Abstract
High-risk human papillomaviruses, such as human papillomavirus type 16 (HPV16), are the primary cause of cervical cancer. The HPV16 E1∧E4 protein associates with keratin intermediate filaments and causes network collapse when expressed in epithelial cells in vitro. Here, we show that keratin association and network reorganization also occur in vivo in low-grade cervical neoplasia caused by HPV16. The 16E1∧E4 protein binds to keratins directly and interacts strongly with keratin 18, a member of the type I intermediate-filament family. By contrast, 16E1∧E4 bound only weakly to keratin 8, a type II intermediate-filament protein, and showed no detectable affinity for the type III protein, vimentin. The N-terminal 16 amino acids of the 16E1∧E4 protein, which contains the YPLLXLL motif that is conserved among supergroup A viruses, were sufficient to target green fluorescent protein to the keratin network. When expressed in the SiHa cervical epithelial cell line, the full-length 16E1∧E4 protein caused an almost total inhibition of keratin dynamics, despite the phosphorylation of keratin 18 at serine 33, which normally leads to 14-3-3-mediated keratin solubilization. Mutant 16E1∧E4 proteins which lack the LLKLL motif, or which have lost amino acids from their C termini, and which were compromised in the ability to associate with keratins did not disturb normal keratin dynamics. 16E1∧E4 was found to exist as dimers and hexamers, whereas a C-terminal deletion mutant (16E1∧E4Δ87-92) existed as monomers and formed multimeric structures only poorly. Considered together, our results suggest that by associating with keratins through its N terminus, and by associating with itself through its C terminus, 16E1∧E4 may act as a keratin cross-linker and prevent the movement of keratins between the soluble and insoluble compartments. The increase in avidity associated with multimeric binding may contribute to the ability of 16E1∧E4 to sequester its cellular targets in the cytoplasm.
Human papillomaviruses (HPVs) are small double-stranded DNA viruses of ∼8,000 bp. They infect stratified epithelium and produce lesions that range in severity from benign warts to invasive carcinomas (24). HPV DNA has been detected in >99.7% of cervical cancers, with HPV16 occurring most frequently (28, 29, 44). HPV16 is a high-risk HPV type which causes cervical lesions that can progress to high-grade neoplasia and cancer (43).
The life cycle of HPVs is closely linked to the differentiation status of the host epithelium. After infecting basal cells through a wound, the viral genome maintains itself episomally at a low copy number (5). As the infected cell migrates toward the epithelial surface and undergoes terminal differentiation, the productive stages of the viral life cycle are triggered. Vegetative viral DNA replication is followed by the expression of capsid proteins and the assembly of infectious virions in the superficial cell layers (24).
The HPV16 E1∧E4 protein is expressed in abundance during the late stages of the virus life cycle in the upper layer of the epithelium and coincides with the onset of viral genome amplification (12, 26, 35). Although the precise role of 16E1∧E4 is unclear, previous work has revealed that 16E1∧E4 can induce cell cycle arrest in G2 (7), can bind to a DEAD box RNA helicase (E4-DBP) (9), and, when expressed in cultured epithelial cells, can interact with keratins and cause the reorganization of the keratin intermediate-filament network (11). Although the mechanism by which 16E1∧E4 mediates keratin filament reorganization is not understood, immunofluorescence staining has shown the LLKLL motif located close to the N terminus to be necessary for filament colocalization and has shown the C terminus to be necessary for filament collapse.
Keratins are major structural proteins in epithelial cells and form the cytoplasmic network of intermediate filaments (17). They contain at least 20 members, called keratin 1 (K1) to K20, which are divided into two types according to the sequence and isoelectric point (pI). K9 to K20 are type I (acidic) keratins. The type II keratins, K1 to K8, are neutral or basic. Type I and type II keratins form noncovalent heteropolymers at a 1:1 ratio (27). The known functions of keratins are to structure the cytoplasm and to resist external stresses (6, 16). Recently, several new functions of keratins have emerged. K8 and K18 prevent Fas- and possibly tumor necrosis factor-induced apoptosis (3, 18, 19, 21). Overexpression of K10 inhibits cell proliferation (32) and tumor progression (40) through the activation of the retinoblastoma protein pRb (32, 33) and the inhibition of Akt/PKB and PKC ζ, which play pivotal roles in the phosphatidylinositol 3-kinase signaling pathway (34).
Keratin intermediate filaments are highly dynamic structures and are reorganized during cellular events such as mitosis and apoptosis (31). Keratin filament reorganization is regulated by posttranslational modifications, such as phosphorylation, glycosylation, transglutamination, and proteolysis, or through the interaction with other proteins, such as the cellular 14-3-3 proteins, which act as solubility cofactors when epithelial cells approach mitosis (31). 14-3-3 binds to phosphorylated K18 at Ser33, leading to solubilization of the K8/K18 network. K18 mutants which lack the 14-3-3 binding site do not assemble into a stable network but form a disorganized perinuclear bundle (20) that is similar to that seen following 16E1∧E4-mediated keratin filament reorganization (11).
Here, we have used cultured epithelial cells which contain K8/K18 intermediate-filament networks as a model to examine the mechanism by which the HPV16 E1∧E4 protein interferes with the structure of the cytokeratin network, and we report that the association of 16E1∧E4 with keratins and the reorganization of the keratin cytoskeleton occur in vivo as well as in vitro. The association between 16E1∧E4 and keratins is mediated through direct interaction and results in inhibition of the movement of keratin subunits between the soluble and insoluble compartments. This occurs despite the phosphorylation of K18 at serine 33, which in normal cells leads to 14-3-3 binding and keratin solubilization. We speculate that the ability of 16E1∧E4 to bind keratins through its N terminus, and to form multimeric complexes through its C terminus, may allow it to act as a keratin cross-linker, facilitating the tethering of E1∧E4 binding proteins in the cytoplasm.
MATERIALS AND METHODS
Cell culture.
SiHa, a human cervical cancer cell line, and Cos7, a simian virus 40-transformed African green monkey epithelial cell line (American Type Culture Collection), were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), penicillin, and streptomycin.
rAd generation and plasmid construction.
Recombinant adenoviruses (rAd) expressing 16E1∧E4, 16E1∧E4ΔLLKLL, and β-galactosidase (β-Gal) have been described previously (7, 9). rAd expressing 16E1∧E4Δ87-92 was prepared using the AdEasy Adenoviral Vector System (Stratagene). The BglII-HindIII fragment of 16E1∧E4Δ87-92 was produced by PCR using primers GTA ACT AGA TCT CCA CCA TGG CTG ATC CTG CAG CAG CAA CGA AGT AT (forward) and ATC GAA GCT TTT ATA CAG TTA ATC C (reverse) and cloned into the pShuttle-CMV vector. pcDNA16E1∧E4, which expresses the wild-type (wt) 16E1∧E4 protein, was prepared by cloning the PCR product generated using the primer pair C ACC ATG GCT GAT CCT GCA GCA GCA (forward) and CTA TGG GTG TAG TGT TAC TAT (reverse) into the vector pcDNA3.1D/V5-His-Topo by directional TOPO cloning (Invitrogen). pcDNA3-16E1∧E4Δ87-92 was prepared by cloning the E1∧E4Δ87-92 fragment obtained using the primer pair ATC GGA ATT CAC CAT GGC TGA TCC TGC AGC AGC A (forward) and ATC GTC TAG ATT ATA CAG TTA ATC C (reverse) between the EcoRI and XbaI sites of pcDNA3 (Invitrogen). Plasmids expressing green fluorescent protein (GFP) fused to 16E1∧E4 (wt) or the 16E1∧E4 fragments MADPAAATKYPLLKLL-GFP (Mut1), MDPAAATKYPLLKLL-GFP (Mut2), and MAPAAATKYPLLKLL-GFP (Mut3) were prepared using the C-terminal GFP fusion plasmid pcDNA3.1/CT-GFP-TOPO (Invitrogen). The primers used to prepare the 16E1∧E4wt-GFP expression plasmid were AGC CAC CAT GGC TGA TCC TGC AGC A (forward) and GTG GGT GTA GTG TTA CTA TTA C (reverse), whereas for Mut1 they were GCC ACC ATG GCT GAT CCT GCA GCA GCA ACG AAG TAT CCT CTC CTG AAA TTA TTA CA (forward) and GTA ATA ATT TCA GGA GAG GAT ACT TCG TTG CTG CTG CAG GAT CAG CCA TGG TGG CA (reverse). For Mut2, the primer pairs were GCC ACC ATG GAT CCT GCA GCA GCA ACG AAG TAT CCT CTC CTG AAA TTA TTA CA (forward) and GTA ATA ATT TCA GGA GAG GAT ACT TCG TTG CTG CTG CAG GAT CCA TGG TGG CA (reverse), whereas for Mut3, GCC ACC ATG GCT CCT GCA GCA GCA ACG AAG TAT CCT CTC CTG AAA TTA TTA CA (forward) and GTA ATA ATT TCA GGA GAG GAT ACT TCG TTG CTG CTG CAG GAG CCA TGG TGG CA (reverse) were used.
Cell synchronization, infection, and treatment with OA.
SiHa cells were synchronized by arresting the cells at G1/S with 0.5% FCS for 48 h, followed by incubation in DMEM with 5% FCS and 1 μg of the DNA polymerase α inhibitor aphidicolin (Sigma)/ml for 24 h. The drug was then washed off, and the cells were incubated with fresh DMEM containing 10% FCS for 10 to 12 h to enrich for cells in G2/M. Sixteen hours prior to release, SiHa cells were infected at a multiplicity of infection of 100 rAd particles per cell. Two hours prior to being harvested, the cells were treated with 1 μg of okadaic acid (OA) (Sigma)/ml.
Fractionation, immunoprecipitation, and Western blotting.
Cells (2 × 106) were pelleted, resuspended in 500 μl of phosphate-buffered saline (PBS) fractionation buffer (10 mM EDTA, 0.5 μg of OA/ml, 1 tablet of miniprotease inhibitor cocktail [Roche] per 10 ml of PBS), homogenized with 15 strokes, and centrifuged at 10,000 × g for 15 min to obtain the cytosolic fraction. The pellet was resuspended in 500 μl of 1% NP-40 in PBS fractionation buffer and placed on a rotary inverter for 15 min before being respun to obtain the NP-40-soluble fraction. The pellet was resuspended in 500 μl of 1% Empigen (Calbiochem) in PBS fractionation buffer and again placed on a rotary inverter for 15 min before being respun. The pellet was subsequently resuspended in 10% sodium dodecyl sulfate (SDS) in PBS fractionation buffer, incubated at 95°C for 15 min, and respun to obtain the SDS-soluble fraction. Equal volumes of the different fractions were loaded for Western blotting. NP-40 and Empigen fractions used for immunoprecipitation were incubated for 1 h at 4°C with 10 μl of rabbit anti-16E1∧E4 polyclonal antibody (10) or 10 μl of normal rabbit serum (Sigma). Complexes were pulled down on protein G-Sepharose (Amersham Pharmacia Biotech). Samples were separated on SDS-10 or 15% polyacrylamide gels and transferred to Immobilon membranes (Millipore) for Western blotting. Detection was done with anti-16E1∧E4 rabbit polyclonal (9) or monoclonal antibody, clone TGV402 (9); anti-K18, clone CY-90 (Sigma); or anti-α tubulin, clone B-5-1-2 (Sigma), followed by anti-rabbit (NA9340)- or anti-mouse (NA931)-horseradish peroxidase conjugates (Amersham Pharmacia Biotech) and development using enhanced chemiluminescence (Amersham Pharmacia Biotech).
Far-Western and dot blotting.
Empigen extracts from E1∧E4-expressing or non-E1∧E4-expressing SiHa cells were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to an Immobilon-P membrane (Millipore). After being blocked with 5% skim milk in PBS for 1 h at room temperature, the membrane was incubated with 1 μg of purified K18 protein (Cymbus Biotechnology Ltd., Chandlers Ford, United Kingdom)/ml in PBS for 2 h at room temperature. After three washes, the membrane was probed with anti-K18, clone Cy-90 (Sigma), or anti-16E1∧E4, clone TVG402, using the detection procedure outlined above. For dot blot analysis, 3 μg of purified K18, K8, or vimentin (Cymbus Biotechnology Ltd.) was loaded onto the nitrocellulose transfer membrane. After being blocked with 3% bovine serum albumin (BSA) in PBS for 1 h at room temperature, the membrane was incubated with 1 μg of purified 16E1∧E4/ml in 1% BSA in PBS for 2 h at room temperature. After being washed, the membrane was probed (overnight at 4°C) with anti-16E1∧E4, clone TVG402, and antibody binding was visualized using enhanced chemiluminescence as described above.
Immunofluorescence and immunohistochemistry.
Cells were grown on 13-mm-diameter coverslips before being fixed for 5 min in 5% formaldehyde and permeabilized in 0.5% Triton X-100 for 10 min; 6-μm-thick sections of formalin-fixed, paraffin-embedded cervical tissue were dewaxed in xylene and rehydrated in graded alcohols to water (41). Antigen retrieval was achieved by microwaving the sections in 1 M citric acid buffer for 18 min. After cooling for 20 min in buffer, the sections were blocked in 20% normal goat serum in PBS for 15 min before being incubated in the presence of mouse anti-K13 (1:100; Novocastra) for 1 h at room temperature and in the presence of anti-mouse Alexa 594. 16E1∧E4 was then detected using Alexa 488-conjugated TVG 405 (human Fab fragment) diluted 1:100 for 1 h, and the nuclei were counterstained with 1 μg of DAPI (4′-6-diamidino-2-phenylindole; Sigma)/ml. Estimation of the number of cells showing some evidence of filament reorganization was carried out following immunostaining of HPV16-induced cervical intraepithelial neoplasia grade 1 lesions (5) using antibody TVG 405. 16E1∧E4 immunostaining of monolayer cells was carried out using either TVG 402 or TVG 405 (12) directly conjugated to Alexa 488 or Alexa 594 (Molecular Probes). Keratins were stained with anti-K8/K18 rabbit polyclonal antibody 8592 (20) or antipankeratin monoclonal antibody, clone c2562 (Sigma), followed by fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (Amersham Pharmacia Biotech) or Alexa 594-conjugated anti-mouse antibody (Molecular Probes). Immunostained cells were examined using an Olympus 1X70 inverted epifluorescence microscope. Immunofluorescence images were recorded with an air-cooled charge-coupled-device camera.
Preparation of recombinant proteins.
Glutathione S-transferase-16E1∧E4 fusion proteins were expressed in BL21 cells from recombinant pGEX-6P1, and the proteins were cleaved using preScission protease (Amersham Pharmacia Biotech) as described by the manufacturer. The cells were grown at 37°C to an optical density of 0.6 at 600 nm in the presence of 0.1 mg of ampicillin/ml before being induced by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Growth was allowed to continue at 30°C for a further 3 h before the cells were pelleted and lysed by sonication in 50 mM Tris-Cl (pH 7.4), 1 mM EDTA, 300 mM NaCl, and 10 μM β-mercaptoethanol. The glutathione S-transferase fusion protein was bound to preequilibrated glutathione-Sepharose beads and washed with 10 bed volumes of cleavage buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT]) at 4°C. Cleavage was carried out by resuspending the column in 80 U of preScission protease in 960 μl of cleavage buffer overnight at 4°C. The cleaved protein was collected by gravity flow, and the concentration was determined by absorbance at 280 nm.
In vitro transcription-translation and gel filtration analysis.
[3H]leucine-labeled proteins were generated from recombinant pcDNA-16E1∧E4 and pcDNA-16E1∧E4Δ87-92 by in vitro transcription and translation (TNT-coupled wheat germ system; Promega) using mixed amino acids supplemented with 3H-labeled leucine according to the manufacturer's instructions. The transcription-translation products were either examined by SDS-PAGE or separated on a Superdex 75 column using the column buffer (20 mm Tris-Hcl, pH 7.0, 140 mM NaCl). Fractions (0.4 ml) were collected and immobilized on a Hybond-C extra membrane (Amersham Life Science). Fractions that contained labeled proteins were identified by scintillation counting. The column was calibrated using globular standards of known molecular masses (12.4 to 66 kDa). The results from gel filtration analysis were used to obtain a calibration curve by plotting Kav (the gel phase distribution coefficient) versus log Mr (molecular weight). The Kavs for the individual proteins were calculated as follows: Kav = (VR − V0)(VC − V0), where V0 is the void volume of the column, VR is the elution volume of the protein, and VC is the geometric bed volume in milliliters. The void volume of the column was determined using Blue Dextran 2000.
RESULTS
The localization of 16E1∧E4 to the keratin cytoskeleton occurs in vivo as well as in vitro.
Previous work has shown that the HPV16 E1∧E4 protein associates with keratin filaments and causes keratin network collapse when expressed in human keratinocytes in vitro (11). To establish whether 16E1∧E4 associates with the keratin network in vivo, tissue sections from naturally occurring HPV16-infected cervical tissue (i.e., cervical intraepithelial neoplasia grade 1) were double stained using anti-pankeratin or anti-K13 (the predominant keratin type found in the upper layers of the cervix) antibodies and antibodies to 16E1∧E4 (E1∧E4-K13 staining is shown in Fig. 1). In cells expressing 16E1∧E4 in the upper layers of infected epithelium, colocalization with keratins was apparent (Fig. 1A, B, and D). Cells expressing 16E1∧E4 sometimes contained partially reorganized or bundled keratin cytoskeletons (Fig. 1C and E), similar to the collapsed keratin networks found in cultured cervical epithelial cells in vitro (Fig. 1F). The proportion of cells showing clear evidence of keratin bundling was typically between 3 and 8% (see Materials and Methods). The L1 and L2 proteins could occasionally be detected in these cells but were also apparent in cells that did not show any obvious signs of keratin reorganization. Even so, virus structural proteins were found only in cells that also contained E4, and although gross reorganization was not always apparent, it is possible that keratin structure may be disturbed to a lesser degree in these cells. Keratin association and collapse of the keratin network to perinuclear bundles occurs in many cell types, including HaCat, SiHa, HeLa, and Cos (reference 11 and our unpublished data). Keratin bundles were not seen in normal cervical tissue adjacent to the area of HPV infection (Fig. 1D) and were not found in regions of infected tissue where 16E1∧E4 was not apparent (Fig. 1A and D). Interestingly, keratin association was also apparent when the E1∧E4 proteins of other papillomaviruses from supergroup A were examined, including HPV2, HPV11, HPV18, HPV31, HPV35, and HPV45 (data not shown). These results show that 16E1∧E4 can interact with keratins in vivo (K4/K13), as well as in vitro (K8/K18), and that the association may interfere with keratin network organization during natural infection of cervical tissue.
FIG. 1.
HPV16E1∧E4 is associated with reorganized keratin network in vivo and in vitro. (A and D) Tissue sections from cervical lesions caused by HPV16 were triple stained for 16E1∧E4 (green), K13 (red), and DNA (blue). The dotted lines indicate the position of the basal layer. The images were taken using a 10× objective. The cells enclosed within boxes are shown at higher magnification in panels B, C, and E (images captured using a 100× objective). (B) Colocalization of 16E1∧E4 and K13 filaments. The image corresponds to the right-hand box in panel A. (C) Association of reorganized keratin networks with 16E1∧E4. The image corresponds to the left-hand box in panel A. (E) Association of reorganized keratin networks with 16E1∧E4. The image corresponds to the left-hand box in panel D. The small box in the top right-hand corner of panel D, which contains cells with normal keratin staining, is shown enlarged in the inset. (F) 16E1∧E4-associated keratin network collapse in cultured epithelial cells is shown for comparison 24 h after infection of SiHa cells with rAd16E1∧E4. The cells were stained to show the presence of 16E1∧E4 (green), K18 (red), and DNA (blue). The image was taken using a 40× objective.
Interaction of HPV16 E1∧E4 with keratins is mediated by direct binding.
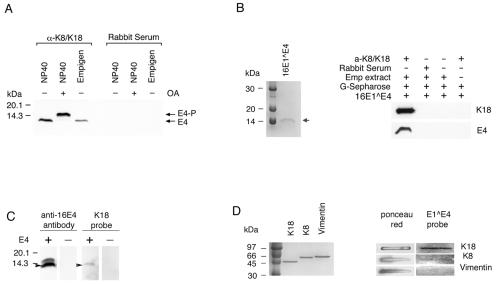
To determine how 16E1∧E4 interacts with keratins, a series of coimmunoprecipitation experiments were carried out. Cells expressing K8/K18 were chosen for the experiments, as these keratins are more widely used to assess keratin function than K4/K13. NP-40 or Empigen extracts from 16E1∧E4-expressing cells, with or without treatment with the protein phosphatase inhibitor OA, were immunoprecipitated using either anti-K8/K18 rabbit serum or normal rabbit serum prior to Western blotting using the anti-16E1∧E4 antibody TVG402. As shown in Fig. 2A, both the unphosphorylated and phosphorylated E1∧E4 proteins were coprecipitated using anti-K8/K18 rabbit serum but not using normal rabbit serum, indicating that 16E1∧E4 forms a stable complex with keratins.
FIG. 2.
HPV16 E1∧E4 binds to keratins. (A) Phosphorylated and unphosphorylated 16E1∧E4 was coimmunoprecipitated using anti-K8/K18 rabbit serum but not using normal rabbit serum. NP-40 and Empigen extracts from 16E1∧E4-expressing cells (either treated [+] or untreated [−] with OA) were immunoprecipitated with anti-K8/K18 rabbit serum (α-K8/K18) or normal rabbit serum, and the 16E1∧E4 protein was detected by Western blotting. 16E1∧E4 could be immunoprecipitated using antibodies to keratins. (B) Cellular keratins bind to purified 16E1∧E4 protein prepared in E. coli. Two micrograms of purified 16E1∧E4 protein is shown in the Coomassie blue-stained gel on the left. The position of the 16E1∧E4 protein is indicated by the arrow. The image on the right shows the results of the keratin binding experiment. Empigen (Emp) extract from SiHa cells was immunoprecipitated using anti-K8/K18 rabbit serum or normal rabbit serum. Following binding to protein G-Sepharose beads, the immobilized K8/K18 proteins were incubated with purified wt 16E1∧E4 protein, separated by SDS-PAGE, and Western blotted using an anti-16E1∧E4 antibody or an antibody to keratins. Keratins were isolated from epithelial cells in association with recombinant 16E1∧E4. (C) Purified K18 binds to denatured 16E1∧E4. Empigen extracts from 16E1∧E4-expressing or non-16E1∧E4-expressing cells were separated by SDS-PAGE and transferred to an Immobilon-P membrane. The membrane was incubated with purified K18 and blotted using anti-K18 antibody (two right-hand lanes). Subsequently, the blot was reprobed with anti-16E1∧E4 antibody (two left-hand lanes). (D) 16E1∧E4 binds to keratins directly. Three micrograms of purified K8, K18, or vimentin was loaded on a nitrocellulose transfer membrane, incubated with purified 16E1∧E4 protein, and probed using anti-16E1∧E4 antibody. Three micrograms of purified K8, K18, or vimentin is shown in the Coomassie blue-stained gel on the left. Equal loading was confirmed by Ponceau red staining. Purified 16E1∧E4 is able to bind directly to purified K18 but not to vimentin, and only weakly to K8.
Next, we tested whether recombinant 16E1∧E4 expressed in Escherichia coli could bind to keratins extracted from simple epithelial cells. Empigen is a zwitterionic detergent which solubilizes ∼40% of the total keratin pool and maintains the proteins in their native conformation (14, 23). The use of Empigen allowed us to partially purify K8/K18 from SiHa cells by immunoprecipitation using anti-K8/K18 rabbit serum. The partially purified keratins were immobilized on protein G-Sepharose and incubated with purified wt 16E1∧E4 protein expressed in E. coli (Fig. 2B) before being analyzed by Western blotting (Fig. 2B). A strong 16E1∧E4 band was present following precipitation using anti-K8/K18 rabbit serum but was absent when normal rabbit serum was used for immunoprecipitation (Fig. 2B). Neither anti-K8/K18 rabbit serum itself nor protein G-Sepharose could bind to purified 16E1∧E4. These results indicate that purified 16E1∧E4 binds to cellular K8/K18.
To examine whether the association of 16E1∧E4 with keratins was mediated by direct binding, far-Western blotting was carried out. Empigen fractions from E1∧E4-expressing or non-E1∧E4-expressing SiHa cells were separated by SDS-PAGE and transferred to an Immobilon-P membrane. The membrane was incubated with purified K18 before being blotted with anti-K18 antibody. Subsequently, the blot was reprobed with antibody to 16E1∧E4. A K18 protein band was apparent at the position where the 16E1∧E4 protein was found (Fig. 2C), suggesting that 16E1∧E4 has the ability to associate directly with K18. To confirm this finding and to demonstrate that the interaction is mediated through direct association, purified K8, K18, or vimentin (a type III intermediate filament found in cells of mesenchymal origin, such as fibroblasts) (15), was directly loaded onto a nitrocellulose transfer membrane using a slot blot apparatus (Fig. 2D). After being blocked with 3% BSA, the membrane was incubated with purified E1∧E4 protein and blotted using the anti-16E1∧E4 antibody TVG402. In contrast to vimentin, which has no association with 16E1∧E4, K18 showed strong and reproducible binding to 16E1∧E4. A very weak association was seen with K8 (Fig. 2D).
The 16 N-terminal amino acids of 16E1∧E4 are sufficient to localize GFP to the keratin cytoskeleton.
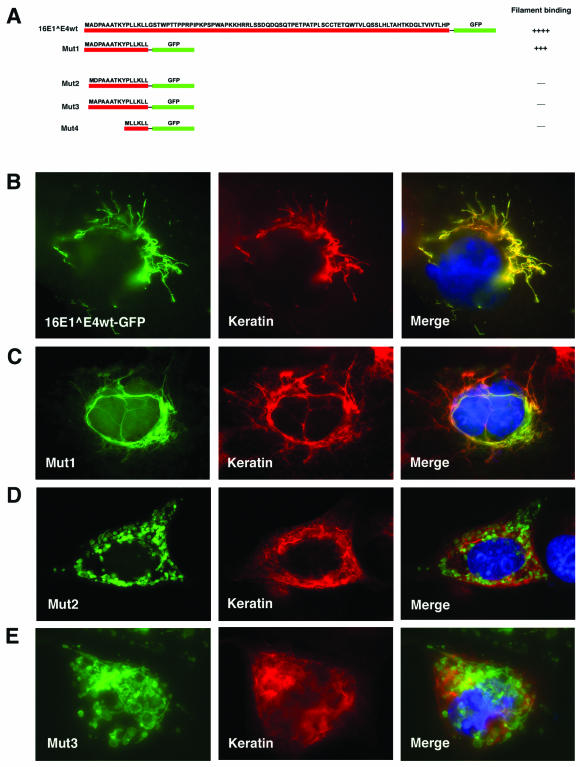
Immunofluorescence studies have suggested a role for the 16E1∧E4 LLKLL motif in keratin association (37, 38). To examine this further, wt 16E1∧E4 and a panel of 16E1∧E4 N-terminal fragments were fused to GFP (Fig. 3A) and analyzed by direct fluorescence and indirect immunofluorescence for the ability to target GFP to the keratins. Cos-7 cells were transfected for 24 h with GFP-fused 16E1∧E4wt or GFP-fused deletion mutants before being fixed with 5% formaldehyde and stained using a pankeratin antibody. As with the wt 16E1∧E4 protein (Fig. 3B), a deletion mutant (Mut1; Fig. 3A) which contains only the N-terminal 16 amino acids of 16E1∧E4 (including the LLKLL motif) showed a predominantly filamentous appearance and colocalized with keratins when fused to GFP (Fig. 3C), although the filamentous pattern was generally finer than with wt E1∧E4 (compare Fig. 3B with C). N-terminal sequence variants that had lost only one amino acid at position 2 (Mut2) or 3 (Mut3) were unable to efficiently target GFP to the keratin network and produced a punctate pattern in the cytoplasm (Fig. 3D and E). An identical pattern was obtained when the LLKLL motif was fused directly to GFP (data not shown). These results demonstrate that the LLKLL motif does not mediate keratin interaction on its own and that association may depend on the integrity of the N-terminal 16 amino acids of 16E1∧E4.
FIG. 3.
The N-terminal 16 amino acids of 16E1∧E4 are sufficient for keratin association. (A) The fragments of 16E1∧E4 that were fused to GFP are shown diagrammatically (red line, E1∧E4; green line, GFP). Mut1 contains the N-terminal 16 amino acids of 16E1∧E4, including the LLKLL motif. Mut2 has a deletion at amino acid 2, whereas Mut3 has a deletion at amino acid 3. Mut4 contains only the LLKLL motif fused to GFP. Cos7 cells were transfected for 24 h with the GFP-fused 16E1∧E4 wt protein (B), Mut1 (C), Mut2 (D), or Mut3 (E). After transfection, the cells were fixed with 5% formaldehyde and double stained with antibody to keratins (middle column; red) before being counterstained with DAPI (blue), which is visible in the merged images.
Expression of the 16E1∧E4 protein inhibits keratin solubilization, leading to the retention of 14-3-3 isoforms in the insoluble fraction.
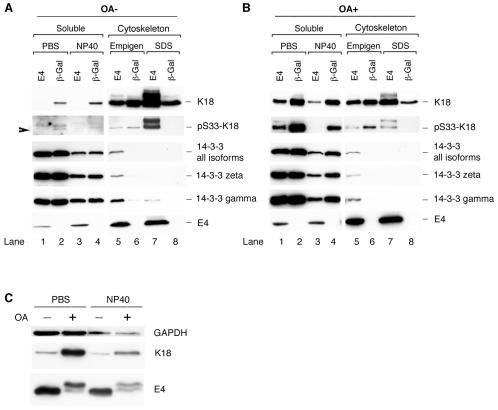
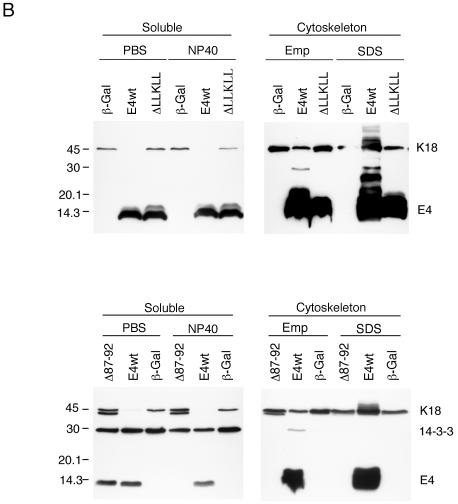
Having established that 16E1∧E4 can associate directly with keratin filaments, that the N-terminal 16 amino acids are important for binding, and that filament association occurs in vivo, we next wished to establish the mechanism of 16E1∧E4-mediated keratin filament reorganization. As keratin dynamics are linked to cellular events such as mitotic progression (31), and since 16E1∧E4 arrests cells in G2 (7), we used cells that were G2/M enriched. Four cellular fractions (PBS [cytosol], NP-40 [membrane associated], Empigen, and SDS [keratin-cytoskeleton associated]) were extracted from SiHa cells expressing 16E1∧E4 or β-Gal and analyzed by Western blotting using antibodies to 16E1∧E4, K18, or the 14-3-3 proteins. The K18 distribution in the four fractions in β-Gal-expressing cells was as expected, with ∼5% of the total keratin found in the soluble fraction (4) (Fig. 4A, K18). In 16E1∧E4-expressing cells, the soluble keratin pool was almost completely lost (Fig. 4A, compare lanes 1 and 3 with lanes 2 and 4).
FIG. 4.
Distribution of K18 and 14-3-3 proteins in 16E1∧E4- or β-Gal-expressing cells in the presence (OA+) or absence (OA−) of OA. (A) G2/M-enriched SiHa cells expressing 16E1∧E4 (E4) or β-Gal were harvested and fractionated to obtain four fractions (PBS-cytosol, NP-40 membrane-associated, Empigen-cytoskeletal, and SDS-cytoskeletal). Equal volumes of the four fractions were analyzed by Western blotting using antibodies to 16E1∧E4, K18, pS33 K18, and the 14-3-3 proteins. Note that the loss of the soluble keratin pool (lanes 1 and 3) and the appearance of 14-3-3 in the cytoskeletal fraction (lane 5) occur in 16E1∧E4-expressing cells. (B) G2/M-enriched SiHa cells expressing 16E1∧E4 or β-Gal were treated with the protein phosphatase inhibitor OA 2 h prior to harvest. The cells were harvested for fractionation using buffers containing PBS or NP-40 (soluble fraction), or Empigen or SDS (cytoskeleton), before being blotted to detect the presence of K18, pS33 K18, 14-3-3 proteins, and 16E1∧E4. The levels of 14-3-3 (ζ, γ, or all isoforms) were not significantly reduced in 16E1∧E4-expressing cells after OA treatment. (C) Equal amounts of protein from the soluble fractions of 16E1∧E4-expressing cells with or without OA treatment were Western blotted using antibodies to 16E1∧E4 and K18. Equal loading was confirmed using an antibody to GAPDH. The level of 16E1∧E4 protein did not increase in the soluble fraction after OA treatment despite an increase in the level of soluble keratins.
14-3-3 proteins consist of seven members: β, ɛ, γ, η, σ, τ, and ζ, which are abundantly expressed in eukaryotic cells and which are usually found in the cytoplasmic compartment. They act by binding to target proteins, which can lead to their altered subcellular localization (30, 42). The association of the 14-3-3 proteins with phosphorylated K8/K18 occurs in the cytosolic and membrane-associated compartments during the S and G2/M phases of the cell cycle (20). As expected, the 14-3-3 proteins were located exclusively in the soluble fraction in β-Gal-expressing cells, but following 16E1∧E4 expression, they were also found in the insoluble cytoskeletal fraction and could be extracted using Empigen (Fig. 4, lane 5). The association of 14-3-3 with keratins depends on the presence of a phosphoserine residue at position 33 of K18 (20). S33-phoshorylated keratins were detected (using antibody 8250 [20]) in the soluble fractions in cells expressing β-Gal, in agreement with previously published results (20), but they were concentrated in the insoluble cytoskeletal compartment in cells expressing 16E1∧E4 (present in the SDS fraction). Although the 14-3-3 proteins were found in the Empigen fraction rather than the SDS fraction, where the majority of the S33-phosphorylated keratins were found, this is most likely because the noncovalent association between 14-3-3 and phosphorylated keratins is destroyed during Empigen solubilization. To determine which isoforms of 14-3-3 were sequestered into the insoluble cytoskeletal compartment, antibodies that recognize the different isoforms of 14-3-3 were used. While we cannot rule out the involvement of additional 14-3-3 isoforms in vivo, only 14-3-3 ζ and γ were sequestered into the insoluble fraction following expression of 16E1∧E4 in monolayer culture (Fig. 4A and data not shown). Our results suggest that the 16E1∧E4 protein can inhibit keratin solubilization and cause a redistribution of 14-3-3 to the cytoskeleton despite the presence of serine 33 phosphorylation.
OA treatment increases the level of soluble keratins but does not significantly affect 14-3-3 and 16E1∧E4 redistribution.
Since the binding of phospho-K8/K18 to 14-3-3 increases keratin solubility (20), we asked whether 16E1∧E4 is still able to inhibit keratin solubilization when keratins are hyperphosporylated. G2/M-enriched SiHa cells expressing 16E1∧E4 or β-Gal were treated with the protein phosphatase inhibitor OA 2 h prior to harvest, followed by fractionation and Western blotting as described above. The extent of keratin phosphorylation at serine 33 was examined by Western blotting using antibody 8250 (20) (Fig. 4B). In agreement with previous findings (20), an increase in keratin solubility was apparent after OA treatment in cells expressing β-Gal (compare Fig. 4B, row 2, lanes 2 and 4, with A, lanes 2 and 4). However, in 16E1∧E4-expressing cells, although some K18 returned to the soluble fraction (compare Fig. 4B, row 1, lanes 1 and 3, with A, lanes 1 and 3), much of the S33-phosphorylated keratins and cytoskeleton-associated 14-3-3 protein remained in the cytoskeletal fractions, where the majority of the 16E1∧E4 was found (Fig. 4B). To assess whether the solubilization of keratins by phosphorylation affects 16E1∧E4 distribution, the levels of 16E1∧E4 protein in the soluble fraction before and after OA treatment were examined more precisely. Equal amounts of total protein from the soluble extracts of 16E1∧E4-expressing cells, with or without OA treatment, were loaded for Western blotting (Fig. 4C). While a significant proportion of K18 was relocalized to the cytosolic compartment after OA treatment, the levels of 16E1∧E4 protein in these fractions did not increase (Fig. 4C). Our results suggest that only those keratins that are not associated with 16E1∧E4 can be solubilized after OA treatment and that 16E1∧E4-associated keratins are not efficiently solubilized, despite their hyperphosphorylation and potential to associate with 14-3-3. Taken together, these results suggest that 16E1∧E4 inhibits the solubilization of keratins and blocks the ability of 14-3-3 to function as a solubility cofactor.
16E1∧E4 mutants which lack the leucine cluster motif or which have lost C-terminal residues have no effect on keratin dynamics.
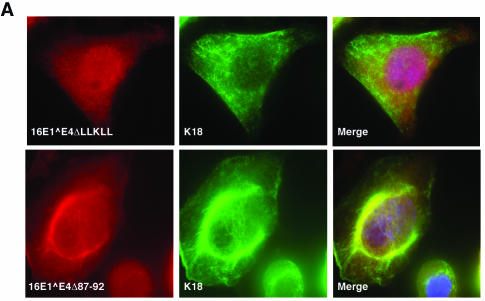
Immunofluorescence staining has previously implicated amino acids at the C terminus of 16E1∧E4 in keratin network collapse (38). The N-terminal region, which contains the LLXLL motif, is involved in keratin association (Fig. 3) (38). To examine the role of these regions in the dynamics of keratin movement, rAd encoding 16E1∧E4ΔLLKLL (rAd16E1∧E4ΔLLKLL, which encodes an E1∧E4 protein that lacks the leucine cluster) and 16E1∧E4Δ87-92 (rAd16E1∧E4Δ87-92, which encodes an E1∧E4 protein that lacks the extreme C terminus) were constructed. Twenty-four hours after infection with rAd16E1∧E4 ΔLLKLL or rAd16E1∧E4Δ87-92, SiHa cells were fixed, permeabilized, and stained for 16E1∧E4 and K8/K18. Whereas 16E1∧E4Δ87-92 showed partial association with keratins and was located in the cytoplasm, 16E1∧E4 ΔLLKLL showed little association with keratins and was distributed diffusely throughout the cell (Fig. 5A). To examine the effects of these mutants on keratin dynamics, G2/M-enriched SiHa cells expressing 16E1∧E4wt, 16E1∧E4ΔLLKLL, 16E1∧E4Δ87-92, or β-Gal were fractionated (as described above) and Western blotted using antibodies to 16E1∧E4, K18, and 14-3-3. As shown in Fig. 5B, the distribution of keratins in cells expressing the two 16E1∧E4 mutants (ΔLLKLL and Δ87-92) resembled that seen in β-Gal-expressing cells, indicating that both mutants are unable to affect keratin solubilization. Interestingly, 16E1∧E4Δ87-92, which can be shown by immunostaining to partially associate with the keratin network (Fig. 5A) (38), is found primarily in the cytosolic compartment following fractionation, suggesting that it associates with the keratin network only poorly.
FIG. 5.
Effects of 16E1∧E4 mutants on keratin association and keratin dynamics. (A) Twenty-four hours after infection with rAd16E1∧E4ΔLLKLL or rAd16E1∧E4Δ87-92, SiHa cells were fixed, permeabilized, and stained with anti-16E1∧E4 antibody (red), anti-K8/K18 antibody (green), and DAPI (blue). The images were taken using 40× (upper panels) or 60× (lower panels) objectives. Whereas 16E1∧E4Δ87-92 showed evidence of keratin association, 16E1∧E4ΔLLKLL did not. (B) The 16E1∧E4 mutants, 16E1∧E4ΔLLKLL and 16E1∧E4Δ87-92, did not disturb keratin dynamics. G2/M-enriched SiHa cells expressing 16E1∧E4wt (E4wt), 16E1∧E4ΔLLKLL (ΔLLKLL), 16E1∧E4Δ87-92 (Δ87-92), or β-Gal were harvested and fractionated as described in the legend to Fig. 4. Equal volumes of the four cellular fractions were loaded and Western blotted to detect 16E1∧E4, K18, and 14-3-3 (all isoforms). In contrast to wt 16E1∧E4, in which the soluble keratin pool was almost completely lost, in cells expressing β-Gal or the mutant 16E1∧E4 proteins, keratin dynamics were unaltered. Emp, Empigen.
wt HPV16 E1∧E4, but not 16E1∧E4Δ87-92, forms dimers and hexamers in vitro.
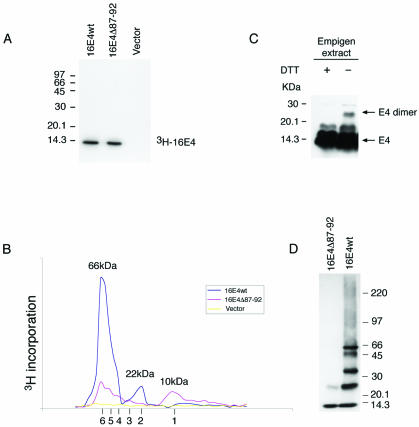
Based on the results described above and previous studies of the E1∧E4 protein of HPV11 (2), we speculated that 16E1∧E4, which can associate with keratins through its N terminus, may associate with itself through its C terminus. To investigate this, we examined the multimeric state of 16E1∧E4 using column chromatography. [3H]leucine-labeled 16E1∧E4wt or 16E1∧E4Δ87-92 was generated from the plasmid pcDNA-16E1∧E4 or pcDNA-16E1∧E4Δ87-92 by in vitro transcription-translation using the TNT wheat germ extract system (Promega). The in vitro transcription-translation reactions were separated by SDS-PAGE, and the 3H-labeled proteins were visualized by autoradiography. Only one band was apparent in the total reaction contents prepared using pcDNA-16E1∧E4 or pcDNA-16E1∧E4Δ87-92, whereas no band was apparent in the vector control sample (data not shown). Immunoprecipitation of these proteins using specific anti-16E1∧E4 rabbit polyclonal antibody confirmed that the bands were 16E1∧E4 or 16E1∧E4Δ87-92 (Fig. 6A). The products from the in vitro transcription-translation reactions were subjected to gel filtration chromatography followed by fractionation and scintillation counting to detect 3H incorporation. Both the 3H-labeled 16E1∧E4 protein and the 3H-labeled truncated 16E1∧E4Δ87-92 protein were eluted with two main peaks (Fig. 6B). According to the elution profile of protein molecular mass standards, the estimated molecular mass of the first peak eluted from the wt 16E1∧E4 protein is 66 kDa, whereas the second is 22 kDa (Fig. 6B). The protein molecular mass corresponding to the first peak eluted from the 16E1∧E4Δ87-92 reaction mixture was 66 kDa, and the second was 10 kDa (Fig. 6B). Since the predicted molecular mass of the 16E1∧E4 protein monomer is 10 kDa, the 10-, 22-, and 66-kDa species are consistent with their being monomeric, dimeric, and hexameric forms, respectively. Interestingly, the elution profile of the truncated 16E1∧E4Δ87-92 protein suggested the existence of additional smaller peaks whose molecular masses were consistent with their being dimeric, trimeric, tetrameric, and pentameric forms (Fig. 6B; the positions of the multimeric forms are shown beneath the graph). The hypothesis that 16E1∧E4 can produce multimers up to the size of hexamers was further supported by the finding that highly purified 16E1∧E4 protein migrates as species of increasing molecular mass following gel electrophoresis in the presence of low levels of SDS (Fig. 6D). Interestingly, hexameric E1∧E4 complexes have also been detected following analysis of the multimeric state of the HPV1 E1∧E4 protein, although in that case additional higher-order structures (possibly 24-mers) were also apparent (13). To determine whether the 16E1∧E4 protein can self-associate and form high-molecular-mass complexes in cultured epithelial cells, the Empigen extracts from 16E1∧E4-expressing SiHa cells were analyzed by SDS-PAGE in the presence or absence of the reducing agent DTT. Under nonreducing conditions, an ∼25-kDa band was detected in the cell extracts (Fig. 6C), suggesting that 16E1∧E4 forms dimers that are stabilized by disulfide bonds. Hexamers were not detected following Empigen extraction, which suggests that they are unlikely to be stabilized disulfide bonds. Taken together, these results suggest that the wt 16E1∧E4 protein forms hexamers and disulfide-stabilized dimers and that the ability of the 16E1∧E4Δ87-92 mutant to form such complexes is impaired.
FIG. 6.
16E1∧E4 self-associates to form multimers of 66 kDa. (A) The 3H-labeled 16E1∧E4 protein generated by in vitro transcription-translation was immunoprecipitated using anti-16E1∧E4 rabbit polyclonal antibody and separated by SDS-PAGE. Both wt and mutant (Δ87-92) E1∧E4 proteins gave a single band of ∼10 kDa. Molecular mass standards are shown on the left. (B) The 3H-labeled 16E1∧E4 proteins shown in panel A were separated on a Superdex 75 column, and the elution profile was obtained following scintillation counting. The column was calibrated using globular standards, and the molecular masses of the major 16E1∧E4 peaks are shown above the profiles. wt 16E1∧E4 eluted as a major peak of 66 kDa, which corresponds to the predicted size of 16E1∧E4 hexamers. A minor peak of 22 kDa was also apparent. Mutant 16E1∧E4 (Δ87-92) eluted as peaks of increasing molecular mass, with the smallest peak corresponding to the size of 16E1∧E4 monomers (10 kDa). The expected elution positions of E1∧E4 multimers (monomers to hexamers) are shown beneath the profile. (C) Empigen extracts from 16E1∧E4-expressing SiHa cells were analyzed by SDS-PAGE and Western blotted following treatment with DTT. The positions of monomeric 16 E1∧E4 and disulfide-stabilized 16E1∧E4 dimers are shown on the right. Complexes larger than dimers were not apparent following extraction using Empigen, which is necessary to solubilize the cytoskeleton-associated E1∧E4 protein. Molecular mass standards are shown on the left. +, present; −, absent. (D) wt 16E1∧E4 protein purified from E. coli migrates as a series of bands of increasing molecular mass when analyzed by gel electrophoresis in the presence of low levels of SDS. The largest band has a molecular mass of 66 kDa, in agreement with the results obtained using fast protein liquid chromatography. The C-terminal truncation (16E1∧E4Δ87-92) exists only as monomers and dimers.
DISCUSSION
Although several functions have been proposed for the HPV16 E1∧E4 protein, its precise role in the virus life cycle is unclear. The HPV16 E1∧E4 protein is expressed at low levels as the infected cell leaves the basal layer and migrates toward the epithelial surface, but it increases in abundance in the suprabasal cell layers (25). The high-level expression of E1∧E4 marks the onset of late events in the virus life cycle, in which vegetative viral DNA replication occurs and the viral structural proteins L1 and L2 are produced. The timing of E1∧E4 expression suggests a potential role in viral DNA replication, new progeny virion assembly, and final virus release (24).
More than 100 papillomavirus types have been DNA sequenced, allowing the classification of different virus types into supergroups (8). The E1∧E4 proteins encoded by supergroup A viruses share the conserved LLXLL motif, which is important for keratin association (37, 38), and for many viruses from this group, including HPV16, -31, and -2 (11, 25, 36-39) and -11, -18, -35, and -45 (data not shown), the predicted association with keratins has been shown experimentally. Although we do not know whether the E1∧E4 proteins of all papillomaviruses are able to associate with keratins, these results suggest that keratin association may be a general feature of the E1∧E4 proteins encoded by viruses from supergroup A. With 16E1∧E4, the association with keratins causes keratin network collapse through an unknown mechanism (11). Our data demonstrate that the 16E1∧E4 protein is associated with cellular keratins when expressed in tissue sections from HPV16-infected cervical epithelium. The perinuclear bundle pattern of reorganized keratin networks was apparent in some of the 16E1∧E4-expressing cells, indicating that 16E1∧E4-mediated keratin network reorganization can occur in vivo as well as in vitro.
The importance of the leucine cluster in keratin binding was supported by our observation, and by the observations of others (38), that 16E1∧E4ΔLLKLL no longer localizes efficiently to the keratin network. Interestingly, the N-terminal 16 amino acids, including the LLKLL motif of 16E1∧E4, when fused to GFP (Mut1) behaved in a manner similar to that of the wt 16E1∧E4 protein, showing a filamentous pattern and colocalizing with keratins when expressed in Cos7 cells. Deletion of only one amino acid at position 2 or 3 in this region, or expression of just the LLKLL motif fused to GFP, resulted in the E1∧E4 protein having a punctate staining pattern rather than keratin association (Fig. 3 and data not shown). This suggests that 16E1∧E4 has a sequence requirement in addition to the LLKLL motif for association with cellular keratins. The failure of Mut1 to localize exclusively to the keratin network may reflect lower binding affinity than full-length E4 or a failure to form multimeric structures that can associate with greater avidity.
Whether the association of 16E1∧E4 with keratins is through direct interaction or whether it involves other unknown proteins has not been established. In this study, we used coimmunoprecipitation to show that K8 and K18 can form a complex with phosphorylated and unphosphorylated 16E1∧E4. Since Empigen solubilizes keratins without destroying their antigenicity, we were able to partially purify the cellular K8 and K18 from the Empigen fraction using an anti-K8/K18 rabbit polyclonal antibody and to show that they bound purified 16E1∧E4 expressed in bacteria. Direct binding of 16E1∧E4 protein to K8/K18 was confirmed by dot blotting using purified K8, K18, vimentin, and 16E1∧E4, as well as by far-Western analysis. Considered together, these results suggest that the association of 16E1∧E4 with keratins is through direct physical interaction. Previous work on the E1∧E4 protein of HPV1, a virus from supergroup E, failed to show a direct association with keratins (13), and we speculate that the E1∧E4 proteins of such viruses, which form inclusion granules in vivo (rather than filamentous networks), may have a lower affinity for keratins than the E1∧E4 proteins of A group viruses, such as HPV16. Although 1E1∧E4 and 16E1∧E4 both contain LLXLL motifs, their upstream amino acids differ, and for HPV16 E1∧E4, the preservation of these amino acids is important for keratin binding (Fig. 3). Differences in keratin affinity and in the ability to form structures such as granules may explain the transient association with keratins reported for 1E1∧E4 (13, 39), rather than the more apparent association seen with 16E1∧E4.
The C terminus of 16E1∧E4 has previously been implicated in 16E1∧E4-mediated keratin network collapse (38), and although the multimerization state of 16E1∧E4 was not established, for 1- and 11E1∧E4, the C terminus was shown to be important (1, 2). By in vitro transcription-translation and gel filtration, it appears that 16E1∧E4 exists primarily as a hexameric complex with a fraction of the protein forming complexes the size of dimers (Fig. 6B). By contrast, the C-terminal deletion mutant (16E1∧E4Δ87-92) exists as monomers and forms hexamers at a low level compared to the wt protein. Our work is in agreement with previous studies, which have shown that the ability of a similar mutant (16E1∧E4Δ86-92) to dimerize is compromised (38). Interestingly, purified 16E1∧E4 produced from bacteria also formed high-molecular-mass complexes (in the absence of DTT) with molecular masses similar to those seen following the expression of 16E1∧E4 using a wheat germ in vitro translation system. These data suggest that 16E1∧E4 forms hexamers in vitro and that high-molecular-mass E1∧E4 structures may also exist in epithelial cells. When it is considered that K8 and K18 form heteropolymers at a 1:1 ratio in epithelial cells, it seems possible that the 16E1∧E4 protein, which appears to bind to K8/K18 through its N terminus and to form hexamers through its C terminus, may be able to cross-link the K8/K18 network (Fig. 7). Interestingly, the HPV1 E1∧E4 protein was previously shown to form multimers of hexameric size (13), as well as structures of higher molecular mass that may represent tetrameric complexes of hexamers (13). The change in the apparent size of 1E1∧E4 complexes, and in particular the loss of hexameric species when keratins were present, may result from keratin binding (13). Unlike 16E1∧E4, however, 1E1∧E4 is thought to be only briefly associated with keratin prior to the formation of inclusion granules in the cytoplasm (13, 39). There is no evidence that 1E1∧E4 induces keratin network collapse, and it is possible that the ability to form inclusion granules may be an evolutionary adaptation that is not necessary for viruses of supergroup A. Our hypothesis suggests that by acting as a cross-linker, 16E1∧E4, and possibly also other HPV types from supergroup A, can interfere with normal keratin network reorganization (Fig. 7), a possibility that is supported by our studies of keratin dynamics.
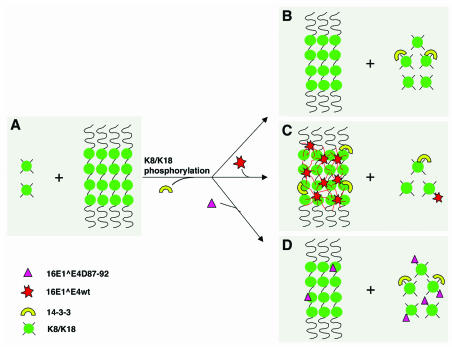
FIG. 7.
Model for the mechanism of HPV 16E1∧E4-mediated keratin network reorganization. (A) Soluble K8/K18 tetramers represent ∼5% of the total cellular keratin during G0/G1 (20). (B) When cells approach mitosis or are treated with phosphatase inhibitor, 14-3-3 binds to hyperphosphorylated K18 and increases their solubility (20). (C) wt 16E1∧E4 associates with keratins through sequences at its N terminus and is able to form multimers (hexamers) through sequences at its C terminus. This may allow the 16E1∧E4 protein to act as a keratin cross-linker and to inhibit 14-3-3-mediated keratin solubilization. (D) 16E1∧E4Δ87-92, which retains its keratin-targeting motif but has a diminished ability to multimerize (and thus to act as a cross-linker), has no effect on keratin solubilization. Multimeric 16E1∧E4 complexes are expected to associate with keratin more tightly than monomeric 16E1∧E4 due to an increase in avidity.
Previous work has shown that K8/K18 network dynamics are modulated by the cellular 14-3-3 proteins, which function as keratin solubility cofactors during the cell cycle. When K18 is phosphorylated at S33 during progression through the G2/M phase or following OA treatment, 14-3-3 proteins bind to phosphorylated K18, leading to solubilization of the keratin network and the redistribution of keratins within the cytoplasm (20, 22). If S33 within K18 is replaced by an alanine (which is unable to be phosphorylated and therefore fails to bind to 14-3-3), K8/K18 network dynamics are disturbed and the network collapses into a perinuclear bundle (20). This is similar to the pattern seen in cells expressing 16E1∧E4. In such cells, the soluble keratin pool is almost completely lost (Fig. 4A) and the 14-3-3 proteins are sequestered into the cytoskeletal compartment. Hyperphosphorylation of K18 by OA treatment did not affect the distribution of 14-3-3 and 16E1∧E4, although the level of K18 in the soluble fraction was increased (Fig. 4B and C). 16E1∧E4ΔLLKLL, which has lost the ability to bind to keratin, and 16E1∧E4Δ87-92, whose ability to self-associate is compromised, did not affect keratin dynamics, indicating that these two regions play an important role in 16E1∧E4-mediated keratin reorganization. Interestingly, 16E1∧E4Δ87-92 was present predominantly in the soluble fractions following extraction, despite appearing by immunostaining to associate with the keratin network. It seems likely that this results from its association with keratins as a monomer or dimer rather than a higher-order multimer. The increase in avidity that is associated with multimeric binding is predicted to be necessary for tight keratin binding and subsequent disruption of the intact keratin network. Interestingly, the cellular protein filaggrin, which can aggregate keratin filaments in vivo, also causes filament reorganization when expressed in vitro, although in that case the effect on keratin solubility was not examined (22).
The data presented here raise the questions of why the HPV16 E1∧E4 protein binds to keratin and what role keratin binding plays during the HPV16 life cycle. One possibility is that the 16E1∧E4 protein that is present in the cytoplasm, and which is tethered by keratins, interacts with important cellular proteins, thereby facilitating viral DNA replication or virus synthesis. 16E1∧E4-induced G2 arrest (7) is a possible example of such a function. Activated cyclin B/CDK1 is found in the cytoplasm together with 16E1∧E4 and keratins, and it has been speculated that this may create an environment conducive to HPV DNA replication rather than replication of the cellular genome (C. Davy, unpublished observations).
Taken together, our results suggest a model that may explain keratin filament reorganization (Fig. 7). Although the affinity of the N-terminal region of 16E1∧E4 for keratins is unlikely to be high, multimerization would be expected to increase the avidity of binding, leading to an inhibition of keratin solubilization despite keratin hyperphosphorylation in G2 and the binding of 14-3-3. The inhibition of keratin solubilization results in keratin collapse following the expression of 16E1∧E4 in monolayer culture and, to a lesser extent, in differentiating epithelial cells, which express the more robust K4/K13 network. Differences in the ability of 16E4 to reorganize different keratin networks have been suggested previously (11), and it appears that the K4/K13 network present in differentiating keratinocytes may be less susceptible to reorganization than the K8/K18 network found in simple epithelial cells. The precise reasons for keratin binding in vivo remain speculative, but it is possible that, as for cyclin B/CDK1, there are other proteins (both viral and cellular) that are functionally regulated by their association with E1∧E4 and their subsequent tethering to the cytoplasmic keratin network. The elucidation of such interactions should shed further light on the role of 16E1∧E4 in the virus life cycle and on the significance of its in vivo association with keratins.
Acknowledgments
We thank John Skehel, FRS, and Jonathan Stoye for their support and encouragement during the course of this work. We also thank Kate Sullivan, Stamatis Pagakis, and Jeremy Adlen for technical help in using the Deltar Vision microscope.
This work was supported largely by the United Kingdom Medical Research Council (Q.W., H.G., D.J., A.M., C.D., P.A.W., and J.D.), but also by the Association for International Cancer Research (S.S.).
REFERENCES
- 1.Ashmole, I., P. H. Gallimore, and S. Roberts. 1998. Identification of conserved hydrophobic C-terminal residues of the human papillomavirus type 1 E1∧E4 protein necessary for E4 oligomerisation in vivo. Virology 240:221-231. [DOI] [PubMed] [Google Scholar]
- 2.Bryan, J. T., K. H. Fife, and D. R. Brown. 1998. The intracellular expression pattern of the human papillomavirus type 11 E1∧E4 protein correlates with its ability to self associate. Virology 241:49-60. [DOI] [PubMed] [Google Scholar]
- 3.Caulin, C., C. F. Ware, T. M. Magin, and R. G. Oshima. 2000. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J. Cell Biol. 149:17-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, C. F., C. L. Riopel, L. S. Rott, and M. B. Omary. 1993. A significant soluble keratin fraction in ‘simple' epithelial cells. Lack of an apparent phosphorylation and glycosylation role in keratin solubility. J. Cell Sci. 105:433-444. [DOI] [PubMed] [Google Scholar]
- 5.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 6.Coulombe, P. A., and M. B. Omary. 2002. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 14:110-122. [DOI] [PubMed] [Google Scholar]
- 7.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. Millar, and J. Doorbar. 2002. Identification of a G2 arrest domain in the E1∧E4 protein of human papillomavirus type 16. J. Virol. 76:9806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Villiers, E. M. 2001. Taxonomic classification of papillomaviruses. Papillomavirus Rep. 12:57-63. [Google Scholar]
- 9.Doorbar, J., R. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. Griffin, P. Masterson, S. Stacey, Y. Mengitsu, and J. Dunlop. 2000. The E1∧E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 74:10081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doorbar, J., S. Ely, N. Coleman, M. Hibma, D. H. Davies, and L. Crawford. 1992. Epitope-mapped monoclonal antibodies against the HPV16E1-E4 protein. Virology 187:353-359. [DOI] [PubMed] [Google Scholar]
- 11.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 12.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 13.Doorbar, J., E. Medcalf, and S. Napthine. 1996. Analysis of HPV1 E4 complexes and their association with keratins in vivo. Virology 218:114-126. [DOI] [PubMed] [Google Scholar]
- 14.Dyke, S. F., W. Williams, and J. T. Seto. 1978. Glycoproteins of representative paramyxoviruses: isolation and antigenic analysis using a zwitterionic surfactant. J. Med. Virol. 2:143-152. [DOI] [PubMed] [Google Scholar]
- 15.Evans, R. M. 1998. Vimentin: the conundrum of the intermediate filament gene family. Bioessays 20:79-86. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, E., and D. W. Cleveland. 1998. A structural scaffolding of intermediate filaments in health and disease. Science 279:514-519. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, E., and K. Weber. 1994. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 63:345-382. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, S., A. Loranger, N. Daigle, and N. Marceau. 2001. Simple epithelium keratins 8 and 18 provide resistance to Fas-mediated apoptosis. The protection occurs through a receptor-targeting modulation. J. Cell Biol. 154:763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inada, H., I. Izawa, M. Nishizawa, E. Fujita, T. Kiyono, T. Takahashi, T. Momoi, and M. Inagaki. 2001. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J. Cell Biol. 155:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ku, N. O., J. Liao, and M. B. Omary. 1998. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 17:1892-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ku, N. O., R. M. Soetikno, and M. B. Omary. 2003. Keratin mutation in transgenic mice predisposes to Fas but not TNF-induced apoptosis and massive liver injury. Hepatology 37:1006-1014. [DOI] [PubMed] [Google Scholar]
- 22.Liao, J., and M. B. Omary. 1996. 14-3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol. 133:345-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowthert, L. A., N. O. Ku, J. Liao, P. A. Coulombe, and M. B. Omary. 1995. Empigen BB: a useful detergent for solubilization and biochemical analysis of keratins. Biochem. Biophys. Res. Commun. 206:370-379. [DOI] [PubMed] [Google Scholar]
- 24.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In P. M. Howley (ed.), Virology. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 25.Middleton, K. 2003. Analysis of papillomavirus E1∧E4 expression with respect to epithelial proliferation and differentiation in productive and neoplastic papillomavirus lesions. Ph.D. thesis. University of London, London, United Kingdom.
- 26.Middleton, K., W. Peh, S. A. Southern, H. M. Griffin, K. Sotlar, T. Nakahara, A. El-Sherif, L. Morris, R. Seth, M. Hibma, D. Jenkins, P. F. Lambert, N. Coleman, and J. Doorbar. 2003. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 77:10186-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moll, R., W. W. Franke, D. L. Schiller, B. Geiger, and R. Krepler. 1982. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 31:11-24. [DOI] [PubMed] [Google Scholar]
- 28.Munoz, N., and F. X. Bosch. 1997. Cervical cancer and human papillomavirus: epidemiological evidence and perspectives for prevention. Salud Publica Mex. 39:274-282. [DOI] [PubMed] [Google Scholar]
- 29.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 30.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 31.Omary, M. B., N. O. Ku, J. Liao, and D. Price. 1998. Keratin modifications and solubility properties in epithelial cells and in vitro. Subcell. Biochem. 31:105-140. [PubMed] [Google Scholar]
- 32.Paramio, J. M., M. L. Casanova, C. Segrelles, S. Mittnacht, E. B. Lane, and J. L. Jorcano. 1999. Modulation of cell proliferation by cytokeratins K10 and K16. Mol. Cell. Biol. 19:3086-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paramio, J. M., S. Lain, C. Segrelles, E. B. Lane, and J. L. Jorcano. 1998. Differential expression and functionally co-operative roles for the retinoblastoma family of proteins in epidermal differentiation. Oncogene 17:949-957. [DOI] [PubMed] [Google Scholar]
- 34.Paramio, J. M., C. Segrelles, S. Ruiz, and J. L. Jorcano. 2001. Inhibition of protein kinase B (PKB) and PKCζ mediates keratin K10-induced cell cycle arrest. Mol. Cell. Biol. 21:7449-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 76:10401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pray, T. R., and L. A. Laimins. 1995. Differentiation-dependent expression of E1-E4 proteins in cell lines maintaining episomes of human papillomavirus type 31b. Virology 206:679-685. [DOI] [PubMed] [Google Scholar]
- 37.Roberts, S., I. Ashmole, L. J. Gibson, S. M. Rookes, G. J. Barton, and P. H. Gallimore. 1994. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J. Virol. 68:6432-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts, S., I. Ashmole, S. Rookes, and P. Gallimore. 1997. Mutational analysis of the human papillomavirus type 16 E1∧E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J. Virol. 71:3554-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts, S., M. L. Hillman, G. L. Knight, and P. H. Gallimore. 2003. The ND10 component promyelocytic leukemia protein relocates to human papillomavirus type 1 E4 intranuclear inclusion bodies in cultured keratinocytes and in warts. J. Virol. 77:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos, M., C. Ballestin, R. Garcia-Martin, and J. L. Jorcano. 1997. Delays in malignant tumor development in transgenic mice by forced epidermal keratin 10 expression in mouse skin carcinomas. Mol. Carcinog. 20:3-9. [DOI] [PubMed] [Google Scholar]
- 41.Southern, S. A., I. W. McDicken, and C. S. Herrington. 2000. Evidence for keratinocyte immortalization in high-grade squamous intraepithelial lesions of the cervix infected with high-risk human papillomaviruses. Lab. Investig. 80:539-544. [DOI] [PubMed] [Google Scholar]
- 42.van Hemert, M. J., H. Y. Steensma, and G. P. van Heusden. 2001. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. Bioessays 23:936-946. [DOI] [PubMed] [Google Scholar]
- 43.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 44.Walboomers, J. M., and C. J. Meijer. 1997. Do HPV-negative cervical carcinomas exist? J. Pathol. 181:253-254. [DOI] [PubMed] [Google Scholar]