Abstract
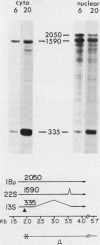
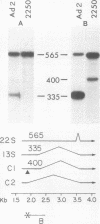
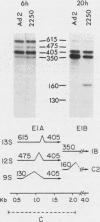
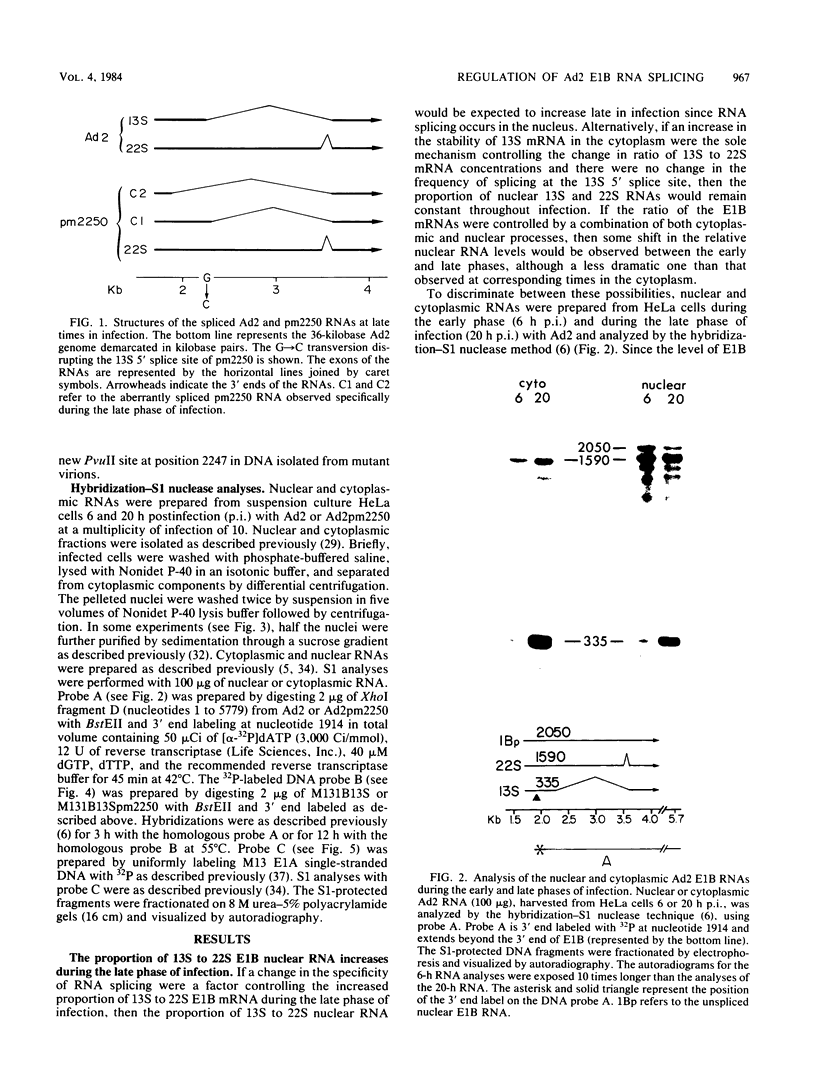
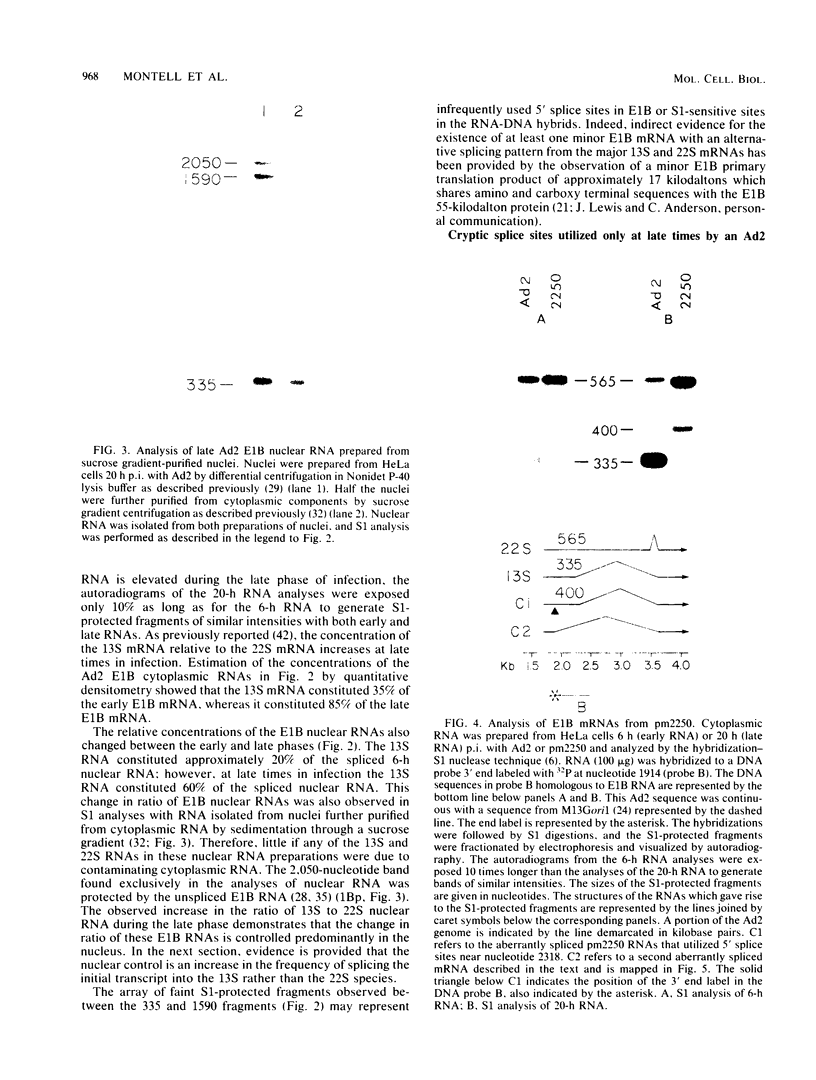
The primary transcript from adenovirus 2 early region 1B (E1B) is processed by differential RNA splicing into two overlapping mRNAs, 13S and 22S. The 22S mRNA is the major E1B mRNA during the early phase of infection, whereas the 13S mRNA predominates during the late phase. In previous work, it has been shown that this shift in proportions of the E1B mRNAs is influenced by increased cytoplasmic stability of the 13S mRNA at late times in infection. Two observations presented here demonstrate that the increase in proportion of the 13S mRNA at late times is also regulated by a change in the specificity of RNA splicing. First, the relative concentrations of the 13S to 22S nuclear RNAs were not constant throughout infection but increased at late times. Secondly, studies with the mutant, adenovirus 2 pm2250 , provided evidence that there was an increased propensity to utilize a 5' splice in the region of the 13S 5' splice site at late times in infection. Adenovirus 2 pm2250 has a G----C transversion in the first base of E1B 13S mRNA intron preventing splicing of the 13S mRNA but not of the 22S mRNA. During the early phase of a pm2250 infection, the E1B primary transcripts were processed into the 22S mRNA only. However, during the late phase, when the 13S mRNA normally predominates, E1B primary transcripts were also processed by RNA splicing at two formerly unused or cryptic 5' splice sites. Both cryptic splice sites were located much closer to the disrupted 13S 5' splice site than to the 22S 5' splice site. Thus, the temporal increase in proportion of the 13S mRNA to the 22S mRNA is regulated by two processes, an increase in cytoplasmic stability of the 13S mRNA and an increased propensity to utilize the 13S 5' splice site during the late phase of infection. Adenovirus 2 pm2250 was not defective for productive infection of HeLa cells or for transformation of rat cells.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiello L., Guilfoyle R., Huebner K., Weinmann R. Adenovirus 5 DNA sequences present and RNA sequences transcribed in transformed human embryo kidney cells (HEK-Ad-5 or 293). Virology. 1979 Apr 30;94(2):460–469. doi: 10.1016/0042-6822(79)90476-8. [DOI] [PubMed] [Google Scholar]
- Akusjärvi G., Persson H. Controls of RNA splicing and termination in the major late adenovirus transcription unit. Nature. 1981 Jul 30;292(5822):420–426. doi: 10.1038/292420a0. [DOI] [PubMed] [Google Scholar]
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Amara S. G., Jonas V., Rosenfeld M. G., Ong E. S., Evans R. M. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982 Jul 15;298(5871):240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Lee F., Harrison T., Williams J., Sharp P. A. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell. 1979 Aug;17(4):935–944. doi: 10.1016/0092-8674(79)90333-7. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Bos J. L., Polder L. J., Bernards R., Schrier P. I., van den Elsen P. J., van der Eb A. J., van Ormondt H. The 2.2 kb E1b mRNA of human Ad12 and Ad5 codes for two tumor antigens starting at different AUG triplets. Cell. 1981 Nov;27(1 Pt 2):121–131. doi: 10.1016/0092-8674(81)90366-4. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Darnell J. E., Jr Variety in the level of gene control in eukaryotic cells. Nature. 1982 Jun 3;297(5865):365–371. doi: 10.1038/297365a0. [DOI] [PubMed] [Google Scholar]
- Dunsworth-Browne M., Schell R. E., Berk A. J. Adenovirus terminal protein protects single stranded DNA from digestion by a cellular exonuclease. Nucleic Acids Res. 1980 Feb 11;8(3):543–554. doi: 10.1093/nar/8.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Felber B. K., Orkin S. H., Hamer D. H. Abnormal RNA splicing causes one form of alpha thalassemia. Cell. 1982 Jul;29(3):895–902. doi: 10.1016/0092-8674(82)90451-2. [DOI] [PubMed] [Google Scholar]
- Fisher E. F., Caruthers M. H. Color coded triarylmethyl protecting groups useful for deoxypolynucleotide synthesis. Nucleic Acids Res. 1983 Mar 11;11(5):1589–1599. doi: 10.1093/nar/11.5.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher P. B., Babiss L. E., Weinstein I. B., Ginsberg H. S. Analysis of type 5 adenovirus transformation with a cloned rat embryo cell line (CREF). Proc Natl Acad Sci U S A. 1982 Jun;79(11):3527–3531. doi: 10.1073/pnas.79.11.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor R. B., Berk A. J. Cis-acting induction of adenovirus transcription. Cell. 1983 Jul;33(3):683–693. doi: 10.1016/0092-8674(83)90011-9. [DOI] [PubMed] [Google Scholar]
- Gaynor R. B., Tsukamoto A., Montell C., Berk A. J. Enhanced expression of adenovirus transforming proteins. J Virol. 1982 Oct;44(1):276–285. doi: 10.1128/jvi.44.1.276-285.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Green M., Brackmann K. H., Cartas M. A., Matsuo T. Identification and purification of a protein encoded by the human adenovirus type 2 transforming region. J Virol. 1982 Apr;42(1):30–41. doi: 10.1128/jvi.42.1.30-41.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison T., Graham F., Williams J. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology. 1977 Mar;77(1):319–329. doi: 10.1016/0042-6822(77)90428-7. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Sloan J. S., Kelly J. D. A Drosophila metabolic gene transcript is alternatively processed. Cell. 1983 Sep;34(2):405–414. doi: 10.1016/0092-8674(83)90374-4. [DOI] [PubMed] [Google Scholar]
- Hines J. C., Ray D. S. Construction and characterization of new coliphage M13 cloning vectors. Gene. 1980 Nov;11(3-4):207–218. doi: 10.1016/0378-1119(80)90061-x. [DOI] [PubMed] [Google Scholar]
- Hutchison C. A., 3rd, Phillips S., Edgell M. H., Gillam S., Jahnke P., Smith M. Mutagenesis at a specific position in a DNA sequence. J Biol Chem. 1978 Sep 25;253(18):6551–6560. [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- King C. R., Piatigorsky J. Alternative RNA splicing of the murine alpha A-crystallin gene: protein-coding information within an intron. Cell. 1983 Mar;32(3):707–712. doi: 10.1016/0092-8674(83)90056-9. [DOI] [PubMed] [Google Scholar]
- Kitchingman G. R., Westphal H. The structure of adenovirus 2 early nuclear and cytoplasmic RNAs. J Mol Biol. 1980 Feb 15;137(1):23–48. doi: 10.1016/0022-2836(80)90155-2. [DOI] [PubMed] [Google Scholar]
- Kumar A., Lindberg U. Characterization of messenger ribonucleoprotein and messenger RNA from KB cells. Proc Natl Acad Sci U S A. 1972 Mar;69(3):681–685. doi: 10.1073/pnas.69.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki R., Roeder W., Traunecker A., Sidman C., Wabl M., Raschke W., Tonegawa S. The role of DNA rearrangement and alternative RNA processing in the expression of immunoglobulin delta genes. Cell. 1981 May;24(2):353–365. doi: 10.1016/0092-8674(81)90325-1. [DOI] [PubMed] [Google Scholar]
- Mariman E. C., van Beek-Reinders R. J., van Venrooij W. J. Alternative splicing pathways exist in the formation of adenoviral late messenger RNAs. J Mol Biol. 1983 Jan 15;163(2):239–256. doi: 10.1016/0022-2836(83)90005-0. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F., Jr, Murphy E. C., Jr, Huang R. C. Transcription of ribonucleic acid in isolated mouse myeloma nuclei. Biochemistry. 1973 Aug 28;12(18):3440–3446. doi: 10.1021/bi00742a013. [DOI] [PubMed] [Google Scholar]
- McGrath J. P., Capon D. J., Smith D. H., Chen E. Y., Seeburg P. H., Goeddel D. V., Levinson A. D. Structure and organization of the human Ki-ras proto-oncogene and a related processed pseudogene. Nature. 1983 Aug 11;304(5926):501–506. doi: 10.1038/304501a0. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Inhibition of RNA cleavage but not polyadenylation by a point mutation in mRNA 3' consensus sequence AAUAAA. Nature. 1983 Oct 13;305(5935):600–605. doi: 10.1038/305600a0. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Wilson M. C. Regulation of adenovirus-2 gene expression at the level of transcriptional termination and RNA processing. Nature. 1981 Mar 12;290(5802):113–118. doi: 10.1038/290113a0. [DOI] [PubMed] [Google Scholar]
- Nomura N., Ray D. S. Expression of a DNA strand initiation sequence of ColE1 plasmid in a single-stranded DNA phage. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6566–6570. doi: 10.1073/pnas.77.11.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perricaudet M., Le Moullec J. M., Pettersson U. Predicted structure of two adenovirus tumor antigens. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3778–3782. doi: 10.1073/pnas.77.7.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Hirose T., Itakura K., Riggs A. D. Efficient correction of a mutation by use of chemically synthesized DNA. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4268–4270. doi: 10.1073/pnas.75.9.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozek C. E., Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983 Jan;32(1):23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D. The infectivity of adenovirus genomes lacking DNA sequences from their left-hand termini. Nucleic Acids Res. 1982 Sep 11;10(17):5105–5119. doi: 10.1093/nar/10.17.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Proudfoot N. J., Shander M., Maniatis T. A single-base change at a splice site in a beta 0-thalassemic gene causes abnormal RNA splicing. Cell. 1982 Jul;29(3):903–911. doi: 10.1016/0092-8674(82)90452-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wilson M. C., Darnell J. E., Jr Control of messenger RNA concentration by differential cytoplasmic half-life. Adenovirus messenger RNAs from transcription units 1A and 1B. J Mol Biol. 1981 May 25;148(3):231–251. doi: 10.1016/0022-2836(81)90537-4. [DOI] [PubMed] [Google Scholar]