19: 76–77 [Google Scholar]
- Chen, X.H., Liu, Q.Q., Wang, Z.Y., Wang, X.W., Cai, X.L., Zhang, J.L., and Gu, M.H. (2002) Introduction of antisense Waxy gene into main parent lines of indica hybrid rice. Chinese Sci. Bulletin 47: 1192–1197 [Google Scholar]
- Chen, Y.J. (2002) Molecular genetic studies on amy lose contents in rice. Journal of Hunan Agricultural University (Natural Sciences) 28: 195–198 [Google Scholar]
- Cheng, P., Liu, Q., Peng, Q., Zhao, Y., and Gong, X. (2010) A preliminary study on co-transformation parent-lines of indica hybrid rice with antisense Waxy gene. Hunan Agric. Sci. 15: 1–3 [Google Scholar]
- Ding, S., Liu, Q., Huang, Z., Zhou, J., and Xiao, L. (2009) Genetic transformation of indica rice 9311 with antisense Waxy gene. Journal of Hunan Agricultural University (Natural Sciences) 35: 366–368, 382 [Google Scholar]
- Ho, M.W., Ryan, A., and Cummins, J. (1999) Cauliflower mosaic viral promoter—a recipe for disaster? Microbial Ecology in Health and Disease 11: 194–197 [Google Scholar]
- Huang, C.W., and Li, Y. (1990) The genetic analysis of amylose content of rice (Oryza sativa L.). Journal of South China Agricultural University 11: 23–29 [Google Scholar]
- Jefferson, R.A. (1987) Assaying chimeric genes in plants: the gus gene fusion system. Plant Mol. Biol. Rep. 5: 387–405 [Google Scholar]
- Josef, V., Michal, S., Adam, P., Daniela, P., and Jindrich, B. (2003) Comparison of hCMV immediate early and CaMV 35S promoters in both plant and human cells. J. Biotec. 103: 197–202 [DOI] [PubMed] [Google Scholar]
- Juliano, B.O. (1971) A simplified assay for milled rice amylose. Cereal Science Today 16: 334–340 [Google Scholar]
- Li, G.S., Dong, S.Y., Li, C.S., and Zhang, Y.K. (1994) Combining ability for characters of yield and grian quality in early indica hybrid rice. Acta Agric. Zhejiangensis 6: 71–75 [Google Scholar]
- Li, J.Y., Mao, W.X., Yang, L.J., Zhou, G.Y., Liu, J., Yan, Q.T., and Mi, D. (2005) Introducing antisense Waxy gene into rice seeds reduces grain amylose contents using a safe transgenic technique. Chinese Sci. Bull. 50: 39–44 [Google Scholar]
- Li, J.Y., Mao, W.X., Fan, S.J., and Lv, Y.H. (2007) Improving tasting and nutritional quality of rice by introducing anti-Waxy gene. Bull. Botani. Res. 27: 94–98 [Google Scholar]
- Li, J.Y., Lv, Y.H., Yang, L.J., Zhou, Y.G., and Li, S.Y. (2008) Reducing amylose content of “Xiang Qing” seeds and improving eating quality of hybrid rice by introducing anti-Waxy Gene. Acta Bot. Boreal. 28: 1082–1087 [Google Scholar]
- Li, J.Y., Xu, S.Z., Yang, L.J., Zhou, Y.G., Fan, S.J., and Zhang, W. (2009) Breeding elite japonica-type soft rice with high protein content through the introduction of the anti-Waxy gene. African J. Biotec. 8: 161–166 [Google Scholar]
- Liao, F.M., Zhou, K.L., Yang, H.H., and Xu, Q.S. (2003) Comparison of grain quality between F1 hybrids and their parents in indica hybrid rice. Chinese J. Rice Sci. 17: 134–140 [Google Scholar]
- Liu, Q., Wang, Z.Y., Cheng, X.H., Cai, X.L., Tang, S.Z., Yu, H.X., Zhang, J.L., Hong, M.M., and Gu, M.H. (2003) Stable inherence of the anti-sense Waxy gene in transgenic rice with reduced amylose level and improved quality. Transgenic Res. 12: 71–82 [DOI] [PubMed] [Google Scholar]
- Liu, Q., Xiao, L.T., Wu, S., Zhu, Y.H., Liu, S.C., Shi, Q., and Kang, D.L. (2006) Antisense Waxy gene introduced into the calli in parents of hybrid rice by Agrobacterium tumefaciens. Mol. Pl. Breed. 4: 500–505 [Google Scholar]
- Okagaki, R. J., and Wessler, S.R. (1988) Comparison of non-mutant and mutant waxy genes in rice and maize. Genetics 120: 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, G.Z., Yin, L.Q., Wang, X.Q., Cai, X.L., and Wang, Z.Y. (2003) Transgenic rice with markerless antisense rice Waxy gene obtained by Agrobacterium-mediated co-transformation. J. Plant Physiol. Mol. Biol. 29: 569–575 [Google Scholar]
- Shen, G.Z., Wang, X.Q., Yin, L.Q., and Wang, H.M. (2004a) Determination of amylose content in transgenic progenies containing an anti-sense Waxy gene and identifying of pure lines in indica rice. Acta Biologiae Experimentalis Sinica 37: 429–435 [PubMed] [Google Scholar]
- Shen, G.Z., Wang, X.Q., Yin, L.Q., Cai, X.L., and Wang, Z.Y. (2004b) Rapidly obtaining the markerless transgenic rice with reduced amylose content by co-transformation and anther culture. J. Plant Physiol. Mol. Biol. 30: 637–643 [PubMed] [Google Scholar]
- Shimada, H., Tada, Y., Kawasaki, T., and Fujimura, T. (1993) Antisense regulation of the rice Waxy gene expression using a PCR-amplified fragment of the rice genome reduce the amylose content in grain starch. Theor. Appl. Genet. 86: 665–672 [DOI] [PubMed] [Google Scholar]
- Terada, R., Nakajima, M., Isshiki, M., Okagaki, R.J., Wessler, S.R., and Shimamoto, K. (2000) Antisense Waxy gene with highly active promoters effectively suppress Waxy gene expression in transgenic rice. Plant Cell Physiol. 41: 881–888 [DOI] [PubMed] [Google Scholar]
- Tong, Y.Z. (1986) Biological Statistics, Hunan Science and Technology Press, Changsha, pp. 176–215 [Google Scholar]
- Wang, X.Q., Shen, G.Z., Cheng, L., and Yin, L.Q. (2003) The genetic stanility of antisense Waxy gene segment in the progeny of transgenic plants in rice. Acta Agricul. Shanghai 19: 12–16 [Google Scholar]
- Wu, S., Xiao, L.T., Liu, Q., Shen, G.Z., and Wang, R.Z. (2008) Transgenic plants of markerless antisense Waxy indica rice obtained by Agrobacterium-mediated co-transformation. Scienfia Agricultura Sinica 41: 2909–2915 [Google Scholar]
- Yang, L.J., Rong, Y.Q., Zhang, S.M., and Li, J.Y. (2009) Strategy for building safe plant express vector, an example of construct for expressing rice anti-Waxy gene. Molec. Plant Breed. 7: 1027–1031 [Google Scholar]
- Yin, L.Q., Wang, X.Q., Han, Z.Y., Cheng, L., and Shen, G.Z. (2001) Transformation of Waxy antisense gene mediated by Agrobacterium in japonica rice. Journal of Shanghai Jiaotong University (Agricultural Science ) 19: 285–289 [Google Scholar]
- Yin, Z.M., Zhou, Y.G., Xu, S.Z., and Li, J.Y. (2007) Reduction of amylose contents in transgenic rice with different copies of anti-Waxy gene. Acta Bot. Boreal. 27: 2108–2111 [Google Scholar]