Abstract
Despite the effectiveness of currently available human immunodeficiency virus type 1 (HIV-1) therapies, a continuing need exists for new drugs to treat HIV-1 infection. We investigated the mechanism by which 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid (DSB) inhibits HIV-1 replication. DSB functions at a late stage of the virus life cycle but does not inhibit the HIV-1 protease in vitro or interfere with virus assembly or release. DSB specifically delays the cleavage of Gag between the capsid (CA) and p2, resulting in delayed formation of the mature viral core and reduced HIV-1 infectivity. Replication of simian immunodeficiency virus (SIV) was resistant to DSB; however, a chimeric SIV carrying CA-p2 sequences from HIV-1 was inhibited by the drug, indicating that susceptibility to DSB maps to the CA-p2 region of the HIV-1 Gag protein. A single point mutation at the CA-p2 cleavage site of HIV-1 conferred strong resistance to DSB, confirming the target of the drug. HIV-1 strains that are resistant to a variety of protease inhibitors were sensitive to DSB. These findings indicate that DSB specifically protects the CA-p2 cleavage site from processing by the viral protease during virion maturation, thereby revealing a novel mechanism for pharmacologic inhibition of HIV-1 replication.
Treatment of human immunodeficiency virus (HIV) infection has been dramatically enhanced by the advent of highly active antiretroviral therapy (HAART), including the combined use of drugs targeting the viral enzymes reverse transcriptase (RT) and protease (PR). Despite the therapeutic success of HAART, effective treatment requires long-term administration of the drugs. HAART is associated with undesirable side effects, and HIV type 1 (HIV-1) eventually becomes resistant to the drugs. Therefore, the identification of compounds with novel viral and cellular targets remains a high-level research priority.
Mature infectious HIV-1 particles contain an electron-dense conical core with a shell composed of the viral capsid (CA) protein. Virions mature late in the virus replication cycle through cleavage of the Gag polyprotein by PR, resulting in release of the virion proteins MA, CA, NC, and p6. Incomplete Gag processing results in virions that lack a functional core and are noninfectious (1, 3, 24). Cleavage of Gag occurs in a temporally regulated manner, with scission of the junction between CA and p2 representing the final event required for condensation of the CA shell (for a review, see reference 22). Particles containing unprocessed CA-p2 exhibit reduced core stability and are likely impaired at an early postentry step in infection in analogy to point mutations in CA that destabilize the HIV-1 core and result in impaired reverse transcription in target cells (10). These studies underscore the importance of complete processing of Gag, with formation of the mature CA protein, for HIV-1 replication.
Here we report that 3-O-{3′,3′-dimethylsuccinyl}-betulinic acid (DSB), a derivative of betulinic acid previously referred to as YK-FH312 (14), inhibits HIV-1 replication through a novel mechanism. DSB acts late in the HIV-1 life cycle to specifically retard cleavage at the CA-p2 junction of Gag by the viral PR without affecting processing at other sites in Gag. Our data suggest that the HIV-1 Gag protein is a viable target for antiviral therapy.
MATERIALS AND METHODS
Cells and viruses.
Primary CD4+ T cells were purified by positive selection from human blood drawn from healthy normal donors and were activated and grown as previously described (19). CEM cells were grown in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and antibiotics. 293T cells were grown in Dulbecco's modified Eagle medium supplemented with 10% FBS and antibiotics. The wild-type HIV-1 molecular clones R9 (11) and pNL4-3 (2) were used for these studies. Both clones carry full-length open reading frames for all HIV-1 genes. R9 contains the sequences spanning the BssHII-BamHI sites of pNL4-3 (nucleotides 712 to 8474) in the background of R7HXB2 and is therefore highly similar to pNL4-3. The PIR-1 and PIR-2 viruses were generated from the proviral clones pM46I/L63P/V82T/I84V and pL10R/M46I/L63P/V82T/I84V, respectively, obtained through the National Institutes of Health AIDS Research and Reference Reagent Program. These viruses contain multiple mutations in the PR coding sequence, resulting in resistance to a variety of HIV-1 PR inhibitors (7). Virions were produced using a calcium phosphate coprecipitation method by transfection of 293T cells, as previously described (6). HeLa-CD4/LTR-lacZ (P4) cells were used as target cells in single-cycle infection assays, as previously described (25). HIV-1 cores were isolated as previously reported (17), and yields were quantified by a p24 antigen capture enzyme-linked immunosorbent assay (ELISA) (23).
Synthesis of DSB.
DSB was synthesized and purified as previously described (13). The compound (formula weight = 585 g/mol) was dissolved in dimethyl sulfoxide at a concentration of 1 mg/ml and added directly to culture medium at the desired concentrations.
HIV-1 virus-cell fusion assay.
HIV-1 particles carrying a β-lactamase reporter protein were produced by cotransfection of 293T cells with cloned HIV-1 proviral DNA and pMM310, a construct encoding β-lactamase (BlaM) fused to the amino terminus of the virion protein Vpr (BlaM-Vpr), thereby targeting BlaM to the virion. Virus dilutions (100 μl) were added to SupT1-CCR5 cells (100,000 cells per well in 96-well plates), and cultures were incubated at 37°C for 2 h to allow virus-cell fusion. Cells were loaded with the cell-permeative, fluorescent BlaM substrate CCF2/AM (26). Upon excitation at 409 nm, CCF2/AM fluoresces green (520 nm) and the BlaM cleavage product emits at a blue wavelength (447 nm). Fusion of HIV-1 reporter particles results in delivery of active BlaM into the cytosol; thus, the ratio of blue to green fluorescence of the culture is directly correlated with the efficiency of virus-cell fusion. Cells were pelleted and resuspended in phosphate-buffered saline, and the fluorescence was measured at 447 nm and 520 nm using a microplate fluorometer (BMG FluoStar) following excitation at 409 nm. A similar assay has recently been reported (5).
Isolation of DSB-resistant HIV-1.
Selection for drug resistance was performed by growing HIV-1NL4-3 in CEM cells in the presence of increasing concentrations of DSB. CEM cells were inoculated with HIV-1 at a multiplicity of infection of 0.01 as determined by an MAGI assay (16). For the first round of selection, the drug was maintained at a concentration of 0.12 μM. The cells were split every 3 days until a mass cytopathic effect was observed (day 8). The culture supernatant was then used to inoculate fresh CEM cells, and this process was repeated in the presence of DSB concentrations of 0.34, 1.2, and 3.4 μM (cycles 2, 3, and 4). Culture supernatants were collected at day 9 following inoculation for selection cycles 2 to 4. To identify mutations conferring resistance, chromosomal DNA was extracted from the infected CEM cells, the HIV-1 gag sequence was amplified by PCR (forward primer, 5′-AAAGTAAAGCCAGAGGAGATCTCTC-3′; reverse primer, 5′-gcccttctttgccacaattg-3′), and the product was cloned into pCR3.1 (Invitrogen, Carlsbad, Calif.) for sequence analysis.
Mutagenesis.
The wild-type HIV-1NL4-3 5′ clone (12) was used together with a site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) to create the gag L363F mutation detected in the DSB-resistant virus. The mutant HIV-1NL4-3 5′ clone and the wild-type HIV-1NL4-3 3′ half-plasmids were cotransfected into CEM cells to generate the infectious mutant viruses. A full-length mutant proviral clone was also generated by transferring the SpeI-ApaI fragment containing the mutation into pNL4-3 (2).
Quantitation of HIV-1 reverse transcription in vivo.
HIV-1NL4-3 was harvested from transfected 293T cells grown in the presence and absence of DSB (2.7 μM). Virus stocks were treated with DNaseI (20 μg/ml) for 30 min at 37°C to remove contaminating plasmid DNA, and equal quantities of viruses (corresponding to 100 ng of p24) were used to inoculate P4 cells. Cells were harvested by trypsinizing at various times after infection, and DNA was isolated using a DNeasy purification kit (Qiagen, Inc.). The levels of early and late reverse transcripts in the samples were quantified by quantitative real-time PCR using Taqman probes on an ABI 7700 apparatus, as previously described (4, 20). Early RT products were detected using primers 5′-GTGCCCGTCTGTTGTGTGAC-3′ and 5′-GGCGCCACTGCTAGAGATTT-3′ and probe 5′-(FAM)-CTAGAGATCCCTCAGACCCTTTTAGTCAGTGTGG-(TAMRA)-3′. Late RT products were detected using primers 5′-TGTGTGCCCGTCTGTTGTGT-3′ and 5′-GAGTCCTGCGTCGAGAGAGC-3′ and probe 5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′. Reactions were performed using Taqman universal master mix (Perkin-Elmer), 300 nM primers, 100 nM probe, and 100 to 500 ng of template DNA in 50-μl volumes. Reaction mixtures were incubated at 50°C for 2 min and 95°C for 10 min followed by 40 cycles of amplification at 95°C for 15 s and 60°C for 1 min.
Pulse-chase assay of HIV-1 maturation.
Transfected 293T cells or HIV-1-infected CEM cells were grown overnight in the presence or absence of DSB (4.3 μM). The cells were washed with phosphate-buffered saline, starved for 20 min in cysteine- and methionine-free medium containing 5% dialyzed FBS, and pulse labeled for 30 min in the same medium after [35S]cysteine and methionine (Pro-Mix; Amersham) (0.2 mCi/ml) had been added. DSB was present throughout the procedure in the drug-treated cultures. The cells were then washed and grown in unlabeled complete medium with and without DSB, and portions of the cultures were harvested at various times following the chase. Cells were lysed in a buffer containing NP-40 (20 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 0.5% NP-40), nuclei were removed by centrifugation, and postnuclear supernatants were collected and assayed for protein content by the bicinchoninic acid method (Pierce). Viral lysates were prepared by addition of 2× radioimmunoprecipitation assay buffer to clarified culture supernatants. Viral lysates, and quantities of cell lysates normalized for protein, were subjected to immunoprecipitation in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 0.15 M NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], pH 7.5) with rabbit polyclonal antiserum specific for HIV-1 CA. Immune complexes were immobilized to protein A-conjugated agarose beads (Santa Cruz Biotechnology), and bound proteins were separated by SDS-polyacrylamide gel electrophoresis. The radioactive bands were visualized and quantified using a Fuji FLA-2000 phosphorimager.
Assay of HIV-1 core stability.
HIV-1 particles were produced by transfection of 293T cells with the R9 proviral clone. At 1 day following transfection, cells were washed to remove budded virions and grown in fresh medium. Supernatants were harvested 1 h later and placed on ice or incubated at 37°C for 4 h. To isolate HIV-1 cores, the virions were sedimented through a thin layer of 0.5% Triton X-100 into a 20 to 70% linear sucrose density gradient, as previously described (17). Fractions were collected, and core yields were determined as a percentage of the total CA in the gradient that was found in fractions with the expected density of HIV-1 cores.
RESULTS
DSB specifically inhibits cleavage of Gag at the CA-p2 junction.
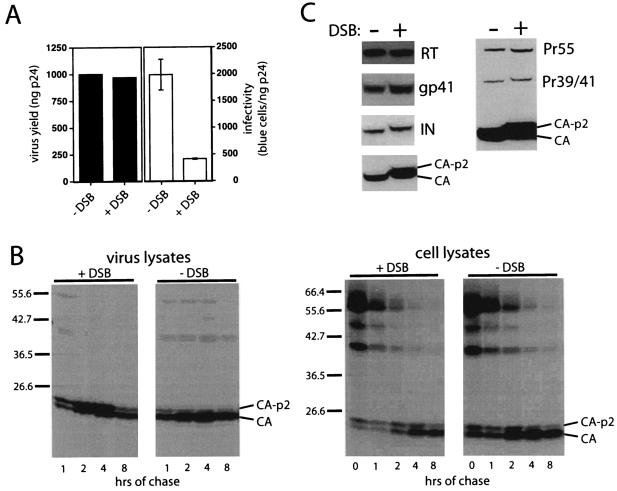
DSB is a derivative of betulinic acid that potently inhibits HIV-1 replication (15). A previous study has shown that DSB is effective at reducing viral infectivity only when the drug was present at the time of virus production. The compound does not inhibit the early steps of infection when added at the time of virus inoculation (14). In addition, we observed no effect of the drug on HIV-1 release from transfected 293T cells (Fig. 1A) or infected CEM T cells (data not shown). To determine whether DSB affects HIV-1 morphogenesis, we used a pulse-chase assay of HIV-1 maturation to compare the rates of Pr55Gag processing in the presence and absence of the drug (Fig. 1B). DSB markedly delayed the processing of the CA-p2 intermediate in virions without detectably affecting other Gag cleavage events, as revealed by analysis of intracellular levels of Gag processing intermediates. Processing of the Gag-Pol precursor was not detectably affected by DSB, as revealed by the accumulation of normal quantities of RT and integrase (IN) proteins in virions produced in the presence of the drug (Fig. 1C). The slight shift in mobility of IN demonstrated in Fig. 1C was not reproducible and was likely due to a gel artifact. DSB did not affect the levels of virion-associated gp41 or gp120 (Fig. 1C and data not shown); however, HIV-1 particles harvested from DSB-treated cells contained significant quantities of uncleaved CA-p2, as determined by Western blot analysis. These particles were fivefold-less infectious than those of control untreated virions (Fig. 1A and data not shown). Immunoblot analysis of viral lysates revealed no apparent effect of DSB on cleavage at the NC-p6 junction (data not shown). These results demonstrate that DSB specifically slows the processing of the CA-p2 junction during HIV-1 particle maturation. Accordingly, electron-microscopic analysis of HIV-1-infected cells revealed an increase in the percentage of immature progeny virions when cells were grown in the presence of DSB (data not shown).
FIG. 1.
DSB delays processing of CA-p2 during HIV-1 maturation. (A) DSB inhibits HIV-1 infectivity but not virus production. 293T cells were transfected with a wild-type HIV-1 proviral clone and grown in the presence and absence of DSB (4.3 μM). Viral supernatants were harvested and assayed for p24 content by ELISA, and infectious titers were determined by titration on P4 cells. (B) 293T cells were transfected with the wild-type HIV-1 proviral clone R9, and cells were pulse labeled with [35S]cysteine and [35S]methionine. At the indicated times, cells and virus supernatants were harvested. Cell lysates and viral lysates were immunoprecipitated with antiserum to HIV-1 CA, and immunoprecipitates were analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. (C) HIV-1 particles (harvested from infected H9 cells grown in the presence [+DSB] and absence [−DSB] of DSB) were analyzed by immunoblotting using antisera specific for RT, gp41, CA, and IN. The right panel shows an image produced by a longer exposure of the CA immunoblot, allowing detection of the incompletely processed Gag cleavage products.
DSB inhibits formation of a stable HIV-1 core.
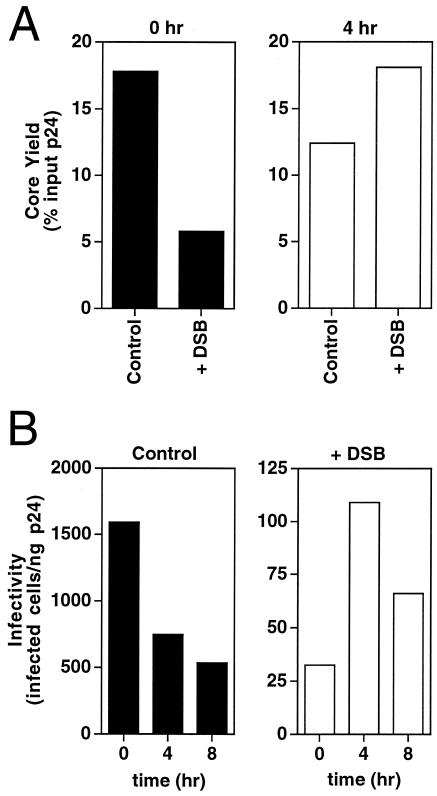
To determine whether DSB affects HIV-1 core formation, we examined the kinetics of core formation during virion maturation. Cores were isolated from fresh HIV-1 particles harvested during a 1-h culture period from cells maintained in the presence and absence of DSB, and cores were recovered by equilibrium density gradient centrifugation following a brief exposure to nonionic detergent, as previously described (10). Core yields were determined by quantifying the percentage of recovery of CA in the dense fractions of the gradients. Immediately following harvest, virions produced in the presence of DSB gave a yield of cores approximately one-third the size of the yield seen with control untreated virions (Fig. 2A). Incubation of freshly harvested drug-treated virions for 4 h at 37°C resulted in a threefold increase in core yield, while the yield of cores from the corresponding control virions was reduced by 30% during the incubation (Fig. 2A). Thus, the presence of DSB during HIV-1 budding resulted in delayed formation of stable viral cores. Freshly harvested HIV-1 particles produced in the presence of DSB were 50-fold-less infectious than corresponding untreated virions, and incubation of the viruses for 4 h reduced this difference to 7-fold (Fig. 2B). Although incubation of drug-treated virus for 4 h resulted in core yields similar to those seen with untreated HIV-1, the DSB-treated virions remained poorly infectious even upon extended incubation at 37°C (Fig. 2B). Because DSB was present in the medium of the virions, it is likely that the DSB-induced block to cleavage of Gag is not absolute. We conclude that DSB delays the formation of a stable viral core, resulting in impaired HIV-1 infectivity.
FIG. 2.
HIV-1 particles produced in the presence of DSB are delayed in the formation of stable cores. (A) Yields of HIV-1 cores recovered from freshly harvested virions (0 h) or following incubation of the virions for 4 h. (B) DSB results in delayed acquisition of HIV-1 infectivity. HIV-1 virions produced in the presence (+DSB) and absence (Control) of DSB (4.3 μM) were incubated for the indicated time periods and assayed for infectivity by titration on HeLa-CD4 indicator target cells. The data shown are representative of four independent experiments with similar outcomes.
DSB results in an early postentry defect in target cells.
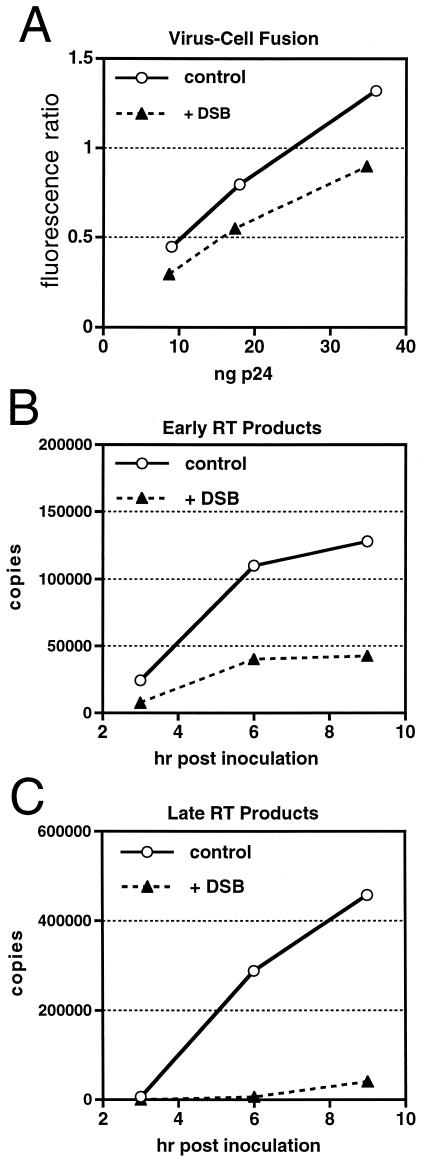
To identify the functional defect in DSB-treated HIV-1 particles, we assayed the ability of HIV-1 particles to enter target cells and to undergo reverse transcription. Virions produced in the presence of DSB exhibited only a modest reduction in fusion with target cells relative to control virions produced in the absence of drug, as determined using a fluorescent reporter-based assay of HIV-1 virus-cell fusion (Fig. 3A). However, drug-treated particles were profoundly impaired for reverse transcription in target cells, as determined by quantitative PCR assays of viral DNA synthesis. We observed a moderate reduction in synthesis of early products (minus-strand strong-stop DNA) of reverse transcription (Fig. 3B), and the defect was most pronounced at a late stage of reverse transcription occurring after the second strand-transfer event (Fig. 3C). The observed block in reverse transcription is reminiscent of the phenotype of HIV-1 particles containing aberrant or unstable cores (9, 10, 21). In agreement with these findings, we have observed that an HIV-1 mutant containing uncleaved CA-p2 is impaired for reverse transcription in target cells (our unpublished observations). We conclude that DSB specifically inhibits the terminal step of Gag processing, resulting in the formation of incompletely matured virions that are impaired for reverse transcription in target cells.
FIG. 3.
HIV-1 particles produced in the presence of DSB are impaired at an early postentry step in infection. (A) Analysis (using the BlaM-Vpr reporter assay) of control and DSB-treated (+DSB) virions for fusion with SupT1 target cells. (B and C) Quantitative analysis of intracellular reverse transcription. P4 cells were inoculated with equal quantities of HIV-1 produced in the presence (+DSB) and absence (Control) of DSB (2.7 μM). Cell lysates were prepared at the indicated times following infection, and the levels of early (B) and late (C) HIV-1 reverse transcripts were determined by quantitative PCR.
The target of DSB maps to the CA-p2 region of Gag.
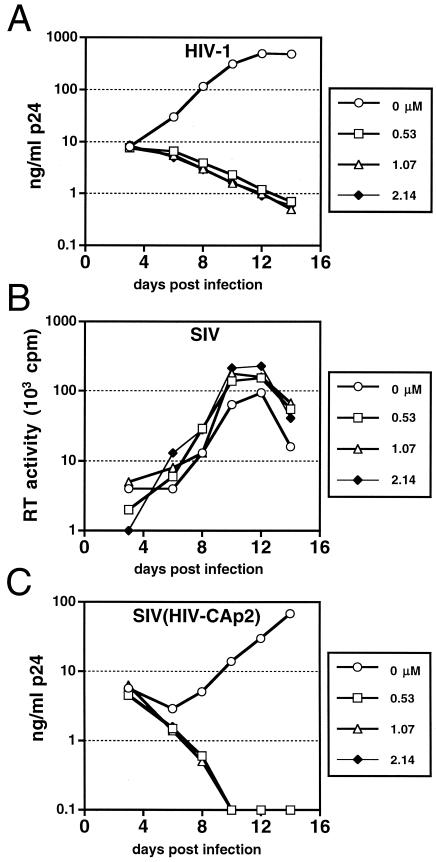
To evaluate the specificity of DSB, we assayed the ability of the compound to inhibit replication of HIV-1 and SIVmac239 in activated primary human CD4+ T cells (Fig. 4). DSB at a concentration of 0.53 μΜ completely inhibited HIV-1 replication (Fig. 4A). In contrast, the growth of simian immunodeficiency virus (SIVmac239) was not inhibited by DSB at concentrations up to 2.14 μM (Fig. 4B). To determine whether the CA-p2 region of HIV-1 was sufficient to confer drug sensitivity, we tested a chimeric virus, carrying CA and p2 sequences derived from HIV-1 in the background of SIVmac239, designated SIV(HIV-CAp2) (8). DSB potently inhibited the replication of the chimeric virus (Fig. 4C), indicating that the CA-p2 region of HIV-1 is required for drug sensitivity. Single-cycle infection assays using a variety of cloned viruses confirmed that inhibition is specific for the HIV-1 CA-p2 region and does not require Nef, the envelope glycoproteins, or the PR of HIV-1 (data not shown). Furthermore, DSB did not inhibit PR-mediated cleavage of recombinant Gag or of Gag in permeabilized immature HIV-1 particles in vitro (data not shown). These results indicate that DSB inhibits HIV-1 replication through a novel mechanism distinct from those of known HIV-1 antiviral compounds.
FIG. 4.
HIV-1 sensitivity to DSB maps to the CA-p2 region of gag. Primary CD4+ T cells were inoculated at low multiplicities of infection with HIV-1 (1 ng of p24), SIVmac239, or SIV(HIV-CAp2), and cultures were maintained in the presence and absence of the indicated (micromolar) concentrations of DSB. Virus replication was monitored by quantifying p24 (A and C) or RT (B) activity in the culture supernatants at the indicated times. (A) HIV-1; (B) SIVmac239; (C) SIV(HIV-CAp2). In this experiment, complete inhibition of replication was observed as a declining p24 level in the culture over time due to dilution of the initial virus inoculum as the cultures were replenished with fresh medium and drug.
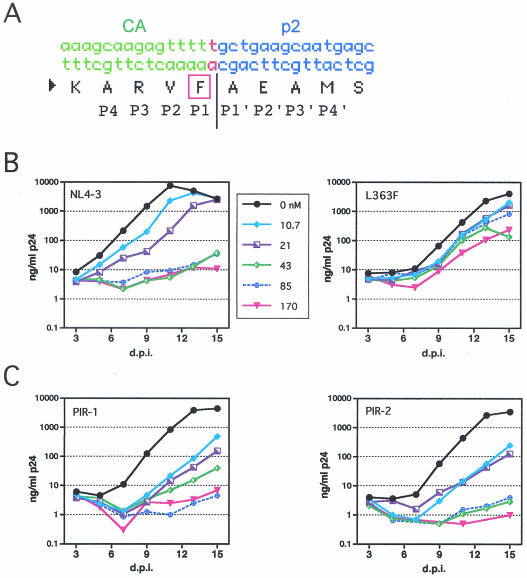
A mutation at the CA-p2 cleavage site confers resistance to DSB.
To more precisely identify the viral target of DSB, we isolated a drug-resistant HIV-1 mutant. Wild-type HIV-1 was serially passaged in CEM cells in the presence of increasing concentrations of DSB until efficient viral growth was observed. To identify mutations that contribute to drug resistance, we cloned and sequenced regions of the gag and pol genes of the resulting virus isolate. Two mutations were detected, including a single missense mutation at codon 363 of Gag corresponding to the P1 position of the CA-p2 junction (Fig. 5A). This substitution results in a Phe codon for Leu at that position. The second mutation was a G-to-A transition at position 2097, which results in a silent mutation in p6. Because this region is shared by the pol open reading frame, it also produces a substitution of Asp for Asn in p6pol. However, no substitutions in PR were detected. To determine whether the L363F substitution in Gag was responsible for the observed resistance to DSB, we engineered this single mutation into the wild-type NL4-3 proviral clone and assayed the resulting mutant for replication in the presence of DSB. The L363F recombinant virus replicated in the presence of drug concentrations that completely inhibited the replication of wild-type HIV-1 (Fig. 5B). Thus, a single amino acid substitution at the CA-p2 junction conferred strong resistance to DSB.
FIG. 5.
A mutation at the CA-p2 cleavage site confers resistance to DSB. A DSB-resistant HIV-1 mutant was isolated by repeated virus passage in the presence of the drug, and regions of gag and pol were cloned and sequenced. (A) A point mutation at the CA-p2 junction was detected which results in the substitution of Phe for Leu at codon 363 of gag. (B) The L363F mutation at the CA-p2 junction results in resistance to DSB. Cultures of CEM cells were inoculated in parallel with HIV-1NL4-3 and the L363F mutant and were maintained in the indicated concentrations of DSB (values are expressed in nanomoles). Replication was monitored by a p24 ELISA. (C) PR inhibitor-resistant viruses are sensitive to DSB. Replication of the PIR-1 and PIR-2 viruses was assayed in CEM cells in the presence of the indicated concentrations of DSB.
The observation that a single mutation in gag confers resistance to DSB suggests that the target of DSB is the Gag protein itself. The drug does not inhibit HIV-1 PR in vitro (reference 14 and our unpublished observations). However, it remains possible that the compound targets the viral PR by altering the substrate specificity of the enzyme to affect cleavage of only the CA-p2 junction. As an additional approach to probe the viral target of DSB, we tested the sensitivity of mutants that are resistant to known inhibitors of HIV-1 PR. The two viruses, PIR-1 and PIR-2, carry multiple mutations in pol that confer resistance to a variety of HIV-1 PR inhibitors (7). These viruses remained highly sensitive to DSB; however, the PIR-1 virus replicated to a limited extent in the presence of 43 nM DSB (Fig. 5C). These results suggest that PR inhibitors and DSB act through distinct mechanisms. On the basis of these results and the observation that DSB does not inhibit PR-mediated cleavage of Gag in vitro, we conclude it is unlikely that DSB acts directly on the HIV-1 PR.
Resistance to DSB is associated with normal processing of the CA-p2 junction of Gag.
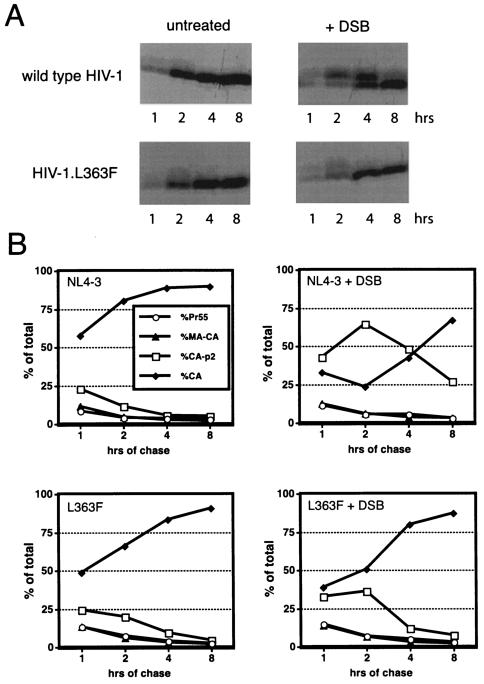
To further correlate the effects of DSB on maturation and infectivity, we performed pulse-chase studies to determine whether DSB alters the kinetics of maturation of the drug-resistant mutant (L363F). In contrast to the results seen with wild-type HIV-1, cleavage of the L363F mutant Gag protein was only slightly affected by the drug (Fig. 6). Thus, the L363F mutation resulted in normal processing of the CA-p2 junction in the presence of DSB. Resistance to DSB is therefore correlated with restored processing of Gag, reinforcing the mechanistic link between the effects of the drug on cleavage of the CA-p2 junction and inhibition of HIV-1 replication.
FIG. 6.
Resistance to DSB correlates with normal cleavage of CA-p2. CEM cells were infected with wild-type and L363F virions, and cells were subsequently grown in the presence (+DSB) and absence (−DSB) of DSB (4.3 μM). (A) Pulse-chase analysis of virion maturation. (B) Phosphorimager quantitation of the radioactivity in the bands shown in panel A. Values shown are percentages of the total Gag proteins detected in each lane.
DISCUSSION
In this report, we describe a novel mechanism for pharmacologic inhibition of HIV-1 replication. The betulinic acid derivative DSB acts late during HIV-1 maturation to specifically inhibit processing of the CA-p2 Gag intermediate, resulting in elevated accumulation of unprocessed CA-p2 and, consequently, aberrant maturation of the viral core. A previous study had suggested that DSB impairs the release of HIV-1 particles (14); however, our results indicate that the drug exhibits little, if any, inhibition of virus production. Rather, virions produced in the presence of the drug are delayed for formation of a stable core and are poorly infectious in a single cycle of infection due to impaired reverse transcription in target cells. In this respect, the observed phenotype is reminiscent of the effects of gag mutations inhibiting release of p2 that also result in unstable cores (18, 24).
Pulse-chase analysis demonstrated that DSB slowed but did not completely inhibit cleavage at the CA-p2 junction. Thus, formation of a stable HIV-1 core was delayed in the presence of the drug, but further incubation of the virions resulted in processing of most of the CA-p2 precursor and in formation of stable cores. The infectivity of fresh HIV-1 particles produced in the presence of DSB was increased when the virions were incubated for 4 h at 37°C. Nevertheless, DSB-treated virions were poorly infectious relative to untreated HIV-1 at all time points tested. These results imply that HIV-1 maturation must occur within a narrow temporal window to generate a functional viral core. Further analysis of the cores from DSB-treated virions will be required to identify the specific structural and functional defects that result in impaired reverse transcription.
We propose a mechanism in which DSB binds to the CA-p2 junction of Gag during HIV-1 particle assembly and sterically inhibits cleavage of this site. Several lines of evidence support this hypothesis. First, DSB inhibited cleavage of the CA-p2 junction but not that of other sites in Gag. DSB did not inhibit PR cleavage of Gag in vitro or affect HIV-1 RT activity. Second, DSB inhibited the replication of both HIV-1 and a chimeric SIV containing the CA-p2 segment of HIV-1, but not that of SIV itself, indicating that the inhibition is not specific for the viral PR. Third, a single amino acid substitution at the CA-p2 cleavage site of HIV-1 conferred strong resistance to DSB. Fourth, HIV-1 mutants resistant to PR inhibitors were sensitive to DSB, and the DSB-resistant virus remained sensitive to PR inhibitors (our unpublished observations). Taken together, these data are most consistent with the conclusion that the HIV-1 Gag protein, more specifically the CA-p2 cleavage site, is the likely target of DSB. Our results also suggest that the HIV-1 Gag precursor represents a viable target for the development of small-molecule inhibitors of HIV-1 replication. Despite the attractiveness of this model, we cannot yet unequivocally exclude the possibility that DSB might bind to the HIV-1 PR in an unusual way to inhibit only the final cleavage site in HIV-1 Gag.
HIV-1 readily acquired resistance to DSB through acquisition of a single amino acid substitution at the CA-p2 junction (Leu363 to Phe). A secondary mutation was detected in the p6pol transframe region; this mutation might affect processing of Gag, but we have not yet investigated this possibility since the L363F mutation alone conferred significant resistance. The leucine codon at this position is highly conserved among HIV-1 isolates, and the mutant virus replicated more slowly than the wild type. The DSB resistance was also markedly attenuated in activated primary CD4+ T cells (our unpublished observations). Despite the apparent ease with which HIV-1 acquired resistance to DSB in culture, the high conservation of the Leu residue and the impaired replicative fitness of the DSB-resistant virus together suggest that HIV-1 does not easily acquire resistance to DSB in vivo. DSB may therefore represent an attractive starting compound for the development of novel antiviral drugs.
Acknowledgments
We thank Heinrich Gottlinger for the SIV(HIVCA-p2) construct, Toshiaki Kodama for the full-length SIVmac239 clone, and Luc van Kaer for helpful suggestions for the manuscript. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program: pL10R/M46I/L63P/V82T/I84V and pM46I/L63P/V82T/I84V (from Emilio Emini) and pNL4-3 (from M. Martin).
This work was supported by NIH grants AI50423 to C.A., AI49096 to C.H.C., and AI33066 to K.-H.L.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashorn, P., T. J. McQuade, S. Thaisrivongs, A. G. Tomasselli, W. G. Tarpley, and B. Moss. 1990. An inhibitor of the protease blocks maturation of human and simian immunodeficiency viruses and spread of infection. Proc. Natl. Acad. Sci. USA 87:7472-7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 5.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 8.Dorfman, T., and H. G. Gottlinger. 1996. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70:5751-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 10.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 76:5667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:343-350. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto, F., Y. Kashiwada, L. M. Cosentino, C. H. Chen, P. E. Garrett, and K. H. Lee. 1997. Anti-AIDS agents—XXVII. Synthesis and anti-HIV activity of betulinic acid and dihydrobetulinic acid derivatives. Bioorg. Med. Chem. 5:2133-2143. [DOI] [PubMed] [Google Scholar]
- 14.Kanamoto, T., Y. Kashiwada, K. Kanbara, K. Gotoh, M. Yoshimori, T. Goto, K. Sano, and H. Nakashima. 2001. Anti-human immunodeficiency virus activity of YK-FH312 (a betulinic acid derivative), a novel compound blocking viral maturation. Antimicrob. Agents Chemother. 45:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashiwada, Y., F. Hashimoto, L. M. Cosentino, C. H. Chen, P. E. Garrett, and K. H. Lee. 1996. Betulinic acid and dihydrobetulinic acid derivatives as potent anti-HIV agents. J. Med. Chem. 39:1016-1017. [DOI] [PubMed] [Google Scholar]
- 16.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 73:8824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krausslich, H.-G., M. Facke, A.-M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundquist, C. A., M. Tobiume, J. Zhou, D. Unutmaz, and C. Aiken. 2002. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J. Virol. 76:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munk, C., S. M. Brandt, G. Lucero, and N. R. Landau. 2002. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc. Natl. Acad. Sci. USA 99:13843-13848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 23.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12:288-293. [DOI] [PubMed] [Google Scholar]
- 24.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H.-G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, J., and C. Aiken. 2001. Nef enhances human immunodeficiency virus type 1 infectivity resulting from intervirion fusion: evidence supporting a role for Nef at the virion envelope. J. Virol. 75:5851-5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zlokarnik, G., P. A. Negulescu, T. E. Knapp, L. Mere, N. Burres, L. Feng, M. Whitney, K. Roemer, and R. Y. Tsien. 1998. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science 279:84-88. [DOI] [PubMed] [Google Scholar]