Abstract
A Rho GTPase inactivation domain (RID) has been discovered in the multifunctional, autoprocessing RTX toxin RtxA from Vibrio cholerae. The RID domain causes actin depolymerization and rounding of host cells through inactivation of the small Rho GTPases Rho, Rac and Cdc42. With only a few toxin proteins containing RID domains in the current sequence database, the structure and molecular mechanisms of this domain are unknown. Using comparative sequence and structural analyses, we report homology inference, fold recognition, and active site prediction for RID domains. Remote homologs of RID domains were identified in two other experimentally characterized bacterial virulence factors: IcsB of Shigella flexneri and BopA of Burkholderia pseudomallei, as well as in a group of uncharacterized bacterial membrane proteins. IcsB plays an important role in helping Shigella to evade the host autophagy defense system. RID domain homologs share a conserved diad of cysteine and histidine residues, and are predicted to adopt a circularly permuted papain-like thiol protease fold. RID domains from MARTX toxins and virulence factors IcsB and BopA thus could function as proteases or acyltransferases acting on host molecules. Our results provide structural and mechanistic insights into several important proteins functioning in bacterial pathogenesis.
Keywords: Rho GTPase inactivation, cysteine protease domain, papain-like fold, multifunctional, autoprocessing RTX toxins, Shigella virulence factor IcsB, structure prediction, homology inference
Introduction
Certain Gram-negative bacterial pathogens use type I secretion systems to export large exoproteins called RTX (repeats-in-toxin) toxins, including Escherichia coli α-hemolysin, Pasteurella haefflolytica leukotoxin, and Bordetella pertussis adenylate cyclase toxin. 1 These toxins are characterized by repeats of a glycine and aspartate-rich, calcium-binding sequence motif. A family of multifunctional, autoprocessing RTX toxins (MARTX) such as RtxA from Vibrio cholerae (VcRtxA) 2 has been recently identified. 3 MARTX toxins have several distinct types of repeats present at the N- and C-termini, and a middle mosaic region containing multiple domains with virulence activity. 3 They also possess a cysteine protease domain with a caspase-like fold that is essential for autoprocessing of these toxins to release the virulence activity domains to the cytosol of host cells. 4,5 Two distinct virulence activity domains in VcRtxA have been characterized that each can cause the host cell rounding phenotype: an actin cross-linking domain (ACD) 6,7 and a Rho GTPase inactivation domain (RID). 8 While the ACD domain modifies the cytoskeleton by directly acting on the actin molecules, the RID domain induces actin depolymerization and rounding of host cells through inactivation of the small Rho GTPases Rho, Rac and Cdc42. 8 The structural and molecular mechanisms of RID domains are unknown. In this study, we report distant homology inference, fold recognition, and active site prediction for RID domains through comparative sequence and structural analyses. We found that RID domains are remotely related to two experimentally characterized virulence factors: IcsB of Shigella flexneri 9,10 and BopA of Burkholderia pseudomallei, 11,12 as well as a group of uncharacterized bacterial membrane proteins. RID domain homologs are predicted to adopt a circularly permuted papain-like thiol protease fold with a conserved cysteine and histidine catalytic diad. Our prediction results suggest that RID domains in MARTX toxins and virulence factors IcsB and BopA could function as proteolytic enzymes or acyltransferases acting on host molecules.
Materials and Methods
The PSI-BLAST program 13 was used to search for homologs of the experimentally determined RID domain-containing region of V. cholerae RtxA (residues 2552-3099, gi|153817921) against the NCBI non-redundant database (November 26 2008; 7,365,651 sequences; 2,547,387,767 letters), with an inclusion e-value cutoff of 0.001. Found homologs were clustered and one representative sequence from each group was used to initiate new PSI-BLAST iterations to ensure maximum coverage. Manual inspections of PSI-BLAST hits above the default e-value cutoff were conducted to search for remote homologs of RID domains, and the same PSI-BLAST search strategy was used for these remote homologs. RID homologs were submitted to the web server of HHpred,14 a sequence similarity search method based on profile-profile comparison, 15 to search for distant relationships against a non-redundant structure database (pdb70) and Pfam 16 database (version 23.0). Domain architecture analyses were made by submitting sequences to protein domain database servers such as CDD, 17 Pfam and SMART. 18 Multiple sequence alignments were constructed by using the PROMALS3D program. 19 Manual adjustment of the alignments was made with guidance from available 3D structures and secondary structure predictions by PSIPRED. 20
Results and Discussions
Defining the RID domain region
The RID domain-containing region was originally mapped to residues 2552-3099 in VcRtxA (NCBI gene identification (gi) number 4455065). 8 This region alone can cause Rho GTPase inactivation and cell rounding. 8 The PSI-BLAST 13 program was used to search for homologs of residues 2552-3099 in VcRtxA. The first 100 residues (2552-2651) showed significant sequence similarity to several domains in known structures. These domains are also from bacterial toxins including the Pasteurella multocida toxin PMT (Protein Data Bank (PDB) id: 2ebf) 21 and Clostridium difficile toxin B (PDB id: 2bvl). 22 They adopt a fold of a four helix bundle and have been shown to associate with anionic lipids and contribute to membrane localization of these toxins. This helical domain itself in VcRtxA is unlikely to convey the activity of Rho GTPase inactivation as such an activity has not been reported in its homologs in other toxins. The C-terminal regions of this helical domain in both PMT and toxin B contain putative virulence activity domains. For example, a glycosyltransferase domain responsible for inactivation of RhoA is present right after the helical domain in toxin B. 22 Thus the C-terminal portion (residues 2653-3099) after this helical domain in the experimentally determined RID region in VcRtxA is likely to harbor the virulence activity of Rho GTPase inactivation. We refer to this region (residues 2653-3099) as the RID domain in this study, as opposed to the originally proposed region of residues 2552-3099. 8
Sequence similarity searches for RID domains
PSI-BLAST searches were conducted using the re-defined region of VcRtxA RID domain. With an e-value inclusion threshold of 0.001, these searches converged to 18 domains from 17 proteins in the NCBI non-redundant sequence database. Domain content analysis revealed that these proteins are MARTX toxins from various strains of Vibrio cholerae and a few other bacterial pathogens such as Vibrio vulnificus, Listonella anguillarum, and Proteus mirabilis. The limited number of RID domains in these toxins could have prevented generation of diverse sequence profiles for detection of remote homologs above the significance cutoff.
To investigate if remote homologs of RID domains exist, we inspected PSI-BLAST hits above the default e-value cutoff (0.001). A protein named BopA from Burkholderia thailandensis was found to have a marginal e-value of 0.05. This protein shares a sequence motif present in all RID domains that contains a conserved cysteine residue. PSI-BLAST searches using BopA as the query found closely related sequences in various Burkholderia species as well as an experimentally characterized protein IcsB from Shigella flexaneri. 10 RID domains were also found as significant hits (e-value <0.001) using BopA as the query, suggesting that they are evolutionarily related.
Another marginally significant PSI-BLAST hit of the VcRtxA RID domain is a hypothetical protein from Methylobacterium radiotolerans (gi|170748086, e-value: 0.062). The region of this PSI-BLAST hit (residues 330-442) contains the same sequence motif with a conserved cysteine. PSI-BLAST iterations from this protein detected a group of bacterial proteins as well as the RID domains as significant hits (e-value <0.001). Most of these bacterial proteins are annotated as hypothetical proteins, and a few of them are annotated as ‘membrane-bound protease’ (e.g., gi|154248066) or ‘Zn-dependent proteases’ (e.g., gi|163857530).
The RID domains and the homologous regions in the detected remote homologs do not contain known domains in public protein domain databases, such as CDD, 17 Pfam 16 and SMART. 18 None of their structures has been solved in the current PDB database. We used the HHpred server 14 of profile-profile comparison to search for their remote homologs against a non-redundant structure database (pdb70) and Pfam database. Weak sequence similarities were detected against a protein named PPPDE1 (permuted papain fold peptidases of dsRNA viruses and eukaryotes) with a known structure (PDB id: 3ebq, solved by Structural Genomics Consortium, unpublished results). For example, the hypothetical protein from Methylobacterium radiotolerans (gi|170748086) found 3ebq with a probability score of 47.2. Originally described in a sequence analysis work, 23 PPPDE family proteins have a circularly permuted papain-like fold with a conserved cysteine and histidine diad for protease or acyltransferase activity. In the HHpred alignment, the conserved cysteine residue in RID domain homologs was aligned to the catalytic cysteine in the PPPDE1 protein. Multiple sequence alignments 19 of RID domain homologs also revealed an invariant histidine residue N-terminal to the conserved cysteine residue (Figure 1), which is also reminiscent of the active site arrangement of a circularly permuted papain-like fold.
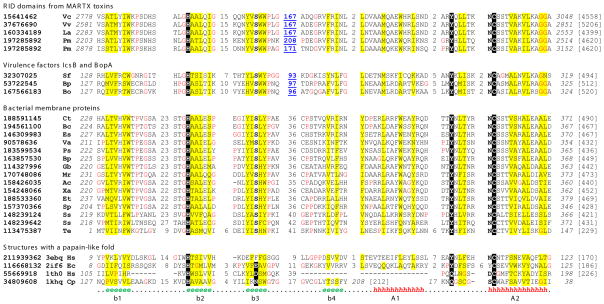
Figure 1. Multiple sequence alignments of RID domain homologs.
Catalytically-important residues are shaded in black. Non-polar residues in positions with mainly hydrophobic residues are shaded in yellow. Glycines and prolines, frequently being in the turn regions, are colored in red. Starting and ending residues numbers (italic), as well as sequence lengths (in brackets), are shown. Insertion regions in the alignment are replaced by the numbers of residues. The long insertion regions in RID domains and IcsB/BopA proteins are represented in blue, bold and underlined numbers. Consensus secondary structures in structural cores are shown for the four structures (h: α-helix; e: β-strand). The proteins are identified by their NCBI gene identification (gi) numbers, followed by the species name abbreviations: Ac, Azorhizobium caulinodans; Bp, Bordetella petrii; Bc, Burkholderia cenocepacia; Bo, Burkholderia oklahomensis; Bp, Burkholderia pseudomallei; Cp, Carica papaya; Ct, Cupriavidus taiwanensis; Es, Enterobacter sp.; Et, Erwinia tasmaniensis; Ec, Escherichia coli; Gb, Granulibacter bethesdensis; Hs, Homo sapiens; La, Listonella anguillarum; Mr, Methylobacterium radiotolerans; Pm, Proteus mirabilis; Ps, Providencia stuartii; Sp, Serratia proteamaculans; Sf, Shigella flexneri; Ss, Synechococcus sp.; Te, Trichodesmium erythraeum; Va, Vibrio angustum; Vc, Vibrio cholerae; Vv, Vibrio vulnificus; Xa, Xanthobacter autotrophicus. PDB ids for sequences with known structures are shown after the gi numbers. The first three structures (3ebq, 2if6 and 1th0) adopt a circularly permuted papain-like fold and the last structure (papain from Carica papaya, 1khq) has a papain-like fold.
Sequence and structure characterization of RID domain homologs
In the MEROPS 24 peptidase database, papain-like thiol proteases belong to clan CA. With 24 distinct families, this clan represents the most divergent group of evolutionarily related cysteine peptidases. The circularly permuted papain-like proteases are grouped in clan CE, currently comprising 6 peptidase families. Several bacterial virulence factors, such as Yersinia YopJ (peptidase family C55), Yersinia YopT and Pseudomonas AvrPphB (peptidase family C58), have been shown to be (circularly permuted) papain-like proteases. 25,26 In the Structural Classification of Proteins (SCOP) database, 27 papain-like proteases and circularly permuted papain-like proteases are grouped in the same superfamily under the ‘cysteine proteinases’ fold, suggesting that they are evolutionarily related. Aside from structural similarities, significant sequence similarities have been observed for papain-like proteases and their circularly permuted versions. 23,28 Catalytically active members of these proteins possess a conserved cysteine residue (in rare cases substituted by a serine residue) that uses the sidechain thiol group as the nucleophile to attack the peptide bond of the substrate. A conserved histidine residue functions as a general base/acid for proton transfer. These domains basically act as acylhydrolases such as peptidases or acyltransferases. For example, the SCOP fold of ‘cysteine proteinases’ also includes transglutaminases that transfer the γ-carboxyl group of glutamine to the ε-amino group of lysine or other primary amines. 29,30
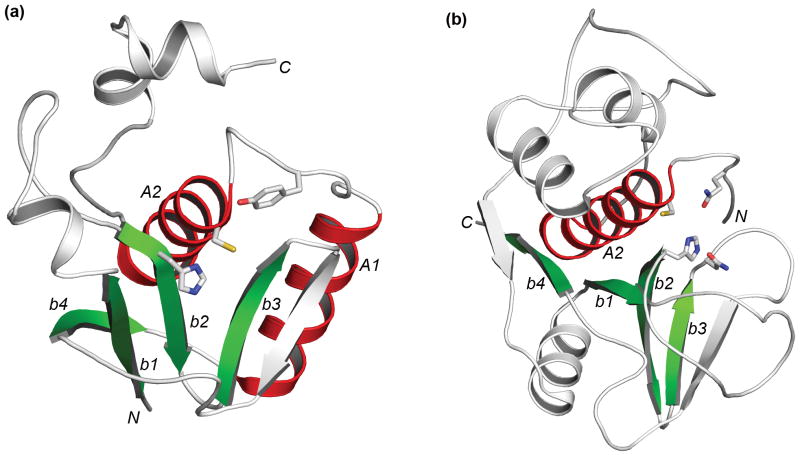
The structural core of the papain-like fold consists of a mainly anti-parallel β-sheet forming a barrel-like sub-domain, and one or several surrounding α-helices. The catalytic histidine and cysteine are located at the beginning of one β-strand and at the beginning of one α-helix, respectively. In papain-like proteases, the catalytic cysteine resides N-terminal to the catalytic histidine (Figure 2b), whereas in circularly-permuted papain-like proteases the order is reversed (Figure 2a). Regions outside the structural core exhibit great divergence in different structures. 28 For example, two circularly permuted papain-like structures PPPDE1 (PDB id: 3ebq) and Senp2 (PDB id:1th0) 31 only share three β-strands (b1, b2, b3 in Figure 1) and one α-helix (A2 in Figure 1) as superimposable core secondary structural elements.
Figure 2. Diagrams of representative structures with (a) a circularly permuted papain-like fold (pdb: 3ebq) and (b) a papain-like fold (pdb: 1khq).
α-Helices and β-strands in structural core regions are colored in red and green respectively. They are labeled in accordance with Figure 1. Other parts of the structures are colored in gray. Sidechains of catalytically important residues (highlighted in black background in Figure 1) are shown as sticks. N- and C- termini are marked. These diagrams are made by PyMOL.
Multiple sequence alignments of RID domain homologs and several known structures suggest the presence of core secondary structural elements of a circularly permuted papain-like fold in RID domains and their remote homologs (Figure 1). The sequences of RID domain homologs are most similar to structures 3ebq (a PPPDE family member) and 2if6 (a bacterial hypothetical protein, solved by New York and Structural Genomix Research Consortium, unpublished results). Among the core secondary structural elements, strands b1, b2 and b3 form a β-meander motif (Figure 2). Strand b4 is anti-parallel to b1 and completes the β-barrel-like structure. Like PPPDE proteases, two α-helices (A1 and A2) follow the β-sheet in RID domain homologs (Figure 1). Catalytic histidine and cysteine are located at the beginning of strand b2 and helix A2, respectively. An asparagine residue before the catalytic cysteine serves as the N-cap for helix A2 in 3ebq. This residue is also conserved in RID domain homologs (Figure 1). For papain-like thiol proteases, usually a conserved polar residue (Asx/Glx) in strand b3 forms an interaction with the catalytic histidine. However, the corresponding regions in the PPPDE family (3ebq) and RID domain homologs do not have such a conserved Asx/Glx residue (Figure 1, a conserved serine in strand b3 is present in RID domains instead), suggesting a catalytic diad (Cys-His) instead of a catalytic triad (Cys-His-Asx/Glx) functioning in these proteins. Another highly conserved residue in both RID homologs and the PPPDE family is a tyrosine (Figure 1) located several residues before the catalytic cysteine. Papain family proteases have a conserved glutamine residue in such a position, which has been shown to be part of the oxyanion hole that stabilizes the main-chain carbonyl group of the P1 residue of the substrate. 21,32
A distinct feature of RID domain homologs is the long insertion between strands b3 and b4 (Figure 1). In particular, these insertion regions in the RID domains of MARTX toxins are more than 150 residues, and mainly consist of predicted α-helices. They could form a (sub)domain that contributes to substrate binding or interactions with other proteins. Corresponding regions in virulence factors IcsB and BopA are also relatively long (>90 residues, Figure 1).
Domain architecture and cellular functions of RID domain homologs
Three groups of RID domain homologs were detected: RID domains in MARTX toxins, bacterial virulence factors IcsB and BopA, and a group of closely related bacterial proteins.
RID domains
RID domains were discovered in several MARTX toxins. MARTX toxins share a similar overall domain organization 3 with several types of repeats present in the N- and C-terminal regions. A cysteine protease domain with a caspase-like fold 4 is present before the C-terminal repeats and is responsible for autoprocessing of these proteins to release domains with virulence activity inside host cell cytosol. 3 The middle region has a mosaic domain structure and contains a variety of putative virulence activity domains in different MARTX toxins. 3 The RID domain is one of the experimentally demonstrated virulence activity domains. 8 Sequence database searches revealed a limited phyletic distribution of RID domains from closely related Gram-negative pathogenic species such as V. cholerae, V. vulnificus, L. anguillarum, and P. mirabilis. RID domain-containing MARTX toxins were also found in the unreleased Xenorhabdus bovienii and Xenorhabdus nematophila genomes. 3 The MARTX from P. mirabilis has two copies of RID domains.
Rho GTPases are targeted by various bacterial virulence factors to modify the plasticity of actin cytoskeleton. 33 As molecular switches, Rho GTPases cycle between GDP-bound inactive state and GTP-bound active state. 34 GAPs (GTPase-activating proteins) facilitate the hydrolysis of GTP to GDP for Rho GTPases. On the other hand, GEFs (GDP-GTP exchange factors) activate Rho GTPases by releasing GDP and allowing binding of GTP. Inactivation of Rho GTPases by the VcRtxA RID domain leads to actin depolymerization and rounding of host cells. 8 Four mechanisms of Rho GTPase inactivation used by bacterial virulence factors have been discovered, including covalent modification of Rho-like GTPases, proteolytic cleavage of Rho-like GTPases, mimicry of host cell GAPs, and dephosphorylation of proteins regulating upstream signaling pathways. 8,33 RID domain-induced cell rounding and actin depolymerization were prevented and rapidly reversed by CNF1-induced constitutive activation of Rho GTPases, suggesting that the RID domain does not directly act on Rho GTPases by proteolytic cleavage or glycosylation. 8 Experiments also showed that the RID domain lacks GAP activity and phosphatase activity. 8 Thus, the RID domain might adopt a different mechanism of Rho GTPase inactivation than the four known inactivation schemes. Several possible mechanisms of the RID domain have been proposed, including regulation of the activation state of Rho GTPases, activation of GAPs, inactivation of GEFs, and interference with signaling pathways upstream of GAPs and GEFs. 8 The prediction of a circularly permuted papain-like fold and the presence of conserved catalytic residues suggest that the RID domain carries out its function by using a peptidase or acyltransferase reaction. For example, one possible mechanism of the RID domain could be inactivation of GEFs by proteolytic cleavage. The substrate(s) of the RID domain remain to be experimentally discovered. The presence of a membrane-binding helical domain just N-terminal to the RID domain in MARTX toxins suggests that the RID substrate(s) have a membrane localization. Interestingly, structural studies revealed that a highly divergent papain-like protease domain is also present C-terminal to such a helical domain in the P. multocida toxin PMT 21.
IcsB and BopA
The Shigella IcsB protein has been characterized as a virulence factor delivered to host cells by the type III secretion system. 9 A subsequent functional study suggests that IcsB is important for Shigella to escape autophagy, 10 a host defense system that engulfs and sequesters bacteria in membrane-bound organelles such as phagosomes. When IcsB is mutated, the VirG protein (also called IcsA) of Shigella induces autophagy by binding to the host autophagy protein Atg5. 10 In non-mutant Shigella, IcsB inhibits such a binding event and thus prevents autophagy. 10 Interestingly, the function of VirG is to induce actin polymerization at one end of the bacteria to facilitate their motility inside the host cells. Therefore IcsB also has a function related to actin cytoskeleton. Contrary to the actin depolymerization phenotype caused by the RID domain, IcsB indirectly facilitates actin polymerization by protecting VirG from being recognized by the host autophagy machinery. The molecular mechanism of IcsB has not been revealed by experimental studies. Our prediction results suggest that IcsB could help protect VirG by proteolysis of Atg5, or via an acyltransferase activity that abolishes the interaction between VirG and Atg5.
The BopA protein from Burkholderia pseudomallei, a relatively close homolog of IcsB, is also a putative type III secreted virulence factor. 11 Recent studies showed that BopA has a similar function to IcsB in mediating bacterial evasion of autophagy. 12 Close homologs of IcsB and BopA were only identified in Shigella and Burkholderia species. IcsB and BopA are about 500 amino acid residues in length. The regions homologous to RID domains are located at the C-termini of these proteins. The N-terminal regions of about 120 residues in these proteins harbor a SicP binding domain (Pfam entry PF09119). SicP is a chaperone that maintains the stability of certain bacterial proteins and assists their secretion. 35 The Shigella flexneri IpgA protein, encoded by a gene immediately downstream of the icsB gene, serves as a chaperone required for stabilization and secretion of IcsB. 9 Similarly, the BicP protein of B. pseudomallei is a putative chaperone for BopA. 12 Thus the SicP binding domains in IcsB and BopA proteins may facilitate chaperone-binding and play a role in their stabilization and secretion.
Uncharacterized bacterial membrane proteins
A third group of RID domain homologs are from bacterial proteins mostly annotated as hypothetical proteins. Although a couple of these proteins are annotated as ‘membrane bound protease’ or ‘Zn-dependent proteases’, we found no experimental studies on them, nor did protein domain database searches reveal any known protease domains in them. These proteins also have a limited phyletic distribution. They are mainly from the proteobacteria species, and a couple of them are from the cyanobacteria species. Most of these proteins have a signal peptide and several transmembrane segments at the N-termini, suggesting that they are secreted membrane proteins. The regions homologous to RID domains are located at the C-termini of these proteins. As an exception, a relatively divergent member in this group from Trichodesmium erythraeum does not have transmembrane regions (gi|113475387, Figure 1). Its open reading frame starts right from the predicted β-strand b1, which indirectly confirms the domain boundaries of other RID domains. The cellular functions of these putative peptidases are yet to be revealed by experimental studies.
Conclusions
We report homology inference, fold recognition, and active site prediction for RID domains present in certain multifunctional, autoprocessing RTX toxins from bacterial pathogens. Remote homologs of RID domains were found in bacterial virulence factors IcsB of Shigella flexneri and BopA of Burkholderia pseudomallei, as well as in a group of uncharacterized bacterial membrane proteins. RID domain homologs are predicted to adopt a circularly permuted papain-like thiol protease fold with a conserved cysteine and histidine catalytic diad. RID domains of MARTX toxins and IcsB/BopA could function as proteolytic enzymes or acyltransferases acting on host molecules. Our computational analyses offer insights into the structural mechanisms of these bacterial virulence factors, generate relevant hypotheses, and facilitate experimental design for them.
Acknowledgments
We would like to thank Lisa Kinch and Dorothee Staber for critical reading of the manuscript and helpful suggestions. This work was supported in part by NIH grant GM67165 and Welch foundation grant I1505 to NVG.
References
- 1.Czuprynski CJ, Welch RA. Biological effects of RTX toxins: the possible role of lipopolysaccharide. Trends Microbiol. 1995;3(12):480–483. doi: 10.1016/s0966-842x(00)89016-2. [DOI] [PubMed] [Google Scholar]
- 2.Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ. Identification of a vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci U S A. 1999;96(3):1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satchell KJ. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect Immun. 2007;75(11):5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lupardus PJ, Shen A, Bogyo M, Garcia KC. Small molecule-induced allosteric activation of the Vibrio cholerae RTX cysteine protease domain. Science. 2008;322(5899):265–268. doi: 10.1126/science.1162403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sheahan KL, Cordero CL, Satchell KJ. Autoprocessing of the Vibrio cholerae RTX toxin by the cysteine protease domain. EMBO J. 2007;26(10):2552–2561. doi: 10.1038/sj.emboj.7601700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordero CL, Kudryashov DS, Reisler E, Satchell KJ. The Actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J Biol Chem. 2006;281(43):32366–32374. doi: 10.1074/jbc.M605275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fullner KJ, Mekalanos JJ. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 2000;19(20):5315–5323. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheahan KL, Satchell KJ. Inactivation of small Rho GTPases by the multifunctional RTX toxin from Vibrio cholerae. Cell Microbiol. 2007;9(5):1324–1335. doi: 10.1111/j.1462-5822.2006.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa M, Suzuki T, Tatsuno I, Abe H, Sasakawa C. IcsB, secreted via the type III secretion system, is chaperoned by IpgA and required at the post-invasion stage of Shigella pathogenicity. Mol Microbiol. 2003;48(4):913–931. doi: 10.1046/j.1365-2958.2003.03489.x. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307(5710):727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 11.Stevens MP, Haque A, Atkins T, Hill J, Wood MW, Easton A, Nelson M, Underwood-Fowler C, Titball RW, Bancroft GJ, Galyov EE. Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology. 2004;150(Pt 8):2669–2676. doi: 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- 12.Cullinane M, Gong L, Li X, Lazar-Adler N, Tra T, Wolvetang E, Prescott M, Boyce JD, Devenish RJ, Adler B. Stimulation of autophagy suppresses the intracellular survival of Burkholderia pseudomallei in mammalian cell lines. Autophagy. 2008;4(6):744–753. doi: 10.4161/auto.6246. [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 16.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36(Database issue):D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37(Database issue):D205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letunic I, Doerks T, Bork P. SMART 6: recent updates andnew developments. Nucleic Acids Res. 2009;37(Database issue):D229–232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36(7):2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292(2):195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 21.Kitadokoro K, Kamitani S, Miyazawa M, Hanajima-Ozawa M, Fukui A, Miyake M, Horiguchi Y. Crystal structures reveal a thiol protease-like catalytic triad in the C-terminal region of Pasteurella multocida toxin. Proc Natl Acad Sci U S A. 2007;104(12):5139–5144. doi: 10.1073/pnas.0608197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler MO, Jank T, Aktories K, Schulz GE. Conformational changes and reaction of clostridial glycosylating toxins. J Mol Biol. 2008;377(5):1346–1356. doi: 10.1016/j.jmb.2007.12.065. [DOI] [PubMed] [Google Scholar]
- 23.Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3(11):1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- 24.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36(Database issue):D320–325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290(5496):1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 26.Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell. 2002;109(5):575–588. doi: 10.1016/s0092-8674(02)00766-3. [DOI] [PubMed] [Google Scholar]
- 27.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 28.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4(2):R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorand L, Conrad SM. Transglutaminases. Mol Cell Biochem. 1984;58(1–2):9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- 30.Makarova KS, Aravind L, Koonin EV. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 1999;8(8):1714–1719. doi: 10.1110/ps.8.8.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reverter D, Lima CD. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure. 2004;12(8):1519–1531. doi: 10.1016/j.str.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Menard R, Carriere J, Laflamme P, Plouffe C, Khouri HE, Vernet T, Tessier DC, Thomas DY, Storer AC. Contribution of the glutamine 19 side chain to transition-state stabilization in the oxyanion hole of papain. Biochemistry. 1991;30(37):8924–8928. doi: 10.1021/bi00101a002. [DOI] [PubMed] [Google Scholar]
- 33.Boquet P, Lemichez E. Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol. 2003;13(5):238–246. doi: 10.1016/s0962-8924(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 34.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 35.Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414(6859):77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]