Abstract
Pathogenic Marek's disease viruses (MDVs) have two head-to-tail copies of a 132-bp repeat. As MDV is serially passaged in cell culture, the virus becomes attenuated and the number of copies of the 132-bp repeat increases from 2 to often more than 20 copies. To determine the role of the repeats in attenuation, we used five overlapping cosmid clones that spanned the MDV genome to reconstitute infectious virus (rMd5). By mutating the appropriate cosmids, we generated clones of infectious MDVs that contained zero copies of the 132-bp repeats, rMd5(Δ132); nine copies of the 132-bp repeats, rMd5(9-132); and nine copies of the 132-bp repeats inserted in the reverse orientation, rMd5(rev9-132). After two passages in cell culture, wild-type Md5, rMd5, and rMd5(Δ132) were stable. However, rMd5(9-132) and rMd5(rev9-132) contained a population of viruses that contained from 3 to over 20 copies of the repeats. A major 1.8-kb mRNA, containing two copies of the 132-bp repeat, was present in wild-type Md5 and rMd5 but was not present in rMd5(Δ132), rMd5(9-132), rMd5(rev9-132), or an attenuated MDV. Instead, the RNAs transcribed from the 132-bp repeat region in rMd5(9-132) and rMd5(rev9-132) closely resembled the pattern of RNAs transcribed in attenuated MDVs. When inoculated into susceptible day-old chicks, all viruses produced various lesions. Thus, expansion of the number of copies of 132-bp repeats, which accompanies attenuation, is not sufficient in itself to attenuate pathogenic MDVs.
One of the most economically devastating infectious diseases of poultry is Marek's disease (MD), caused by Marek's disease virus (MDV), an oncogenic avian herpesvirus. MD in chickens is commonly characterized by a generalized lymphomatosis involving lymphocytic infiltration of nerves and other organs. Clinical manifestations can be numerous and varied, including paralysis, skin lesions, atrophy of the thymus and bursa of Fabricius, immunosuppression, and high mortality. MD commonly appears in 3- to 4-week-old chickens and gradually builds to a peak between 12 and 30 weeks of age (23).
Three MDV serotypes are recognized. Serotype 1 viruses (MDV-1) include the oncogenic MDVs and their cell culture-attenuated variants. The serotype 2 MDVs (MDV-2) include the naturally occurring nononcogenic chicken MDVs, while the nononcogenic turkey herpesviruses (HVT) are classified as serotype 3 viruses (3). The three MDV serotypes have distinctly different restriction endonuclease (RE) patterns (9). However, their genomes are colinear, and they share significant DNA homology (1, 11, 14, 22) as well as several cross-reacting antigens (10), suggesting a common evolutionary origin.
Although it is not known how MDV-1 induces lymphomas, cellular transformation does not appear to occur by a “hit-and-run” mechanism. The viral genome either is maintained as a covalently closed circular episome (21) or is randomly integrated into the chicken chromosome (6). The nature of the T-cell lymphomas; their short latency period; and the existence of oncogenic, attenuated, and nononcogenic MDVs suggest the presence of an MDV-1 oncogene(s). Despite their acute oncogenic nature, MDV-1 viruses can be easily attenuated through serial passages in cell culture. Cell culture-induced attenuation always occurs concurrently with the expansion of a unique 132-bp fragment located in the inverted repeat regions flanking the unique long region of the MDV genome (TRL and IRL in Fig. 1) (20). No comparable 132-bp repeat region exists in either MDV-2 or HVT (19). Concurrently with genomic expansion, a 1.8-kb RNA that contains the 132-bp repeat region within it is altered (13). It has been suggested that the expansion of this region, resulting in head-to-tail concatemers of the 132-bp region, disrupts the 1.8-kb RNA that is essential for oncogenicity, thereby resulting in attenuation (2). To test the hypothesis that expansion of the 132-bp repeat causes attenuation, we constructed recombinant MDVs in which the two copies of the repeats in an oncogenic virus were replaced with nine copies of the repeats.
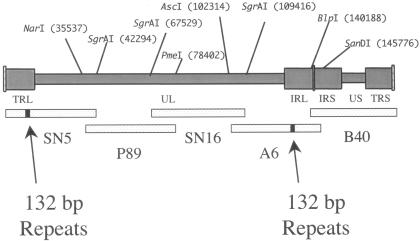
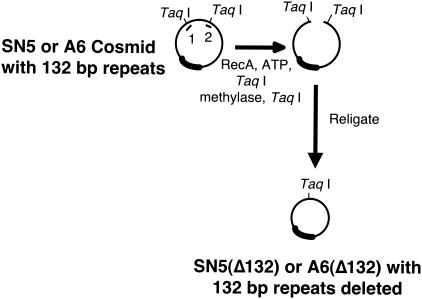
FIG. 1.
Overlapping cosmid clones of Md5. The MDV-1 viruses contain two copies of a 132-bp repeat, located in two separate parts of the genome. One pair of repeats is in the terminal repeat region (TRL) adjacent to the unique long region (UL). The other pair of repeats is inverted and located in the internal repeat (IRL) at the other end of the UL. The five overlapping cosmid clones are shown below the MDV genome and are aligned with the regions of the MDV genome from which they were cloned. The small bar in SN5 and A6 represents the location of the 132-bp repeats in these cosmids and their corresponding location in the TRL and IRL.
MATERIALS AND METHODS
Cells and viruses.
MDV strains used in this study included a very virulent strain, Md5 (24), and the attenuated Rispens strain, cell culture passage 45 (17). Viruses were propagated on primary duck embryo fibroblasts (DEF) maintained in Leibovitz's L-15 medium plus McCoy 5A medium (1:1), supplemented with 2.5% bovine serum and antibiotics.
Cosmids.
The five cosmids (SN5, P89, SN16, A6, and B40) that span the Md5 genome are shown in Fig. 1, and their construction was previously described (16). The RecA-assisted restriction endonuclease (RARE) cleavage procedure was used to delete the two copies of the 132-bp repeat present in SN5 and A6 (7). Briefly, two oligonucleotides were constructed (oligonucleotide 1, 5′-GGA GAA AGT ATG TCG ATT TTA AAT GTA GTT-3′, and oligonucleotide 2, 5′-ACA AG TTC GGT AAC GCT TTC GAT TAG A-3′) that anneal and protect the two TaqI RE sites that flank the 132-bp repeats. The RARE cleavage reaction mixture consists of 8 μl of 5× RARE buffer (125 mM Tris acetate [pH 7.85], 20 mM magnesium acetate, 2.5 mM spermidine, 2.0 mM dithiothreitol), 1 μl of 5 mM EGTA, 4 μl of 10× ATPγ-S (11 mM ADP, 3 mM ATPγ-S), 1 μl of oligonucleotide 1 (1 μg/μl), 1 μl of oligonucleotide 2 (1 μg/μl), 5 μl of RecA (2 μg/μl), and 1 μl of cosmid DNA at 1 μg/μl. The mixture was incubated for 30 min at 37°C. After being annealed with the oligonucleotides, both cosmids were methylated with TaqI methylase (2 μl of TaqI methylase, 18 μl of acetylated bovine serum albumin [20 μg/μl], and 2 μl of S-adenosylmethionine reagent [3.75 μl of 32 mM S-adenosylmethionine, 46.25 μl of water]) for 30 min at 37°C and 30 min at 50°C, phenol extracted, ethanol precipitated, and resuspended in water. The cosmids were digested with TaqI, religated (overnight at 14 to 16°C with T4 DNA ligase), packaged (Gigapack III gold packaging extract; Stratagene, La Jolla, Calif.), and introduced into HB101 Escherichia coli cells. The resulting cosmids were designated SN5(Δ132) and A6(Δ132). By deleting everything between the two flanking TaqI sites, 95 bases on one side and 52 bases on the other side of the repeats were also deleted. The procedure is summarized in Fig. 2.
FIG. 2.
Generation of deletion in cosmid clones. The figure shows an overview of the RARE cleavage procedure used to delete the 132-bp repeats in SN5 and A6. The short lines numbered 1 and 2 represent oligonucleotides 1 and 2, used to protect two TaqI sites. The two TaqI sites shown flank a 411-bp fragment of cosmid DNA that contains two copies of the 132-bp repeat.
The PCR was used to amplify the 132-bp repeats from a virus that had been repeatedly passaged on chicken embryo fibroblasts (CEF). Briefly, we used oligonucleotide 1 as the forward PCR primer and 5′-CTC GTA AGG CTT CCC GTC A-3′ as the reverse primer. The amplicon was cloned into the TA cloning vector from Invitrogen. RE digestion and gel electrophoresis indicated that the clone contained nine copies of the 132-bp repeat. These nine copies were excised with TaqI and cloned into SN5(Δ132) and A6(Δ132). Briefly, we used the same RARE cleavage procedure as used to generate SN5(Δ132) and A6(Δ132), except that we began with the cosmids SN5(Δ132) and A6(Δ132) and used the protecting oligonucleotide 3 (5′-TGG GAG AAA GTA TGT CGA TTA GAA ACT G-3′) to block methylation of the newly created TaqI site. The resulting cosmids were designated SN5(9-132) and A6(9-132) for the two cosmids with the nine copies inserted in the correct orientation and SN5(rev9-132) and A6(rev9-132) for the two cosmids with the nine copies inserted in the reverse orientation. For all the recombinant cosmids, the region in and around the 132-bp repeat region was DNA sequenced in order to confirm that the cloning occurred at the correct insertion site and that no rearrangements had been generated.
Generation of recombinant MDVs.
To generate infectious virus, SN5, P89, SN16, A6, and B40 were cotransfected into DEF by the standard calcium phosphate transfection procedure. Briefly, 0.5 μg of each cosmid was suspended in 1 ml which contained 250 μl of water, 250 μl of 0.5 M calcium chloride, and 500 μl of 2× HEPES buffer (pH 7.05). After incubation for 10 to 15 min, the calcium chloride-precipitated DNA was gently added to DEF in a 60-mm-diameter tissue culture dish containing 4 ml of F-10 medium supplemented with 5% fetal bovine serum and incubated for 4 h at 37°C. The calcium phosphate tissue culture medium was removed, and 5 ml of glycerol solution (15% glycerol in water) was added for 2 to 3 min. The glycerol was removed, and the cells were washed twice with phosphate-buffered saline and refed with Leibovitz's L-15 medium plus McCoy 5A medium (1:1), supplemented with 5% fetal bovine serum. The next day, the cultures were fed with fresh tissue culture medium and were passaged 3 to 4 days later. Plaques usually appeared in 1 to 2 weeks and were expanded following another cell culture passage. The resulting virus was designated rMd5. The 132-bp-deletion virus and the MDVs with nine copies of the 132-bp repeat were generated in the same manner but with use of the appropriately mutated SN5 and A6 cosmids. These recombinant viruses were designated rMd5(Δ132), rMd5(9-132), and rMd5(rev9-132), respectively.
DNA and RNA analysis.
The standard procedure of guanidinium thiocyanate-phenol extraction, as described by Chomczynski and Sacchi (5), was used to extract RNA from heavily infected DEF. The extraction of DNA from infected DEF or tumor tissue followed the standard proteinase K phenol-chloroform extraction protocols (18).
Southern blotting was performed by separating RE-digested DNAs on a 1% agarose Tris-borate-EDTA gel, transferring them to a nylon membrane, and probing them with [32P]dCTP randomly primed probes. Hybridization and autoradiography were carried out using standard protocols. Northern blotting was performed similarly except that the RNA was electrophoresed in a 1% agarose gel containing 2% formaldehyde (18).
Immediately after plates with recombinant MDV plaques had been passaged once more to generate virus stocks, the DNA was extracted from a portion of each virus stock, digested with BamHI, and Southern blotted with a mixture of all five of the cosmids that had been randomly prime labeled with [32P]dCTP. No unexpected rearrangements or deletions were ever detected in any of the recombinant MDVs.
Growth curves.
Each virus was plated on DEF to study its in vitro growth characteristics. Briefly, 100 PFU was inoculated onto freshly seeded DEF in 60-mm-diameter dishes. On days 0, 1, 2, 3, 5, and 7, the infected cultures were trypsinized, diluted, and seeded onto fresh DEF in triplicate. Visible plaques were counted 7 to 8 days later.
In vivo experiments.
Day-old 15 × 7 White Leghorn chickens (F1 progeny of ADOL line 15I5 × 71 chickens) were inoculated intra-abdominally with 1,000 PFU of virus and housed in Horsfall-Bauer isolators for the duration of the experiment. Each Horsfall-Bauer isolator contained 13 inoculated birds that were from dams that had been vaccinated with HVT and consequently contained maternal antibodies (MAb+). Each isolator also contained four noninoculated monitor chickens that were obtained from nonvaccinated dams and were maternal antibody free (MAb−). All birds that died during the course of the experiment or were terminated at 8 weeks were necropsied. Any gross tumors and abnormalities were recorded. Suspicious brachial and sciatic nerves were also examined for histological lesions for confirmation. The data are shown in Table 1.
TABLE 1.
Recombinant viruses in susceptible chickensa
| Group | MAb status | No. of birds
|
|||||
|---|---|---|---|---|---|---|---|
| Total | Healthy (%) | Death (%) | Nerve (%) | Tumor (%) | Atrophy (%) | ||
| Control | MAb− | 4 | 4 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| MAb+ | 13 | 13 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| rMd5 | MAb− | 4 | 0 (0) | 1 (25) | 4 (100) | 4 (100) | 4 (100) |
| MAb+ | 13 | 0 (0) | 3 (25) | 12 (92) | 6 (46) | 8 (62) | |
| rMd5(Δ132) A | MAb− | 4 | 0 (0) | 0 (0) | 3 (75) | 4 (100) | 2 (50) |
| MAb+ | 13 | 2 (15) | 1 (8) | 8 (62)* | 5 (39) | 3 (23)* | |
| rMd5(Δ132) B | MAb− | 4 | 0 (0) | 0 (0) | 4 (100) | 2 (50) | 1 (25) |
| MAb+ | 13 | 5 (39) | 0 (0) | 6 (46)** | 2 (15)* | 4 (31)* | |
| rMd5(9-132) | MAb− | 4 | 0 (0) | 0 (0) | 2 (50) | 3 (75) | 4 (100) |
| MAb+ | 13 | 2 (15) | 0 (0) | 10 (77) | 1 (8)** | 1 (8)** | |
| rMd5(rev9-132) | MAb− | 4 | 1 (25) | 0 (0) | 0 (0) | 3 (75) | 2 (50) |
| MAb+ | 13 | 2 (15) | 0 (0) | 10 (77) | 1 (8)** | 4 (31)* | |
Each Horsfall-Bauer isolator initially contained 17 chicks. Four birds were noninoculated, MAb− chickens and served as monitors for the horizontal spread of MDV. Thirteen MAb+ chicks were inoculated with 1,000 PFU of virus. The column labeled “Healthy” shows the number of birds in each group that had no observable pathogenic lesions. The column labeled “Nerve” shows the number of birds with observable enlargements of brachial and/or sciatic nerves. Birds positive for tumors had one or more macroscopic tumors. Birds positive for atrophy had significant signs of bursal and/or thymic atrophy. rMd5(Δ132) A and rMd5(Δ132) B are two independently constructed deletion mutants. Groups of birds with percentages of pathologies significantly different from those of the rMd5-inoculated birds are marked by asterisks (*, P < 0.05; **, P < 0.01; z score for a one-tailed test).
RESULTS AND DISCUSSION
Generation of mutant MDVs.
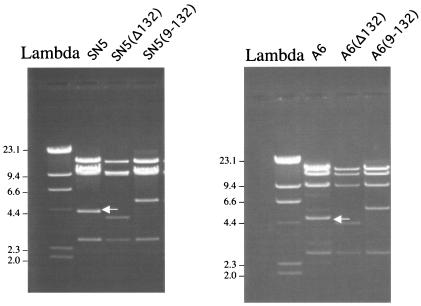
The two copies of the 132-bp repeat region in SN5 and A6 were either deleted or replaced with nine copies. To confirm that the modified cosmids were correctly modified, the cosmids were digested with HindIII and separated on a gel. The arrows in Fig. 3 point to the HindIII fragments in SN5 and A6 that contain the two copies of the 132-bp repeats. The gel demonstrates that the corresponding fragments in the SN5(Δ132), A6(Δ132), SN5(9-132), and A6(9-132) cosmids had the expected shift in their RE pattern that corresponds to either a deletion of the repeats [smaller fragment in SN5(Δ132) and A6(Δ132) of Fig. 3] or the replacement of the two copies with nine copies [larger fragment in SN5(9-132) and A6(9-132) of Fig. 3]. Figure 3 also demonstrates that the cosmids are clonal and do not contain mixtures of mutated and nonmutated cosmids. A similar assay with SN5(rev9-132) and A6(rev9-132) confirmed that these cosmids were also clonal and contained the expected insertion (data not shown).
FIG. 3.
HindIII digestion of cosmids. The DNA from the cosmids was extracted, digested with HindIII, and separated on an agarose gel. The arrows point to the 4.7-kb fragment in SN5 and A6 that contains two copies of the 132-bp repeat. Numbers at left of each panel indicate molecular size in kilobases.
DNA analysis.
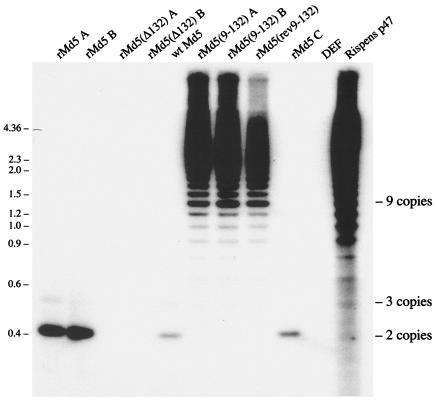
Cotransfection with the modified and unmodified five cosmids resulted in plaques appearing in the tissue culture dishes. Consequently, the 132-bp repeats are not essential for virus replication in cell culture. To determine how stable the 132-bp repeat mutations were, we extracted viral DNA for a Southern blot and probed it with a [32P]dCTP-labeled 132-bp-repeat-specific probe. As expected, three independently derived rMd5 viruses contained primarily two copies of the 132-bp repeats. A small population of viruses appears to contain three copies of the 132-bp repeat (rMd5 A, rMd5 B, and rMd5 C in Fig. 4), similarly to wild-type Md5 (lane 5). Thus, our rMd5 recombinant virus was beginning to generate viruses with expanded numbers of copies of the repeats, just as wild-type viruses normally do. Since duck cells do not contain any sequences homologous to the MDV repeats and the rMd5(Δ132) viruses have no 132-bp repeats, we would not expect passage of rMd5(Δ132) in cell culture to alter this region. As expected, our two independently constructed rMd5(Δ132) viruses lacked any 132-bp repeats (Fig. 4) and a couple of passages in cell culture did not appear to generate any alterations. However, our two independently generated recombinant MDVs that had their two copies of the 132-bp repeats replaced with nine copies of the 132-bp repeats were not stable. In the two cell culture passages required to generate enough virus to analyze, both rMd5(9-132) viruses generated a mixture of viruses that contained anywhere from 3 to over 20 copies of the 132-bp repeat. The 132-bp repeat pattern of both rMd5(9-132) viruses appeared to be identical to the pattern of repeats typically seen in attenuated MDVs (Fig. 4). The rMd5(rev9-132) virus, with the nine copies of repeats inserted in the reverse orientation, was also unstable, generating viruses with a variable number of copies of the 132-bp repeat. The expansion of the number of copies of the 132-bp repeats appears to be a natural consequence of passaging MDVs in cell culture. However, replicating viruses with two copies generate viruses with three or more copies only very slowly. Conversely, MDVs with multiple copies of the repeats (nine copies in our experiment) rapidly generated a population of viruses that contained anywhere from 3 to over 20 copies of the repeats. The fact that the same expansion occurs even when the nine copies of the 132-bp repeats are inserted in the reverse orientation suggests that the generation of multiple copies of head-to-tail repeats is a physical consequence of the tandem direct repeats and not due to any possible coding regions. The nearby MDV origin of replication, located 226 bases from the beginning of the repeats, may also contribute to the expansion process. Regardless of the mechanism, cell culture replication of rMd5(9-132) and rMd5(rev9-132) rapidly results in a population of viruses that appear to be identical to what is seen when wild-type MDVs are repeatedly passaged in cell culture.
FIG. 4.
Southern blot of recombinant MDV. DNA from MDV-infected DEF was digested with TaqI, Southern blotted, and probed with the 132-bp repeats. Viral names that are followed by an A, B, or C represent separate independently generated recombinants. Positions of two, three, and nine copies of the 132-bp repeat are shown on the right. Numbers at left are molecular sizes in kilobases.
RNA analysis.
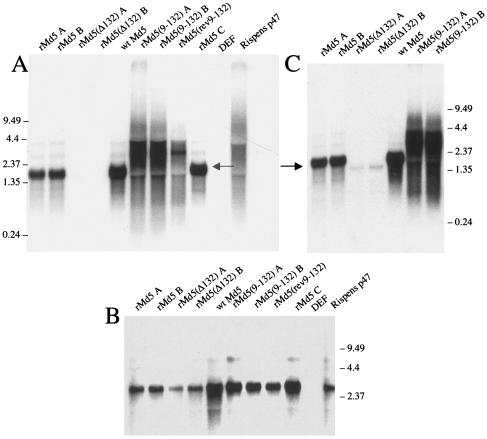
RNA from the 132-bp repeat region in wild-type Md5 (lane 5 in Fig. 5A and C) displayed a complex family of RNAs, identical to the pattern of RNAs that had been reported earlier by other groups (15). The most abundant RNA in the family is the 1.8-kb mRNA (the major RNA band in Fig. 5A and C). The fainter RNA bands above and below the 1.8-kb mRNA represent a series of differentially spliced mRNAs that also encode the 132-bp repeats (15). Figure 5A also confirms that our rMd5(Δ132) viruses lack any RNA that contains 132-bp repeats. Deletion of the 132-bp repeats does not entirely eliminate RNA from this region. RNA slightly smaller than the 1.8-kb RNA can still be detected in rMd5(Δ132) (Fig. 5C). Figure 5B is a control blot demonstrating that nondegraded RNA was isolated from all the virus-infected cells.
FIG. 5.
Northern blot of RNA from infected DEF. RNA extracted from MDV-infected DEF was Northern blotted and probed with a labeled probe from the 132-bp repeat region (A), the gB gene (UL27) (B), or an 834-bp fragment that spans the 132-bp repeat region (C). The arrows in panels A and C point to the 1.8-kb RNA. Numbers at left of panel A and at right of panels B and C are molecular sizes in kilobases.
Replacing the two copies of the 132-bp repeat in rMd5 with nine copies dramatically altered the RNAs from this region (Fig. 5A and C). The single 1.8-kb RNA band is absent, replaced by a high-molecular-weight smear of RNA. Based upon the DNA alterations created in this region, the smear of RNA most likely represents a heterogeneous population of RNAs that contain between 3 and possibly 20 or more copies of the 132-bp repeat. The faint high-molecular-weight RNA smear seen above the first smear in rMd5(9-132) A and rMd5(9-132) B may represent a heterogeneous population of spliced messages that also contain variable copy numbers of the 132-bp repeat. As might be expected if there are no RNA termination signals present, reversing the nine copies of the 132-bp repeats results in the same RNA pattern as seen in rMd5(9-132). To resolve this issue, we are in the process of cDNA cloning and sequencing these large RNAs.
Biological characterization of recombinant MDVs.
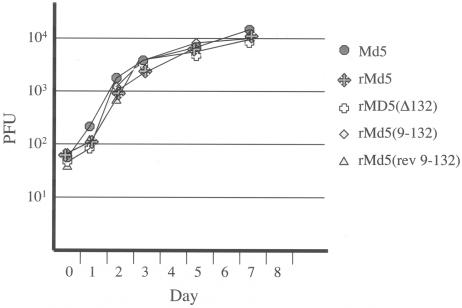
Deletions or modifications in the 132-bp repeats (including 95 and 52 bases of flanking DNA) did not adversely affect the ability of the recombinant MDVs to grow in DEF. All recombinants grew as well as did wild-type Md5 (Fig. 6).
FIG. 6.
Growth curve of recombinant MDVs in DEF. Fresh DEF were infected with the indicated viruses. On days 0, 1, 2, 3, 5, and 7, cells were trypsinized and plated on fresh DEF. The resulting plaques were counted 7 to 8 days later. Each point represents the average from three plates.
We evaluated the ability of the recombinant MDVs to induce disease by inoculating day-old MAb+ chicks with each virus. At 26 days after inoculation, the group inoculated with rMd5 began showing signs of neurological disease, including ataxia and torticollis, indicative of persistent neurological disease (8). By day 28, all the other inoculated groups had birds showing symptoms of persistent neurological disease. Three of the 13 birds inoculated with rMd5 died at days 48, 49, and 56. All three birds showed severe bursal and thymic atrophy, as well as tumor cell infiltrates of the vagus nerve. One noninoculated contact bird, housed with the rMd5-inoculated birds, died at day 26. Upon necropsy, the contact bird had a spleen tumor and mild to moderate thymic and bursal atrophy. Only one other bird died during the course of the experiment. A chicken inoculated with rMd5(Δ132) died at day 45 with tumors in the heart, ovary, and lung. At day 57, the remaining chickens were euthanized and necropsied. At necropsy, we considered any brachial or sciatic nerve enlargements to indicate positive nerve involvement. Later histological examination of these tissues confirmed the earlier diagnosis. Lesions induced by the different virus inocula are summarized in Table 1. As expected, rMd5 was pathogenic, generating assorted tumors, thymic atrophy, and nerve involvement. rMd5(Δ132), lacking any 132-bp repeats, was also pathogenic, generating a similar pattern of pathological lesions, albeit at a slightly lower frequency. Although rMd5(9-132) has a 132-bp repeat DNA and RNA pattern that appears to be identical with the pattern of DNA and RNA in attenuated MDVs, rMd5(9-132) was also pathogenic. However, the incidence of tumors and thymic or bursal atrophy was significantly less (P < 0.01) than what rMd5 induced. rMd5(rev9-132) induced the same spectrum and incidence of lesions as did rMd5(9-132), suggesting that reversing the 132-bp repeats does not disrupt RNA transcription any more than does having the repeats inserted in the correct orientation. None of the 132-bp repeat modifications affected the ability of the viruses to spread horizontally. However, the fact that MDVs with either no 132-bp repeats or nine copies of the repeats are still pathogenic clearly indicates that the expansion of the 132-bp repeat region is not sufficient by itself to attenuate MDV.
In a preliminary experiment, we isolated spleen cells from 8-day-old infected chicks and plated these cells on DEF in culture. By counting the resulting MDV plaques, we found that, in the early cytolytic infection at 8 days postinoculation, we were able to isolate more rMd5 viruses than either rMd5(9-132) or rMd5(rev9-132) viruses. Although this preliminary experiment needs to be repeated, the rMd5(9-132) and rMd5(rev9-132) viruses may be less pathogenic because they do not replicate as efficiently in vivo as rMd5. Furthermore, rather than being strictly associated with pathogenicity, the 1.8-kb mRNA may be more involved in somehow enhancing the replication of MDV in its natural host, the chicken, in contrast to its unnatural tissue culture cell host. Lending credence to this hypothesis is the well-recognized phenomenon that pathogenic viruses, with two copies of the 132-bp repeats, quickly expand the number of copies when the virus is passaged repeatedly in cell culture. Typically, MDVs with expanded copies of the 132-bp repeat replicate more efficiently in cell culture than do viruses with two copies of the repeats. Conversely, when an attenuated MDV with multiple copies of the repeats is repeatedly back-passaged through chickens, the MDVs become more homogeneous, with few viruses containing more than two or three copies of the 132-bp repeat (data not shown). In addition, viruses with two or three copies of the repeats replicate more efficiently in chickens than do viruses with multiple copies of the repeats. Together, these observations suggest that there is strong selective pressure in the chicken to maintain two copies of the 132-bp repeat and consequently to maintain the 1.8-kb mRNA. This selective pressure is not present in cell culture.
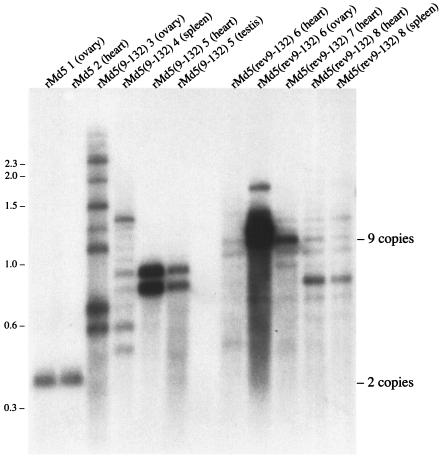
To determine whether tumors induced by rMd5(9-132) or rMd5(rev9-132) contained two copies of the repeats, we isolated DNA from several tumors and digested the DNA with TaqI. A Southern blot of the digested DNA is shown in Fig. 7. In two birds with tumors induced by rMd5, one with an ovarian tumor and the other with a tumor on the heart, the rMd5 remained as a homogenous population and apparently contained only two copies of the repeats. However, tumors induced by either rMd5(9-132) or rMd5(rev9-132) contained a heterogenous population of viruses containing between 3 copies [spleen tumor from bird 4, inoculated with rMd5(9-132)] and over 16 copies [ovarian tumor from bird 3, also inoculated with rMd5(9-132)]. Regardless, none of the tumors from birds inoculated with either rMd5(9-132) or rMd5(rev9-132) contained MDVs with just two copies of the repeats. Thus, the tumors induced by either rMd5(9-132) or rMd5(rev9-132) were not the result of amplifying or selecting for MDVs with two copies of the repeats. Interestingly, the population of viruses present in the tumors varies from bird to bird and sometimes varies between two tumors in the same bird. In Fig. 7, the two tumors from bird 6, rMd5(rev9-132) 6 (heart) and rMd5(rev9-132) 6 (ovary), have different populations of viruses with different copy number of repeats. In contrast, birds 5 and 8 both had two tumors and within each bird the population of viruses appeared to be the same. It is possible that the tumors in birds 5 and 8 arose from a single transforming event whereas the two tumors in bird 6 arose from two separate transforming events.
FIG. 7.
Southern blot of DNA extracted from tumors. Upon death or termination of the animal experiment, DNA was extracted from gross tumors and digested with TaqI. A Southern blot of the RE-digested DNA was probed with a [32P]dCTP-labeled 132-bp repeat fragment. Each lane is labeled with the name of the inoculated virus, followed by the bird number and the tumor source. The positions of two and nine copies of the 132-bp repeat are shown on the right. Numbers at left are molecular sizes in kilobases.
The expansion of the 132-bp repeats may directly influence the nearby MDV origin of replication (4, 25). In fact, it has been shown previously that proper functioning of the MDV origin of replication requires the presence of an intact promoter from the 1.8-kb mRNA (12). We are currently investigating whether the MDV origin of replication functions differently in DEF and lymphoid cells and whether changing the number of copies of the 132-bp repeats will influence any differences.
Acknowledgments
We thank Lonnie Milam for his excellent technical assistance. We also thank Vladimir Zelnik (Slovak Academy of Sciences, Slovak Republic) for critical reading of the manuscript and Huanmin Zhang (Avian Disease and Oncology Laboratory) for helpful suggestions regarding the statistical analysis.
This study was partially supported by a grant awarded to R.F.S. from the USDA National Research Initiative Competitive Grants Program (9702235).
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradley, G., G. Lancz, A. Tanaka, and M. Nonoyama. 1989. Loss of Marek's disease virus tumorigenicity is associated with truncation of RNAs transcribed within BamHI-H. J. Virol. 63:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bülow, V. V., and P. M. Biggs. 1975. Precipitating antigens associated with Marek's disease virus and a herpesvirus of turkeys. Avian Pathol. 4:147-162. [DOI] [PubMed] [Google Scholar]
- 4.Camp, H. S., P. M. Coussens, and R. F. Silva. 1991. Cloning, sequencing, and functional analysis of a Marek's disease virus origin of replication. J. Virol. 65:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski, P., and N. Sacchi. 1987. Single-step method RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 6.Delecluse, H.-J., and W. Hammerschmidt. 1993. Status of Marek's disease virus in established lymphoma cell lines: herpesvirus integration is common. J. Virol. 67:82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrin, L. J., and R. D. Camerini-Otero. 1991. Selective cleavage of human DNA: RecA-assisted restriction endonuclease (RARE) cleavage. Science 254:1494-1497. [DOI] [PubMed] [Google Scholar]
- 8.Gimeno, I. M., R. L. Witter, and W. M. Reed. 1999. Four distinct neurologic syndromes in Marek's disease: effect of viral strain and pathotype. Avian Dis. 43:721-737. [PubMed] [Google Scholar]
- 9.Hirai, K., K. Ikuta, and S. Kato. 1979. Comparative studies on Marek's disease virus and herpesvirus of turkey DNAs. J. Gen. Virol. 45:119-131. [DOI] [PubMed] [Google Scholar]
- 10.Ikuta, K., S. Ueda, S. Kato, and K. Hirai. 1983. Most virus-specific polypeptides in cells productively infected with Marek's disease virus or herpesvirus of turkeys possess cross-reactive determinants. J. Gen. Virol. 64:961-965. [DOI] [PubMed] [Google Scholar]
- 11.Izumiya, Y., H. K. Jang, M. Ono, and T. Mikami. 2001. A complete genomic DNA sequence of Marek's disease virus type 2, strain HPRS24. Curr. Top. Microbiol. Immunol. 255:191-221. [DOI] [PubMed] [Google Scholar]
- 12.Katsumata, A., A. Iwata, and S. Ueda. 1998. Cis-acting elements in the lytic origin of DNA replication of Marek's disease virus type 1. J. Gen. Virol. 79:3015-3018. [DOI] [PubMed] [Google Scholar]
- 13.Kopacek, J., L. J. N. Ross, V. Zelnik, and J. Pastorek. 1993. The 132-bp repeats are present in RNA transcripts from 1.8 kb gene family of Marek disease virus-transformed cells. Acta Virol. 37:191-195. [PubMed] [Google Scholar]
- 14.Lee, L. F., P. Wu, D. Sui, D. Ren, J. Kamil, H. J. Kung, and R. L. Witter. 2000. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc. Natl. Acad. Sci. USA 97:6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng, F., G. Bradley, A. Tanaka, G. Lancz, and M. Nonoyama. 1992. Isolation and characterization of cDNAs from BamHI-H gene family RNAs associated with the tumorigenicity of Marek's disease virus. J. Virol. 66:7389-7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy, S. M., B. Lupiani, I. M. Gimeno, R. F. Silva, L. F. Lee, and R. L. Witter. 2002. Rescue of a pathogenic Marek's disease virus with overlapping cosmid DNAs: use of a pp38 mutant to validate the technology for the study of gene function. Proc. Natl. Acad. Sci. USA 99:7054-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rispens, B. H., H. Van Vloten, N. Mastenbroek, H. J. L. Maas, and K. A. Schat. 1972. Control of Marek's disease in the Netherlands. I. Isolation of an avirulent Marek's disease virus (strain CVI 988) and its use in laboratory vaccination trials. Avian Dis. 16:108-125. [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Silva, R. F., L. F. Lee, and G. F. Kutish. 2000. The genomic structure of Marek's disease virus. Curr. Top. Microbiol. Immunol. 255:143-158. [DOI] [PubMed] [Google Scholar]
- 20.Silva, R. F., and R. L. Witter. 1985. Genomic expansion of Marek's disease virus DNA is associated with serial in vitro passage. J. Virol. 54:690-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka, A., S. Silver, and M. Nonoyama. 1978. Biochemical evidence of the nonintegrated status of Marek's disease virus DNA in virus-transformed lymphoblastoid cells of chicken. Virology 88:19-24. [DOI] [PubMed] [Google Scholar]
- 22.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witter, R. L., and K. A. Schat. 2003. Marek's disease, p. 407-465. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames.
- 24.Witter, R. L., J. M. Sharma, and A. M. Fadly. 1980. Pathogenicity of variant Marek's disease virus isolates in vaccinated and unvaccinated chickens. Avian Dis. 24:210-232. [Google Scholar]
- 25.Wu, T. F., H. H. Chen, and H. Wu. 2001. Functional characterization of Marek's disease virus (MDV) origin-binding protein (OBP): analysis of its origin-binding properties. Virus Genes 23:227-239. [DOI] [PubMed] [Google Scholar]