Abstract
Sporothrix schenckii, previously assumed to be the sole agent of human and animal sporotrichosis, is in fact a species complex. Recently recognized taxa include S. brasiliensis, S. globosa, S. mexicana, and S. luriei, in addition to S. schenckii sensu stricto. Over the last decades, large epidemics of sporotrichosis occurred in Brazil due to zoonotic transmission, and cats were pointed out as key susceptible hosts. In order to understand the eco-epidemiology of feline sporotrichosis and its role in human sporotrichosis a survey was conducted among symptomatic cats. Prevalence and phylogenetic relationships among feline Sporothrix species were investigated by reconstructing their phylogenetic origin using the calmodulin (CAL) and the translation elongation factor-1 alpha (EF1α) loci in strains originated from Rio de Janeiro (RJ, n = 15), Rio Grande do Sul (RS, n = 10), Paraná (PR, n = 4), São Paulo (SP, n = 3) and Minas Gerais (MG, n = 1). Our results showed that S. brasiliensis is highly prevalent among cats (96.9%) with sporotrichosis, while S. schenckii was identified only once. The genotype of Sporothrix from cats was found identical to S. brasiliensis from human sources confirming that the disease is transmitted by cats. Sporothrix brasiliensis presented low genetic diversity compared to its sister taxon S. schenckii. No evidence of recombination in S. brasiliensis was found by split decomposition or PHI-test analysis, suggesting that S. brasiliensis is a clonal species. Strains recovered in states SP, MG and PR share the genotype of the RJ outbreak, different from the RS clone. The occurrence of separate genotypes among strains indicated that the Brazilian S. brasiliensis epidemic has at least two distinct sources. We suggest that cats represent a major host and the main source of cat and human S. brasiliensis infections in Brazil.
Author Summary
Sporotrichosis is a subcutaneous mycosis acquired by traumatic inoculation of soil and plant material contaminated with infectious propagules of the pathogen. The transmission of the disease by cats to other animals and humans occurs by biting or scratching, promoting direct inoculation of yeast cells into host tissue. This may represent an alternative and a successful transmission of the fungus. In order to understand the impact of felines on the epidemiology of sporotrichosis, we evaluated the phenotypic and genotypic features of isolates obtained from animals and humans living in outbreak areas. Although sporotrichosis is caused by a complex of species, in this study we observed that S. brasiliensis is the prevalent etiological agent of feline sporotrichosis, having been recovered from 96.9% of the samples. Moreover, this approach allowed us to recognize that isolates from RJ, SP, PR and MG states are genetically similar among them but different from feline isolates recovered from the RS epidemic. Our study brings new insights into the eco-epidemiology of sporotrichosis in Brazil, clarifying the distribution and prevalence of S. brasiliensis in feline outbreaks. Knowledge about the source and distribution of the etiological agent between outbreak areas may help to establish public strategies for the containment of the epidemic of sporotrichosis in Brazil.
Introduction
Mycotic diseases, particularly those caused by dimorphic fungi such as Sporothrix, can be considered as an emerging threat to various species of animals. Upon introduction of propagules into the mammalian host, the fungus undergoes a thermodimorphic transition to a yeast-like phase, leading to infections varying between fixed localized cutaneous lesions to severe, disseminated sporotrichosis.
The first connection between Sporothrix and animals was made by Lutz and Splendore [1]. Since then sporotrichosis has been reported in dogs, cats, horses, cows, camels, dolphins, goats, mules, birds, pigs, rats, and armadillos, as well as in humans. However, the cat is the animal species most affected by this mycosis [2]. Over the last two decades, Brazil has experienced its largest epidemic of sporotrichosis due to zoonotic transmission, whereby cats were pointed out as key susceptible host. The zoonotic potential of infected cats has been demonstrated by the isolation of S. schenckii s.l. from feline skin lesions, nasal, oral cavities, and claw fragments [3], [4].
In contrast to the classical route of infection by Sporothrix, where soil and plant material loaded with saprophytic hyphae of the pathogen were the source of contamination [5], transmission of Sporothrix spp. by cats to other cats and to humans via direct inoculation of yeast cells represents an alternative and a successful type of dispersal of the disease. The yeast form is more virulent than the mycelial form [6], [7]. Transmission of yeast cells may enhance the appearance of more severe forms of the disease.
Until recently, S. schenckii was considered to be the only species causing sporotrichosis. The infection has a worldwide distribution, mainly in tropical and subtropical countries [8]–[10]. The most common clinical manifestations in humans are the lymphocutaneous and fixed forms, but other clinical types, such as a disseminated form, may also occur [11], [12], partly depending on the immune status of the host.
Multilocus sequencing combined with morphological and physiological data support the separation of at least four distinct Sporothrix species within the S. schenckii complex, uniting the species with high pathogenic potential to mammals. The original taxon S. schenckii (Clades IIa and IIb) and the novel species S. brasiliensis (Clade I), S. globosa (Clade III), and S. luriei (Clade VI) todays are referred to as the S. schenckii complex [13], while the mildly pathogenic species S. mexicana (Clade IV) takes a remote position near the environmental species S. pallida [11], [14]–[19]. The Sporothrix species differ in their pathogenic potential for mammals [20], [21], their geographical distribution [11], [13], [15], [17], and in their sensitivity to antifungal therapy [22]. All species have been reported from Brazil [11].
Endemic areas of sporotrichosis in Brazil are characterized by poor sanitation, substandard housing and little or no access to health services – a challenge to control and eradication of the disease. The oldest outbreaks of sporotrichosis among humans and cats have been reported in the states of Rio de Janeiro [3], [23], [24] and Rio Grande do Sul [25], [26]. Delayed diagnosis and treatment in cats may lead to a rapid spread of the disease through the community members. The increase in the number of cases in cats is followed by higher numbers of human cases, which constitutes a serious public health problem.
Despite the increasing frequency and severity of cases, the eco-epidemiology of feline sporotrichosis in Brazil is still unknown. The aim of the present study was to determine the distribution and prevalence of Sporothrix species among naturally infected felines using phenotypic and molecular phylogenetic approaches.
Methods
Isolates and culture conditions
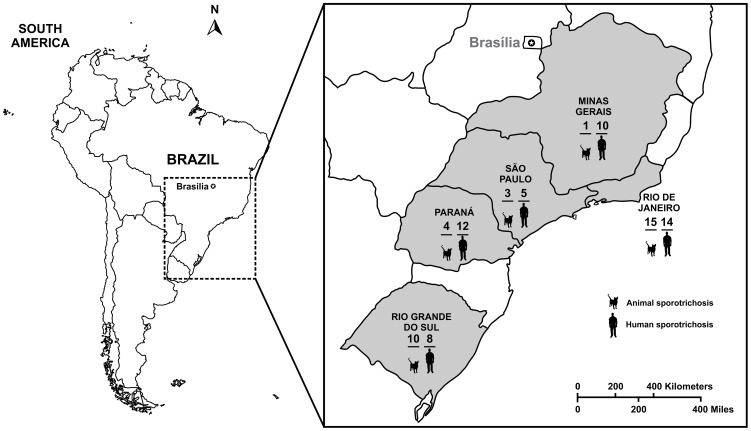
Thirty three (33) Sporothrix isolates from Rio de Janeiro, RJ (n = 15); Rio Grande do Sul, RS (n = 10); Paraná, PR (n = 4); São Paulo, SP (n = 3) and Minas Gerais, MG (n = 1) were obtained from lesions of cats and dogs with sporotrichosis (skin or mucosa lesions) (Fig. 1). Fungal cells were recovered directly from lesions and cultured on Mycosel agar (Difco Laboratories, Detroit, Mich.). Suspected colonies were subcultured on potato dextrose agar (Difco Laboratories, Detroit, Mich.) at room temperature. Isolates were identified phenotypically as S. schenckii s.l. As a control, human clinical isolates (n = 66) inside and outside the Brazilian feline outbreaks areas were included in the study (Table 1).
Figure 1. South America map showing sampling localities in Brazil and total number of animals (n = 33) and humans Sporothrix spp.
(n = 49) isolates evaluated in Rio de Janeiro, Minas Gerais, São Paulo, Paraná and Rio Grande do Sul. = 17) outside the gray area and used as control are not shown in the picture.
Table 1. Strains, species, origin, and GenBank accession numbers of Sporothrix spp. isolates used in this study.
| GenBank | |||||||
| Isolate code | CBS code | Species | Source | Geographic origin | CAL | EF1α | Reference1 |
| Ss05 | CBS 132985 | Sporothrix brasiliensis | Feline sporotrichosis | Belo Horizonte, MG, Brazil | KC693830 | KC576544 | This study |
| Ss53 | CBS 132989 | Sporothrix brasiliensis | Feline sporotrichosis | Rio Grande, RS, Brazil | KC693846 | KC576568 | This study |
| Ss54 | CBS 132990 | Sporothrix brasiliensis | Feline sporotrichosis | Rio Grande, RS, Brazil | JQ041903 | KC576569 | This study |
| Ss152 | CBS 132995 | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693865 | KC576596 | This study |
| Ss153 | CBS 132996 | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693866 | KC576597 | This study |
| Ss154 | - | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693867 | KC576598 | This study |
| Ss155 | - | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693868 | KC576599 | This study |
| Ss156 | CBS 132997 | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693869 | KC576600 | This study |
| Ss157 | CBS 132998 | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693870 | KC576601 | This study |
| Ss171 | CBS 132999 | Sporothrix brasiliensis | Feline sporotrichosis | Londrina, PR, Brazil | KC693871 | KC576602 | This study |
| Ss172 | CBS 133000 | Sporothrix brasiliensis | Feline sporotrichosis | Londrina, PR, Brazil | KC693872 | KC576603 | This study |
| Ss173 | CBS 133001 | Sporothrix brasiliensis | Feline sporotrichosis | Londrina, PR, Brazil | KC693873 | KC576604 | This study |
| Ss174 | CBS 133002 | Sporothrix brasiliensis | Feline sporotrichosis | Londrina, PR, Brazil | KC693874 | KC576605 | This study |
| Ss226 | CBS 133003 | Sporothrix brasiliensis | Feline sporotrichosis | São Paulo, SP, Brazil | KC693875 | KC576616 | This study |
| Ss245 | CBS 133005 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693878 | KC576619 | This study |
| Ss246 | - | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693879 | KC576620 | This study |
| Ss247 | CBS 133006 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693880 | KC576621 | This study |
| Ss248 | CBS 133007 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693881 | KC576622 | This study |
| Ss249 | CBS 133008 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693882 | KC576623 | This study |
| Ss250 | CBS 133009 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693883 | KC576624 | This study |
| Ss251 | CBS 133010 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693884 | KC576625 | This study |
| Ss252 | CBS 133011 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693885 | KC576626 | This study |
| Ss253 | CBS 133012 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693886 | KC576627 | This study |
| Ss254 | CBS 133013 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693887 | KC576628 | This study |
| Ss255 | CBS 133014 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693888 | KC576629 | This study |
| Ss256 | CBS 133015 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693889 | KC576630 | This study |
| Ss257 | CBS 133016 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693890 | KC576631 | This study |
| Ss258 | CBS 133017 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693891 | KC576632 | This study |
| Ss259 | CBS 133018 | Sporothrix brasiliensis | Feline sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693892 | KC576633 | This study |
| Ss260 | CBS 133019 | Sporothrix brasiliensis | Feline sporotrichosis | Pelotas, RS, Brazil | KC693893 | KC576634 | This study |
| Ss151 | CBS 132994 | Sporothrix brasiliensis | Canine sporotrichosis | Pelotas, RS, Brazil | KC693864 | KC576595 | This study |
| Ss227 | CBS 133004 | Sporothrix brasiliensis | Canine sporotrichosis | São Paulo, SP, Brazil | KC693876 | KC576617 | This study |
| Ss07 | CBS 132986 | Sporothrix brasiliensis | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693831 | KC576546 | This study |
| Ss08 | - | Sporothrix brasiliensis | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693832 | KC576547 | This study |
| Ss09 | - | Sporothrix brasiliensis | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693833 | KC576548 | This study |
| Ss10 | CBS 132987 | Sporothrix brasiliensis | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693834 | KC576549 | This study |
| Ss12 | - | Sporothrix brasiliensis | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693835 | KC576550 | This study |
| Ss25 | CBS 132988 | Sporothrix brasiliensis | Human sporotrichosis | Curitiba, PR, Brazil | KC693840 | KC576556 | This study |
| Ss27 | - | Sporothrix brasiliensis | Human sporotrichosis | Curitiba, PR, Brazil | JX077111 | KC576558 | [11] |
| Ss38 | - | Sporothrix brasiliensis | Human sporotrichosis | Curitiba, PR, Brazil | KC693844 | KC576563 | This study |
| Ss52 | - | Sporothrix brasiliensis | Human sporotrichosis | São Paulo, SP, Brazil | KC693845 | KC576567 | This study |
| Ss55 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio Grande, RS, Brazil | KC693847 | KC576570 | This study |
| Ss56 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio Grande, RS, Brazil | KC693848 | KC576571 | This study |
| Ss62 | CBS 132991 | Sporothrix brasiliensis | Human sporotrichosis | Vila Velha, ES, Brazil | JX077113 | KC576572 | [11] |
| Ss69 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693849 | KC576575 | This study |
| Ss70 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693850 | KC576576 | This study |
| Ss71 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693851 | KC576577 | This study |
| Ss72 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693852 | KC576578 | This study |
| Ss79 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693856 | KC576582 | This study |
| Ss82 | CBS 132992 | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693857 | KC576584 | This study |
| Ss87 | CBS 132993 | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693858 | KC576585 | This study |
| Ss125 | - | Sporothrix brasiliensis | Human sporotrichosis | Campinas, SP, Brazil | JX077116 | KC576588 | [11] |
| Ss128 | - | Sporothrix brasiliensis | Human sporotrichosis | Campinas, SP, Brazil | KC693861 | KC576589 | This study |
| Ss149 | - | Sporothrix brasiliensis | Human sporotrichosis | Pelotas, RS, Brazil | KC693862 | KC576593 | This study |
| Ss150 | - | Sporothrix brasiliensis | Human sporotrichosis | Pelotas, RS, Brazil | KC693863 | KC576594 | This study |
| CBS 120339T | CBS 120339T | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | AM116899 | KC576606 | [19] |
| IPEC 16919 | - | Sporothrix brasiliensis | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | AM116898 | KC576607 | [19] |
| Ss261 | - | Sporothrix brasiliensis | Human sporotrichosis | Pelotas, RS, Brazil | KC693894 | KC576635 | This study |
| Ss265 | CBS 133020 | Sporothrix brasiliensis | Human sporotrichosis | Uberaba, MG, Brazil | JN204360 | KC576636 | [12] |
| Ss01 | CBS 132961 | Sporothrix schenckii | Feline sporotrichosis | São Paulo, SP, Brazil | KC693828 | KC576540 | This study |
| Ss02 | CBS 132962 | Sporothrix schenckii | Human sporotrichosis | Porto Alegre, RS, Brazil | KC693829 | KC576541 | This study |
| Ss03 | CBS 132963 | Sporothrix schenckii | Human sporotrichosis | Porto Alegre, RS, Brazil | JX077117 | KC576542 | [11] |
| Ss04 | - | Sporothrix schenckii | Human sporotrichosis | Porto Alegre, RS, Brazil | JX077118 | KC576543 | [11] |
| Ss13 | - | Sporothrix schenckii | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693836 | KC576551 | This study |
| Ss15 | - | Sporothrix schenckii | Human sporotrichosis | Belo Horizonte, MG, Brazil | KC693837 | KC576552 | This study |
| Ss17 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | KC693838 | KC576553 | This study |
| Ss20 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | JX077119 | KC576554 | [11] |
| Ss24 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | KC693839 | KC576555 | This study |
| Ss26 | CBS 132965 | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | KC693841 | KC576557 | This study |
| Ss28 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | JX077121 | KC576559 | [11] |
| Ss31 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | JX077122 | KC576560 | [11] |
| Ss35 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | KC693842 | KC576561 | This study |
| Ss36 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | KC693843 | KC576562 | This study |
| Ss39 | - | Sporothrix schenckii | Human sporotrichosis | Curitiba, PR, Brazil | JQ041899 | KC576564 | This study |
| Ss63 | CBS 132968 | Sporothrix schenckii | Human sporotrichosis | Vila Velha, ES, Brazil | JX077123 | KC576573 | [11] |
| Ss64 | - | Sporothrix schenckii | Human sporotrichosis | Vila Velha, ES, Brazil | JX077124 | KC576574 | [11] |
| Ss73 | - | Sporothrix schenckii | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693853 | KC576579 | This study |
| Ss75 | - | Sporothrix schenckii | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693854 | KC576580 | This study |
| Ss78 | - | Sporothrix schenckii | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693855 | KC576581 | This study |
| Ss80 | CBS 132969 | Sporothrix schenckii | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | JX077125 | KC576583 | [11] |
| Ss90 | - | Sporothrix schenckii | Human sporotrichosis | Rio de Janeiro, RJ, Brazil | KC693859 | KC576586 | This study |
| Ss111 | CBS 132971 | Sporothrix schenckii | Human sporotrichosis | São Paulo, SP, Brazil | KC693860 | KC576587 | This study |
| Ss143 | - | Sporothrix schenckii | Human sporotrichosis | Belém, PA, Brazil | JQ041903 | KC576592 | [11] |
| CBS 359.36T | CBS 359.36T | Sporothrix schenckii | Human sporotrichosis | USA | AM117437 | KC576614 | [19] |
| CBS93872 | CBS 93872 | Sporothrix schenckii | Human sporotrichosis | France | AM490340 | KC576637 | [17] |
| Ss06 | CBS 132922 | Sporothrix globosa | Human sporotrichosis | Belo Horizonte, MG, Brazil | JF811336 | KC576545 | [11] |
| Ss41 | CBS 132923 | Sporothrix globosa | Human sporotrichosis | Fortaleza, CE, Brazil | JF811337 | KC576565 | [11] |
| Ss49 | CBS 132924 | Sporothrix globosa | Human sporotrichosis | Goiânia, GO, Brazil | JF811338 | KC576566 | [11] |
| CBS 120340T | CBS 120340T | Sporothrix globosa | Human sporotrichosis | Spain | AM116908 | KC576608 | [19] |
| CBS 130104 | CBS 130104 | Sporothrix globosa | Human sporotrichosis | Spain | AM116905 | KC576609 | [19] |
| Ss236 | CBS 132925 | Sporothrix globosa | Human sporotrichosis | Minas Gerais, MG, Brazil | KC693877 | KC576618 | This study |
| FMR 8598 | CBS130116 | Sporothrix globosa | Human sporotrichosis | Spain | AM116903 | KC576638 | [19] |
| CBS 937.72T | CBS 937.72T | Sporothrix luriei | Human sporotrichosis | South Africa | AM747302 | KC576615 | [18] |
| Ss132 | CBS 132927 | Sporothrix mexicana | Human sporotrichosis | São Paulo, SP, Brazil | JF811340 | KC576590 | [11] |
| Ss133 | CBS 132928 | Sporothrix mexicana | Human sporotrichosis | Recife, PE, Brazil | JF811341 | KC576591 | [11] |
| CBS 120342 | CBS 120342 | Sporothrix mexicana | Vegetal | Mexico | AM398392 | KC576610 | [17] |
| CBS 120341T | CBS 120341T | Sporothrix mexicana | Soil | Mexico | AM398393 | KC576611 | [17] |
| CBS 302.73T | CBS 302.73T | Sporothrix pallida | Soil | United Kingdom | AM398396 | KC576612 | [17] |
| CBS 111110 | CBS 111110 | Sporothrix pallida | Insect | Germany | AM398382 | KC576613 | [17] |
| CMW 304 | CBS 141.36T | Grosmannia serpens | Environmental | Italy | JN135300 | - | [30] |
| AFTOL-ID 910 | CBS 158.74 | Ophiostoma piliferum | Environmental | Chile | - | DQ471074 | [31] |
Calmodulin literature reference. IPEC, Instituto de Pesquisa Clínica Evandro Chagas, Fiocruz, Brazil; FMR, Facultat de Medicina i Ciències de la Salut, Reus, Spain; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; KMU, Kanazawa Medical University, Ishikawa, Japan; CMW, Culture Collection of the Forestry and Agricultural Biotechnology Institute (FABI); AFTOL, Assembling the Fungal Tree of Life project; NK, not known;
, type strain. All “Ss” strains belong to the culture collection of Federal University of São Paulo (UNIFESP). MG, Minas Gerais; RS, Rio Grande do Sul; PR, Paraná; SP, São Paulo; RJ, Rio de Janeiro; ES, Espírito Santo; PA, Pará; CE, Ceará; GO, Goiás, PE, Pernambuco.
Phenotypic characterization
Morphological identification of cultures was performed according to Marimon et al. [17], [18] including vegetative growth on PDA media at 30, 35, 37 and 40°C, colony colors on corn meal agar (Difco Laboratories, Detroit, Mich.), assimilation profiles of raffinose, ribitol and sucrose, and microscopic morphology in vitro. Growth at different temperatures was measured according to Mesa-Arango et al. [27]: the percent growth inhibition (GI) was calculated at 37°C by the following formula [(colony diameter at 30°C – colony diameter at 37°C)/colony diameter at 30°C]×100. The GI was evaluated by analysis of variance/Tukey test using the GraphPad (GraphPad Prism v. 5.00 for Windows, San Diego California USA, www.graphpad.com), considering statistically significant when p<0.05. Observed data were used for taxonomic characterization applying the dichotomous key to species of the S. schenckii complex proposed by Marimon et al. [18].
DNA extraction, PCR amplification and DNA sequencing
For molecular analysis, genomic DNA was extracted and purified directly from mycelial colonies following the Fast DNA kit protocol (MP Biomedicals, Vista, CA, USA) with the homogenization step repeated three times with a Precellys 24 instrument (Bertin, Montigny le Bretonneux, France). DNA was quantified with NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The calmodulin (CAL) locus region was amplified directly from the genomic DNA by PCR, as described by O'Donnell et al. [28], using the degenerate primers CL1 (5′-GAR TWC AAG GAG GCC TTC TC-3′) and CL2A (5′-TTT TTG CAT CAT GAG TTG GAC-3′), which generated an 800-bp amplicon corresponding to exons 3 through 5. The translation elongation factor-1 alpha (EF1α) locus region was amplified using the newly designed primers EF1-F (5′-CTG AGG CTC GTT ACC AGG AG-3′) and EF1-R (5′-CGA CTT GAT GAC ACC GAC AG-3′) which amplified an 850-bp fragment, covering the last exon of this gene, matching the same region evaluated by the consortium Assembling the Fungal Tree of Life (AFTOL).
Thermal cycling conditions were as follows: one cycle of 5 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 60°C (CAL) or 57°C (EF1α) and 1 min at 72°C, followed by one cycle of 10 min at 72°C.
Amplified products were gel-purified with the Wizard® SV Gel and PCR Clean-Up System (Promega, USA) following the manufacturer instructions. DNA samples were completely sequenced with an ABI 3730 DNA Analyser (Applied Biosystems, Foster City, CA, USA) using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The fragments were sequenced on both strands to increase the quality of sequence data and assembled into single sequences via CAP3 using bases with quality of phred ≥30. Sequences were aligned with MAFFT v. 5.667 [29] and retrieved alignments were manually edited in order to avoid mis-paired bases.
Phylogenetic analysis
Calmodulin sequences deposited at GenBank belonging to the clades of clinical importance in the S. schenckii complex (Table 1) were collected and included in the present alignment as reference strains for the phylogenetic distribution. We choose the saprophytic fungus Grosmannia serpens (Ophiostomataceae), CBS 141.36 [30] as outgroup for CAL analysis [11]. All Sporothrix EF1α sequences used in the phylogenetic analysis were generated in this study (Table 1). The outgroup for the EF1α analysis included the saprophytic fungus Ophiostoma piliferum, CBS 158.74 (AFTOL-ID 910) [31]. This species was chosen because the genus Ophiostoma (Ophiostomataceae) is considered a close related group to Sporothrix species [32].
Phylogenetic analyses were carried out using Neighbor-joining, Maximum Likelihood and Bayesian methods. Neighbor-Joining and Maximum Likelihood trees were constructed using MEGA 5 software [33] and 1000 bootstrap replicates were used to estimate confidence values for individual clades and are shown next to the branches [34]. The evolutionary distances were computed using the Tamura 3-parameters method [35] and the rate variation among sites was modeled with a gamma distribution (shape parameter = 1). For Bayesian analysis by Markov Chain Monte Carlo (MCMC), two independent analyses of four chains each as default were initiated from a random tree and processed for 1.000.000 generations; sample trees were retrieved every 1000 generations. Log-likelihood scores were plotted against its generation number in order to evaluate convergence; samples collected prior to “burn-in” (25%) were ignored. The remaining samples were used to determine the distribution of posterior probability values [36]. The posterior probabilities values of generated clades and overall topology of each replicate were compared in order to verify that each consensus tree converged on a similar phylogeny. Phylograms generated by Bayesian analysis were used to represent the phylogenetic distribution and were produced with the help of the Figtree 1.0 software (available at http://tree.bio.ed.ac.uk/software/figtree/).
Haplotype network
Evolutionary relationships at the intraspecific level were evaluated using haplotype networks in order to visualize differences and diversity among S. brasiliensis sequence data. The number and diversity of CAL and EF1α haplotypes were estimated using the software DNAsp v5.0 [37]. Gaps and missing data were excluded in the calculation. Median-joining networks [38] for the CAL and EF1α dataset were obtained and visualized using the software Network 4.610 (available at www.fluxus-engineering.com).
Recombination event detection
Evidence of recombination in S. brasiliensis population isolated from animals and humans samples was inferred by the split decomposition method [39], implemented by the Splitstrees4 software, version 4.8 [40] which is used to identify branching ambiguities attributable to recombination events. The presence of recombination networks can be detected by bridges between members of the genetically isolated groups. Each isolated group will have an independent branch, showing that it does not share genetic material with the others. This analysis allowed the assessment of recombination possibilities within and between the seven phylogenetic groups considered.
The PHI-test incorporated in the SplitsTree software [40] was used to test signals of recombination (p<0.05, significant evidence of recombination). The test is proven to be a robust calculation and no previous knowledge about population history, recombination rate, mutation rate and rate heterogeneity across sites [41] is necessary. Although large splits in networks do not necessarily imply recombination, split decomposition networks in conjunction with the PHI-test can easily detect which sequences in a given data set contribute the most to the recombination signal [42]. The PHI-test is repeated after possible recombinants are deleted from the alignment until p>0.05 (no evidence of recombination). Also, DNAsp v5.1 [43] was used to evaluate minimum number of recombination events in the history and haplotypic diversity of S. brasiliensis population. The software computes the recombination parameter R = 4Nr, where N is the population size and r is the recombination rate per sequence -or between adjacent sites [44].
Ethics statement
The animals included in this study were examined by a veterinarian with experience in small animal internal medicine. The procedures performed in these animals were approved by the Ethics in Research Committee (CEUA) of the FIOCRUZ, Rio de Janeiro, Brazil, under license number L-041/06.
Results
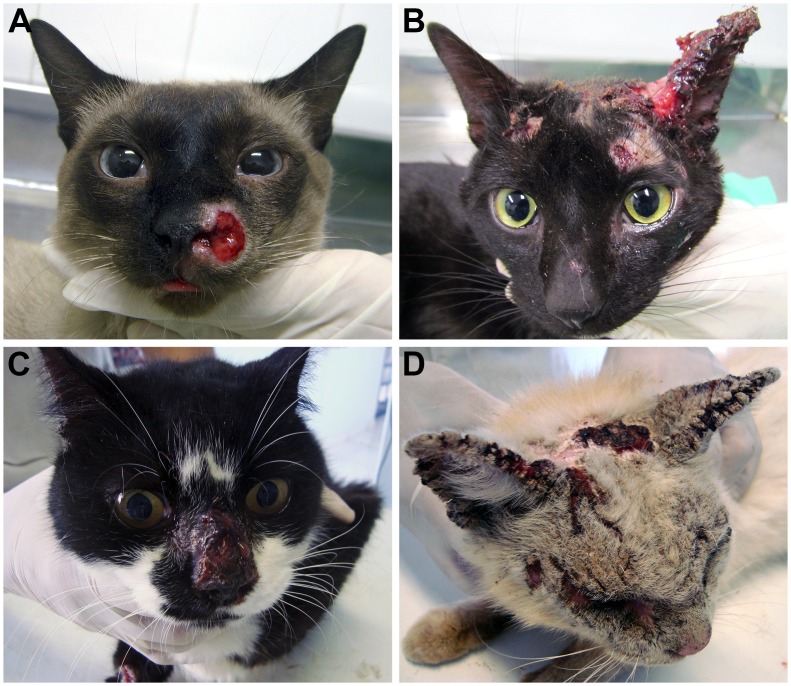
Our study included indoor and feral cats from five different geographic regions in Brazil (RJ, RS, MG, SP, and PR). Diagnosis of sporotrichosis was performed by the clinical evaluation of skin lesions and confirmed by isolation of the pathogen. The suspected colonies of Sporothrix species were grown on Mycosel agar until purification of the pathogen. The fungus was easily isolated from material from the nasal, oral mucosa and skin lesions. Lesions in the cephalic region and/or respiratory tract were observed in most of the animals (Fig. 2).
Figure 2. Clinical aspects of feline sporotrichosis in Brazil.
Cats presenting ulcerated cutaneous lesions in the cephalic region. (A) and (B) felines from Rio de Janeiro; (C) and (D) felines from Paraná.
Phenotypic characterization, i.e. growth at various temperatures, macroscopic and microscopic features, and carbohydrate assimilation, yielded data similar to those found for the reference strains of S. brasiliensis (CBS 120339) and S. schenckii (CBS 359.36) reported by Marimon et al. [17]. Among the 33 strains of Sporothrix isolated from cats (n = 31) and dogs (n = 2) from different geographic regions of Brazil, 32 belonged to S. brasiliensis (96.9%) and 1 to S. schenckii (3%). These phenotypic results showed that S. brasiliensis is highly prevalent among cats with sporotrichosis. The two isolates recovered of canine sporotrichosis (CBS 132994 and CBS 133004 from RS and SP, respectively) were identified as S. brasiliensis.
Using CL1 and CL2A primers we amplified 800 bp of the CAL locus. The complete alignment included 100 strains. Aligned sequences of CAL were 727 bp long, including 366 invariable characters, 214 variable parsimony-informative (29.43%), and 125 singletons. Comparison with sequences available at GenBank revealed a match of 99–100% with the type strain of S. brasiliensis (CBS 120339, AM116899) corroborating our phenotypic data. The single isolate of S. schenckii (CBS 132961) matched 99% with the S. schenckii s. str. strain (FMR 8678, AM117446) from Argentina.
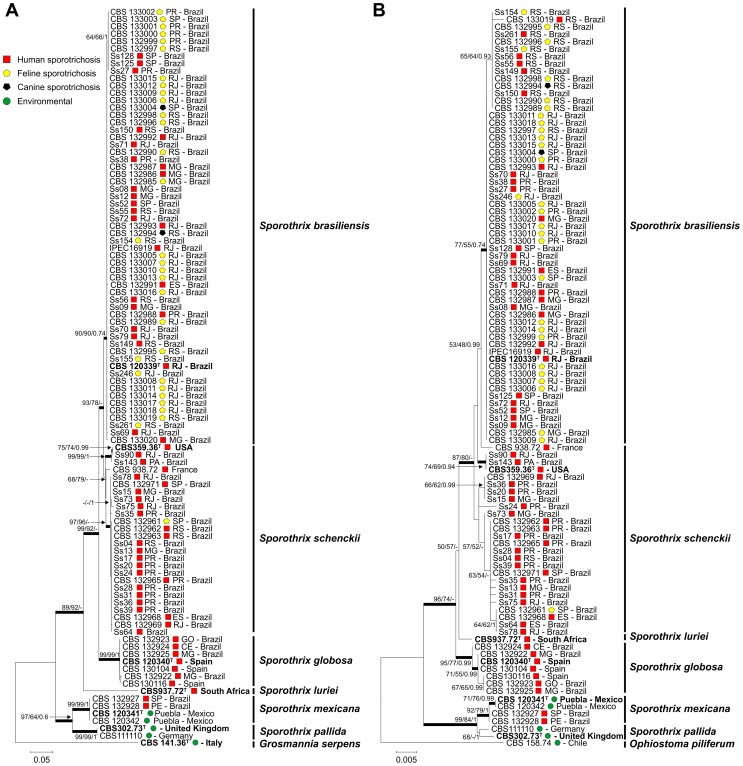
Phylogenetic analysis of isolates from cats and dogs revealed that S. brasiliensis is the prevalent species (32/33); only a single isolate clustered with S. schenckii s. str. The clade of pathogenic Sporothrix species was well supported with high bootstrap and posterior probability values. The S. brasiliensis isolates recovered from animal sources clustered in a single branch together with clinical isolates, indicating that they belonged to the same genotypes and confirming that the disease is transmitted by cats. A cryptic branch was observed in the S. brasiliensis clade composed of the isolates Ss27, Ss125, Ss128, CBS 132997, CBS 132999, CBS 133000, CBS 133001 CBS 133002 and CBS 133003, supported by bootstrap and posterior probabilities values (64/66/1) (Fig. 3A).
Figure 3. Phylogenetic trees generated by Neighbor-joining, Maximum Likelihood and Bayesian analysis using partial nucleotide sequences of the calmodulin-encoding gene (A) and the translation elongation factor-1 alpha (EF1α) locus region (B).
Bootstrap and posterior probabilities values were added to respective branches (NJ/ML/BI). Each species are indicated at each respective position at the phylogenetic tree. Calmodulin and EF1α accessions number are indicated in the Table 1.
Sporothrix brasiliensis presented low genetic diversity compared to its sister taxon S. schenckii when CAL was used as a marker. Elongation factor (EF1α) was used as marker to assess the genetic diversity in the species. All isolates presented similar fragments of 850 bp of the EF1α locus which were amplified and sequenced with primers EF1-F and EF1-R. Aligned sequences of EF1α were 707 bp long, including 639 invariable characters, 34 variable parsimony-informative (5.08%), and 33 singletons. The 100 OTUs were distributed into 7 main groups (Fig. 3B), which were congruent with the CAL phylogeny.
Judging from the EF1α dataset, the S. brasiliensis isolates recovered from animal sources in RJ and RS clustered in two branches with human clinical isolates from the same states, indicating two epidemics with distinct genotypes are concerned (Fig. 3B). Sporothrix brasiliensis presented low genetic diversity in EF1α, in accordance with results obtained for the CAL locus.
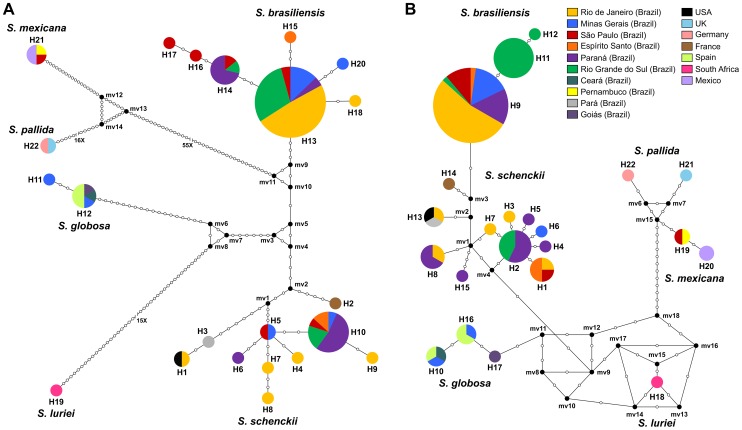
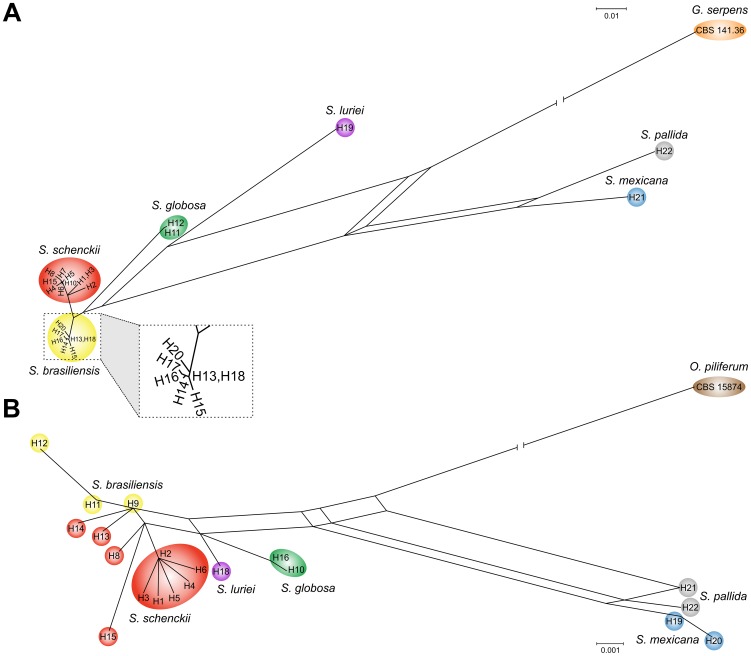
The haplotype diversity of S. brasiliensis species was assessed using the DNASp software. Only 7 haplotypes for CAL (Fig. 4A) and 3 haplotypes for EF1α (Fig. 4B) were found. The low values of haplotype (HdCAL = 0.36 and HdEF1α = 0.37) and nucleotide diversities (πCAL = 0.00152 and πEF1α = 0.00062) lead us to hypothesize that this species is clonal (Table S1). Geographical separation between the RJ and RS epidemics for the EF1α locus was clear. The median-joining network based on the EF1α haplotype showed an intraspecific separation (Fig. 4B, haplotypes H11 and H12) resulting from a nucleotide transition from A to G, between isolates from RJ and RS epidemics (Table S2). The average divergence between S. brasiliensis and its sister species S. schenckii is much higher, suggesting that the species experienced different evolutionary processes.
Figure 4. Median-joining haplotype network of Sporothrix schenckii complex isolates based on partial nucleotide sequences of the calmodulin-encoding gene (A) and the translation elongation factor-1 alpha (EF1α) loci regions (B).
The EF1α haplotype showed a clear intraspecific separation resultant from a nucleotide transition from A to G, between S. brasiliensis isolates recovered from Rio de Janeiro (H9) and Rio Grande do Sul (H11 and H12) feline epidemics. The size of the circumference is proportional to the haplotype frequency. Black dots (median vectors) are hypothetical missing intermediates. Calmodulin and EF1α haplotypes are detailed in the Table S2.
Recombination analysis of S. brasiliensis was first assessed by split decomposition method and no networks linking different isolates were observed in both datasets (Fig. 5), in agreement with the concept of clonal species. Also PHI-test analysis showed no evidence of recombination (pCAL = 0.757 and pEF1α = 0.903), and no recombination events were detected by DNAsp5 software. Taken together, these analyses indicated that S. brasiliensis is a clonal species.
Figure 5. Split decomposition analysis of the Sporothrix brasiliensis isolates from zoonotic epidemic outbreaks in different geographic regions in Brazil according to sequences of the calmodulin-encoding gene (A) and the translation elongation factor-1 alpha (EF1α) locus region (B).
The inset Box represents the S. brasiliensis species alone, showing the absence of recombination possibilities within this species. The absence of reticulated phylogenetic structure in the S. brasiliensis haplotypes suggests a clonality spread of this species among human, cats and dogs in Brazil for both loci.
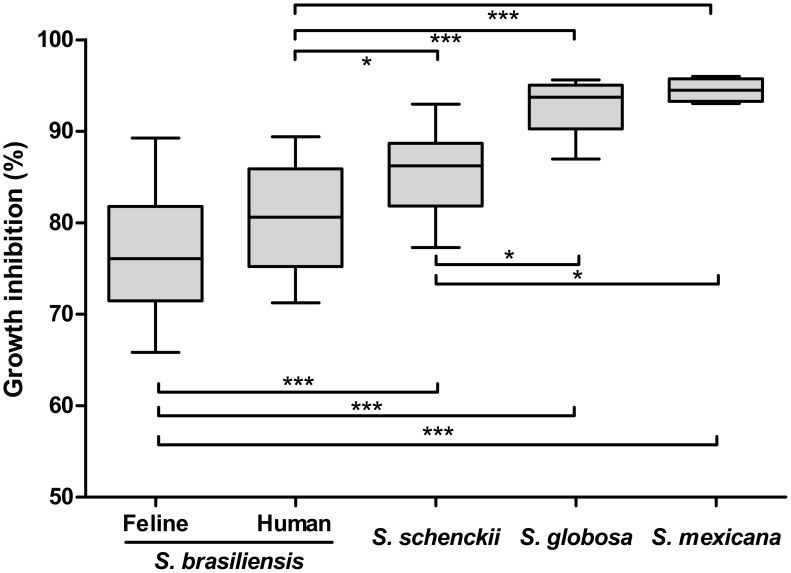
Aiming to evaluate possible phenotypic characteristics that explain the success of this pathogen adaptation to the feline host we evaluated the thermal resistance of strains of clinical interest (human and animal) and environmental strains. Strains of S. brasiliensis from feline origin (n = 30) showed highest temperature tolerance, being inhibited 77.1±6.32% on average at 37°C (Fig. 6). The group differed statistically from other species evaluated herein (S. schenckii s. str., S. globosa, and S. mexicana), suggesting that this factor may confer advantage during the process of infection in the feline host.
Figure 6. In vitro temperature fitness in the Sporothrix species.
Growth inhibition at 37°C compared to 30°C incubation. S. brasiliensis from feline (n = 30) or human source (n = 27) are more resistant to heat incubation and differ statistically when compared to S. schenckii (n = 25), S. globosa (n = 7) and S. mexicana (n = 4). Statistical significance in one-way ANOVAs followed by Tukey's tests: * p<0.05, *** p<0.0001. The line in the boxes and upper and lower bars show the median, maximum and minimum values, respectively. Isolates were not compared at superior temperature (38–40°C) due to low growth observed to S. globosa and S. mexicana. No isolate were able to growth at 40°C.
Supporting information
Supplementary information reported in this section is complementary to the results and describe the genetic diversity of the Sporothrix isolates.
Discussion
Epidemiology of fungal infections can be influenced by several factors, including: (i) biological factors such as fungal virulence and host resistance, (ii) ecological factors such as temperature, atmospheric humidity, ultraviolet radiation, geological conditions, and inter-relationships with other living beings, and (iii) socio-economic factors such as poverty, sanitation, clothing, profession, prophylactic habits and population migrations. In the Brazilian epidemic of feline sporotrichosis a combination of a highly virulent fungus and a susceptible host coupled to low sanitary conditions in the suburbs has made the state of RJ a highly endemic area of this mycosis among animals and humans. The epidemic proportions are noted only since the last two decades.
Little is known about the eco-epidemiology of feline sporotrichosis and its impact on the epidemiology of human sporotrichosis. Cats play a significant role in outbreak areas of sporotrichosis such as RJ and RS. Classically, humans can acquire sporotrichosis by cat scratches or bites, the reason why cats are considered important source of infection in the spread of the disease. In our study we found that S. brasiliensis is the prevalent etiological agent of feline sporotrichosis in Brazil. Among cats, S. brasiliensis was identified in a total of 96.9% of the samples, by isolation of the pathogen from lesions and posterior phenotypic and molecular characterization.
Interestingly, a correlation between cat outbreaks and prevalence of S. brasiliensis among humans was found in the same geographic area, such as in RJ (Table 1). This fact matches with our hypothesis that outbreaks among cats directly influence the prevalence of S. brasiliensis in human cases of sporotrichosis in the same geographic area. A similar situation was observed in the state of RS where S. brasiliensis was isolated with high frequency from cats as well as from humans.
Marimon et al. [17] analyzed 127 Sporothrix isolates using the calmodulin locus and five major clades (I–V) were recognized. The maximum likelihood, neighbor-joining and Bayesian analyses based on the calmodulin (Fig. 3A) or EF1α (Fig. 3B) loci placed our animal Sporothrix isolates in Clade I (S. brasiliensis) composed of clinical samples from the RJ State epidemic, with strong bootstrap and posterior probability support. All pathogenic Sporothrix species are known to occur in Brazil [11], but S. brasiliensis is relatively frequent among feline sporotrichosis outbreaks.
The geographic origin of S. brasiliensis of the Brazilian epidemic is difficult to establish. At least two distinct genotypes occur: one in RS and another in RJ. The latter is the oldest and longest recorded in the literature [3], [4], [23], [24]. Our data show that humans and animals infected in the RS epidemic harbor the same S. brasiliensis genotype, which is distinct from the one of the RJ epidemic. The RJ genotype is also present in the recent outbreaks in PR, MG and SP, which suggests spread of S. brasiliensis from RJ. Additionally, our results showed absence of recombination events in the CAL and EF1α loci, demonstrating that S. brasiliensis is a clonal species. Despite a recent indication of intraspecific variability within the species S. brasiliensis using RAPD [45] we believe that this phenomenon is not frequent or strong enough to break the prevalent pattern of clonal population structure, i.e., recombination or scarce exchange of genetic material may occur in some point of the evolutionary course of the pathogen life without compromise or affect its population structure. This hypothesis has been discussed by Tibayrenc and Ayala [46] through different group of pathogens including fungi.
The existence of clonal populations has repeatedly been proven in fungal pathogens [47]–[50], although most of these species are surmised to have occasional sexuality in any phase of their life cycle. Under permissive conditions, most fungi reproduce very effectively by asexual propagation. Sexual reproduction provides advantages to the pathogen under adverse conditions, generating suitable genotypes that enhance survival. Many fungal epidemics are driven by populations showing low levels of genetic diversity, as demonstrated in Penicillium marneffei [51], [52], Cryptococcus gattii [53], [54] and Batrachochytrium dendrobatidis [55]. Also feline and human sporotrichosis in Brazil caused by S. brasiliensis is driven by the spread of a clonal species. In contrast, outbreaks of other human pathogens such as Coccidioides immitis [56]–[58] and Paracoccidioides brasiliensis [59]–[61], spread by a diversity of genotypes.
The ecological aspects of the pathogenic species within the genus Sporothrix needs to be reevaluated, and this information can be crucial to find the source of S. brasiliensis in nature. Classically, soil [5], thorny plants [62], Sphagnum moss [63]–[65] and hay [66] have been pointed as source of S. schenckii s.l. To date, just a single environmental isolate (FMR 8337) of S. brasiliensis was isolated and reported from domiciliary dust in Brazil [17], [19]. Distant relatives of Sporothrix in the fungal order Ophiostomatales are mainly associates of bark beetles on woody plants [67], [68]. Zhou et al. [13] demonstrated that different ecologies are corroborated by phylogenetic separation.
It is challenging to obtain environmental isolates of S. brasiliensis, and the low number of subjects contaminated with propagules from soil or woody plants is indeed low compared to the high occurrence in warm-blooded hosts [3], [69], [70]. This suggests successful transmission among animals (cat-cat and cat-humans). This scenario is different from epidemics occurring in South Africa [71], [72], India [10], [73], the USA [63], [64], Australia [66], [74], China [75], and Japan [76], where patients are mainly infected through soil and decaying wood. Possibly the Brazilian epidemics of S. brasiliensis are related to the emergence of a pathogenic clone front of a highly susceptible feline host, rather than to an increase in population size of S. brasiliensis in nature. This is corroborated by the high degree of virulence observed in naturally infected cats in the outbreak area [24], as well as demonstrated in a murine model [21]. Besides that, we do not discharge the hypothesis that the emergence of pathogenicity could also be attributed to a recent host-shift from an unknown host to cats as discussed in other groups of pathogens [77]–[79]. Feral cats present a great potential to spread the disease in a short period of time due to their mobility and digging behavior, whereas dispersal from soil or vegetable remains is ineffective.
Classically, accumulation of mutations in fungal populations can lead to speciation processes. However, rapid emergence of a new, highly virulent pathogen which is able to explore different ecological niches may result from other processes than those observed in natural selection. In many plant-pathogenic fungi, such as Fusarium and Alternaria, pathogenicity is determined by mobile, dispensable small chromosomes [80], [81]. Genetic processes such as hybridization of two distinct, sympatric species [82], parasexual recombination [83], [84] or mechanisms of inactivation/activation of virulence genes by insertion of transposons [85] can also drive the emergence of pathogenicity. Hybridization is one of the possible mechanisms of emergence of phytopathogenic fungi [86], [87] as well as fungi pathogenic to animals [88]. It has also been discussed in the genus Ophiostoma, which is phylogenetically related genus to Sporothrix [89]. All these genetic processes, alone or in combination, may be the reason of the emergence of virulence in the species S. brasiliensis. The lack of variation in the populations of S. brasiliensis also may be the result of a strong selective pressure imposed by the feline host. Presence of opposite mating types and sexual reproduction leads to genetic recombination and may increase fitness and widen host ranges. So far, no evidence of sexual recombination was demonstrated experimentally for the species from the S. schenckii complex and this fact, combined with the hostile selective pressure of the cats may provide possible explanations for the lack of diversity in S. brasiliensis.
The association of S. brasiliensis with cats may play an important role in the evolution and spread of this pathogen. The interaction between cats and S. brasiliensis is not an exclusive relationship, since S. schenckii s. str. was also found in the feline host. However, S. brasiliensis has become predominant in this host within less than a decade, indirectly indicating a recent adaptation to the conditions of the feline body. Therefore, cats represent a natural habitat for S. brasiliensis. In contrast to the situation in opportunistic fungi, Sporothrix species are able to escape from the host and be transmitted to the next host, which is one of the hallmarks of a pathogen. Transmission is either direct during fights, or indirect, the fungus returning to soil after the cat has died.
Given the role of the mammal host in Sporothrix evolution, variance in fitness between clonal lineages of S. brasiliensis is expected to lead to populations that are better adapted to host conditions. For example, the body temperature of the feral cat Felis catus is around 38–39°C, depending on its activity [90]. Interestingly, S. brasiliensis has the best rate of vegetative growth when incubated at 37°C, followed by S. schenckii s. str. (Fig. 6). Remaining species of Sporothrix such as S. globosa and S. mexicana appear to be more sensitive to temperature, having a maximum around 35°C. The cat's body temperature could be considered an important selective pressure event, selecting thermo-resistant strains during sporotrichosis outbreak episodes. Transmission of S. brasiliensis by cats promotes inoculation into human hosts of yeast cells of rather than of hyphae and conidia, yeast cells having been reported to be more virulent [6].
The endotherm developed by mammals is a natural defense mechanism against pathogens [91]–[93], and in our study this factor appears to restrict the occurrence of species of the S. schenckii complex that are sensitive to temperatures above 35–37°C [11], [17].
Another important factor in understanding the success of the epidemic of sporotrichosis among cats in RJ, has a socio-economic character. Most cat owners are living in neglected areas and many abandon dead animals in the street [94], favoring contact with other feral cats, or simply bury their pets after death in their backyard or in nearby wastelands. This directly allows the return of the agent into the environment, increasing outbreak risks of the pathogen, and enhancing the spread of the clonal species. In an epidemic scenario, domestic pets such as cats and dogs are the first animals to become infected with the fungus. Subsequently human cases of sporotrichosis are likely to emerge. Thus, we believe that cats can act as sentinel animals for epidemiological services, and its notification should be compulsory by regulatory agencies as the Centers for Zoonosis Control. The predominance of a species that is highly virulent to humans and animals requires fast implementation of public health policies to contain the epidemic, lowering harmful effects to the population.
Supporting Information
Nucleotide diversity (%π) and haplotype diversity (1 – Σfi2) from Brazilian clinical isolates belonging to the Sporothrix schenckii complex.
(DOC)
Identification of the haplotypes in the Sporothrix species according to the calmodulin (CAL) or elongation factor (EF1-α) loci.
(DOC)
Acknowledgments
We gratefully acknowledge Prof. Dr. Mario Augusto Ono, Universidade Estadual de Londrina, Paraná, Prof. Dr. Carlos Pelleschi Taborda, Universidade de São Paulo, São Paulo, Prof. Mario Carlos Araujo Meireles, Universidade Federal de Pelotas, Rio Grande do Sul and Prof. Dr. Júnia Soares Hamdan, Universidade Federal de Minas Gerais, Minas Gerais for providing Sporothrix spp. cultures from cats and dogs.
Funding Statement
AMR is a fellow and acknowledges the financial support of the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2011/07350-1) and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (BEX 2325/11-0). GFF is a fellow of FAPESP (2011/01628-8). ZPdC thanks FAPESP (Proc. 09/54024-2) and CNPq (Proc. 472600/2011-7). TMPS is the recipient of a CNPq fellowship. This work was supported in part by grants from FAPESP (http://www.fapesp.br/), CNPq (http://www.cnpq.br/), and CAPES (http://www.capes.gov.br/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Lutz A, Splendore A (1907) On a mycosis observed in men and mice: Contribution to the knowledge of the so-called sporotrichosis. Revista Médica de São Paulo 21: 443–450 [in Portuguese]. [Google Scholar]
- 2. Pereira SA, Menezes RC, Gremião IDF, Silva JN, de O. Honse C, et al. (2011) Sensitivity of cytopathological examination in the diagnosis of feline sporotrichosis. J Feline Med Surg 13: 220–223 doi: 10.1016/j.jfms.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schubach A, Schubach TM, Barros MB, Wanke B (2005) Cat-transmitted sporotrichosis, Rio de Janeiro, Brazil. Emerg Infect Dis 11: 1952–1954 doi: 10.3201/eid1112.040891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schubach TM, Schubach A, Okamoto T, Barros MB, Figueiredo FB, et al. (2004) Evaluation of an epidemic of sporotrichosis in cats: 347 cases (1998–2001). J Am Vet Med Assoc 224: 1623–1629 doi: 10.2460/javma.2004.224.1623 [DOI] [PubMed] [Google Scholar]
- 5. Mackinnon JE, Conti-Díaz IA, Gezuele E, Civila E, Da Luz S (1969) Isolation of Sporothrix schenckii from nature and considerations on its pathogenicity and ecology. Sabouraudia 7: 38–45 doi:10.1080/00362177085190071 [PubMed] [Google Scholar]
- 6. Fernandes KSS, Coelho ALJ, Bezerra LML, Barja-Fidalgo C (2000) Virulence of Sporothrix schenckii conidia and yeast cells, and their susceptibility to nitric oxide. Immunology 101: 563–569 doi: 10.1046/j.1365-2567.2000.00125.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klein BS, Tebbets B (2007) Dimorphism and virulence in fungi. Curr Opin Microbiol 10: 314–319 doi: 10.1016/j.mib.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pappas PG, Tellez I, Deep AE, Nolasco D, Holgado W, et al. (2000) Sporotrichosis in Peru: Description of an area of hyperendemicity. Clin Infect Dis 30: 65–70 doi: 10.1086/313607 [DOI] [PubMed] [Google Scholar]
- 9.Rippon JW (1988) Medical Mycology– The pathogenic fungi and the pathogenic actinomycetes. Philadelphia, PA : W. B. Saunders Company.
- 10. Verma S, Verma GK, Singh G, Kanga A, Shanker V, et al. (2012) Sporotrichosis in Sub-Himalayan India. PLoS Negl Trop Dis 6: e1673 doi:10.1371/journal.pntd.0001673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rodrigues AM, de Hoog S, de Camargo ZP (2012) Emergence of pathogenicity in the Sporothrix schenckii complex. Med Mycol 51: 405–412 doi:10.3109/13693786.2012.719648 [DOI] [PubMed] [Google Scholar]
- 12. Silva-Vergara ML, de Camargo ZP, Silva PF, Abdalla MR, Sgarbieri RN, et al. (2012) Disseminated Sporothrix brasiliensis infection with endocardial and ocular involvement in an HIV-infected patient. Am J Trop Med Hyg 86: 477–480 doi: 10.4269/ajtmh.2012.11-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou X, Rodrigues AM, Feng P, Hoog GS (2013) Global ITS diversity in the Sporothrix schenckii complex. Fungal Divers 1–13 doi: 10.1007/s13225-013-0220-2 [Google Scholar]
- 14. de Meyer EM, de Beer ZW, Summerbell RC, Moharram AM, de Hoog GS, et al. (2008) Taxonomy and phylogeny of new wood- and soil-inhabiting Sporothrix species in the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 100: 647–661 doi: 10.3852/07-157R [DOI] [PubMed] [Google Scholar]
- 15. Madrid H, Cano J, Gene J, Bonifaz A, Toriello C, et al. (2009) Sporothrix globosa, a pathogenic fungus with widespread geographical distribution. Rev Iberoam Micol 26: 218–222 doi: 10.1016/j.riam.2009.02.005 [DOI] [PubMed] [Google Scholar]
- 16. Madrid H, Gené J, Cano J, Silvera C, Guarro J (2010) Sporothrix brunneoviolacea and Sporothrix dimorphospora, two new members of the Ophiostoma stenoceras-Sporothrix schenckii complex. Mycologia 102: 1193–1203 doi: 10.3852/09-320 [DOI] [PubMed] [Google Scholar]
- 17. Marimon R, Cano J, Gené J, Sutton DA, Kawasaki M, et al. (2007) Sporothrix brasiliensis, S. globosa, and S. mexicana, three new Sporothrix species of clinical interest. J Clin Microbiol 45: 3198–3206 doi: 10.1128/JCM.00808-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marimon R, Gené J, Cano J, Guarro J (2008) Sporothrix luriei: a rare fungus from clinical origin. Med Mycol 46: 621–625 doi: 10.1080/13693780801992837 [DOI] [PubMed] [Google Scholar]
- 19. Marimon R, Gené J, Cano J, Trilles L, dos Santos Lazéra M, et al. (2006) Molecular phylogeny of Sporothrix schenckii . J Clin Microbiol 44: 3251–3256 doi: 10.1128/JCM.00081-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arrillaga-Moncrieff I, Capilla J, Mayayo E, Marimon R, Mariné M, et al. (2009) Different virulence levels of the species of Sporothrix in a murine model. Clin Microbiol Infect 15: 651–655 doi: 10.1111/j.1469-0691.2009.02824.x [DOI] [PubMed] [Google Scholar]
- 21. Fernandes GF, dos Santos PO, Rodrigues AM, Sasaki AA, Burger E, et al. (2013) Characterization of virulence profile, protein secretion and immunogenicity of different Sporothrix schenckii sensu stricto isolates compared with S. globosa and S. brasiliensis species. Virulence 4 3: 1–9 doi: 10.4161/viru.23112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marimon R, Serena C, Gené J, Cano J, Guarro J (2008) In vitro antifungal susceptibilities of five species of Sporothrix . Antimicrob Agents Chemother 52: 732–734 doi: 10.1128/AAC.01012-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Barros MBL, Schubach AdO, do Valle ACF, Galhardo MCG, Conceição-Silva F, et al. (2004) Cat-transmitted sporotrichosis epidemic in Rio de Janeiro, Brazil: Description of a series of cases. Clin Infect Dis 38: 529–535 doi: 10.1086/381200 [DOI] [PubMed] [Google Scholar]
- 24. Barros MBL, Schubach TP, Coll JO, Gremião ID, Wanke B, et al. (2010) Sporotrichosis: development and challenges of an epidemic. Rev Panam Salud Publica 27: 455–460 doi: []10.1590/S1020-49892010000600007 [in Portuguese] [PubMed] [Google Scholar]
- 25. da Rosa ACM, Scroferneker ML, Vettorato R, Gervini RL, Vettorato G, et al. (2005) Epidemiology of sporotrichosis: A study of 304 cases in Brazil. J Am Acad Dermatol 52: 451–459 doi: 10.1016/j.jaad.2004.11.046 [DOI] [PubMed] [Google Scholar]
- 26. Madrid IM, Mattei AS, Fernandes CG, Oliveira Nobre M, Meireles MCA (2012) Epidemiological findings and laboratory evaluation of sporotrichosis: A description of 103 cases in cats and dogs in Southern Brazil. Mycopathologia 173: 265–273 doi: 10.1007/s11046-011-9509-4 [DOI] [PubMed] [Google Scholar]
- 27. Mesa-Arango AC, del Rocío Reyes-Montes M, Pérez-Mejía A, Navarro-Barranco H, Souza V, et al. (2002) Phenotyping and genotyping of Sporothrix schenckii isolates according to geographic origin and clinical form of sporotrichosis. J Clin Microbiol 40: 3004–3011 doi: 10.1128/JCM.40.8.3004-3011.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Donnell K, Nirenberg H, Aoki T, Cigelnik E (2000) A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78 doi: 10.1007/BF02464387 [Google Scholar]
- 29. Katoh K, Misawa K, Kuma Ki, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30: 3059–3066 doi: 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duong TA, de Beer ZW, Wingfield BD, Wingfield MJ (2012) Phylogeny and taxonomy of species in the Grosmannia serpens complex. Mycologia 104: 715–732 doi: 10.3852/11-109 [DOI] [PubMed] [Google Scholar]
- 31. Spatafora JW, Sung G-H, Johnson D, Hesse C, O'Rourke B, et al. (2006) A five-gene phylogeny of Pezizomycotina. Mycologia 98: 1018–1028 doi: 10.3852/mycologia.98.6.1018 [DOI] [PubMed] [Google Scholar]
- 32. Zipfel RD, de Beer ZW, Jacobs K, Wingfield BD, Wingfield MJ (2006) Multi-gene phylogenies define Ceratocystiopsis and Grosmannia distinct from Ophiostoma . Stud Mycol 55: 75–97 doi: 10.3114/sim.55.1.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42: 182–192 doi: 10.1093/sysbio/42.2.182 [Google Scholar]
- 35. Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol 9: 678–687. [DOI] [PubMed] [Google Scholar]
- 36. Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. J Mol Evol 43: 304–311 doi: 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- 37. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452 doi: 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 38. Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16: 37–48. [DOI] [PubMed] [Google Scholar]
- 39. Bandelt H-J, Dress AWM (1992) Split decomposition: A new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol 1: 242–252 doi: 10.1016/1055-7903(92)90021-8 [DOI] [PubMed] [Google Scholar]
- 40. Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23: 254–267 doi: 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 41. Bruen TC, Philippe H, Bryant D (2006) A simple and robust statistical test for detecting the presence of recombination. Genetics 172: 2665–2681 doi: 10.1534/genetics.105.048975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salemi M, Gray RR, Goodenow MM (2008) An exploratory algorithm to identify intra-host recombinant viral sequences. Mol Phylogenet Evol 49: 618–628 doi: 10.1016/j.ympev.2008.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rozas J, Rozas R (1995) DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput Appl Biosci 11: 621–625 doi: 10.1093/bioinformatics/11.6.621 [DOI] [PubMed] [Google Scholar]
- 44. Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. de Oliveira MME, Sampaio P, Almeida-Paes R, Pais C, Gutierrez-Galhardo MC, et al. (2012) Rapid identification of Sporothrix species by T3B fingerprinting. J Clin Microbiol 50: 2159–2162 doi: 10.1128/JCM.00450-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tibayrenc M, Ayala FJ (2012) Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci U S A 109: E3305–3313 doi: 10.1073/pnas.1212452109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Billiard S, López-Villavicencio M, Devier B, Hood ME, Fairhead C, et al. (2011) Having sex, yes, but with whom? Inferences from fungi on the evolution of anisogamy and mating types. Biol Rev Camb Philos Soc 86: 421–442 doi: 10.1111/j.1469-185X.2010.00153.x [DOI] [PubMed] [Google Scholar]
- 48. Gräser Y, de Hoog S, Summerbell RC (2006) Dermatophytes: recognizing species of clonal fungi. Med Mycol 44: 199–209 doi: doi:10.1080/13693780600606810 [DOI] [PubMed] [Google Scholar]
- 49. Henk DA, Eagle CE, Brown K, Van den Berg MA, Dyer PS, et al. (2011) Speciation despite globally overlapping distributions in Penicillium chrysogenum: the population genetics of Alexander Fleming's lucky fungus. Mol Ecol 20: 4288–4301 doi: 10.1111/j.1365-294X.2011.05244.x [DOI] [PubMed] [Google Scholar]
- 50. Henk DA, Shahar-Golan R, Devi KR, Boyce KJ, Zhan N, et al. (2012) Clonality despite sex: The evolution of host-associated sexual neighborhoods in the pathogenic fungus Penicillium marneffei . PLoS Pathog 8: e1002851 doi:10.1371/journal.ppat.1002851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fisher MC, Aanensen D, de Hoog S, Vanittanakom N (2004) Multilocus microsatellite typing system for Penicillium marneffei reveals spatially structured populations. J Clin Microbiol 42: 5065–5069 doi: 10.1128/JCM.42.11.5065-5069.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fisher MC, Hanage WP, de Hoog S, Johnson E, Smith MD, et al. (2005) Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei . PLoS Pathog 1: e20 doi:10.1371/journal.ppat.0010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chowdhary A, Hiremath SS, Sun S, Kowshik T, Randhawa HS, et al. (2011) Genetic differentiation, recombination and clonal expansion in environmental populations of Cryptococcus gattii in India. Environ Microbiol 13: 1875–1888 doi: 10.1111/j.1462-2920.2011.02510.x [DOI] [PubMed] [Google Scholar]
- 54. Halliday CL, Carter DA (2003) Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J Clin Microbiol 41: 703–711 doi: 10.1128/JCM.41.2.703-711.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. James TY, Litvintseva AP, Vilgalys R, Morgan JAT, Taylor JW, et al. (2009) Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog 5: e1000458 doi:10.1371/journal.ppat.1000458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barker BM, Jewell KA, Kroken S, Orbach MJ (2007) The population biology of Coccidioides: epidemiologic implications for disease outbreaks. Ann N Y Acad Sci 1111: 147–163 doi: 10.1196/annals.1406.040 [DOI] [PubMed] [Google Scholar]
- 57. Fisher MC, Koenig GL, White TJ, Taylor JW (2000) Pathogenic clones versus environmentally driven population increase: Analysis of an epidemic of the human fungal pathogen Coccidioides immitis . J Clin Microbiol 38: 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fisher MC, Rannala B, Chaturvedi V, Taylor JW (2002) Disease surveillance in recombining pathogens: Multilocus genotypes identify sources of human Coccidioides infections . Proc Natl Acad Sci U S A 99: 9067–9071 doi: 10.1073/pnas.132178099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Matute DR, McEwen JG, Puccia R, Montes BA, San-Blas G, et al. (2006) Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol 23: 65–73 doi: 10.1093/molbev/msj008 [DOI] [PubMed] [Google Scholar]
- 60. Teixeira MM, Theodoro RC, de Carvalho MJA, Fernandes L, Paes HC, et al. (2009) Phylogenetic analysis reveals a high level of speciation in the Paracoccidioides genus. Mol Phylogenet Evol 52: 273–283 doi: 10.1016/j.ympev.2009.04.005 [DOI] [PubMed] [Google Scholar]
- 61. Theodoro RC, Teixeira MdM, Felipe MSS, Paduan KdS, Ribolla PM, et al. (2012) Genus Paracoccidioides: Species recognition and biogeographic aspects. PLoS ONE 7: e37694 doi:10.1371/journal.pone.0037694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kedes LH, Siemienski J, Braude AI (1964) The syndrome of the alcoholic rose gardener. Sporotrichosis of the radial tendon sheath. Report of a case cured with Amphotericin B. Ann Intern Med 61: 1139–1141. [DOI] [PubMed] [Google Scholar]
- 63. Coles FB, Schuchat A, Hibbs JR, Kondracki SF, Salkin IF, et al. (1992) A multistate outbreak of sporotrichosis associated with Sphagnum moss. Am J Epidemiol 136: 475–487. [DOI] [PubMed] [Google Scholar]
- 64. Dixon DM, Salkin IF, Duncan RA, Hurd NJ, Haines JH, et al. (1991) Isolation and characterization of Sporothrix schenckii from clinical and environmental sources associated with the largest U.S. epidemic of sporotrichosis. J Clin Microbiol 29: 1106–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gastineau F, Spolyar L, Haynes E (1941) Sporotrichosis: Report of six cases among florists. JAMA 117: 1074–1077 doi:10.1001/jama.1941.02820390016005 [Google Scholar]
- 66. Feeney KT, Arthur IH, Whittle AJ, Altman SA, Speers DJ (2007) Outbreak of sporotrichosis, Western Australia. Emerg Infect Dis 13: 1228–1231 doi: 10.3201/eid1308.061462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roets F, Wingfield BD, de Beer ZW, Wingfield MJ, Dreyer LL (2010) Two new Ophiostoma species from Protea caffra in Zambia. Persoonia 24: 18–28 doi: 10.3767/003158510X490392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou X, de Beer ZW, Wingfield MJ (2006) DNA sequence comparisons of Ophiostoma spp., including Ophiostoma aurorae sp. nov., associated with pine bark beetles in South Africa. Stud Mycol 55: 269–277 doi: 10.3114/sim.55.1.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Barros MBL, Schubach AO, Schubach TMP, Wanke B, Lambert-Passos SR (2008) An epidemic of sporotrichosis in Rio de Janeiro, Brazil: epidemiological aspects of a series of cases. Epidemiol Infect 136: 1192–1196 doi: 10.1017/S0950268807009727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. de Lima Barros MB, Schubach AO, de Vasconcellos Carvalhaes de Oliveira R, Martins EB, Teixeira JL, et al. (2011) Treatment of cutaneous sporotrichosis with Itraconazole—Study of 645 patients. Clin Infect Dis 52: e200–e206 doi: 10.1093/cid/cir245 [DOI] [PubMed] [Google Scholar]
- 71. Dangerfield LF, Gear J (1941) Sporotrichosis among miners on the Witwatersrand gold mines. SA Medical Journal April 128–131. [Google Scholar]
- 72. Vismer HF, Hull PR (1997) Prevalence, epidemiology and geographical distribution of Sporothrix schenckii infections in Gauteng, South Africa. Mycopathologia 137: 137–143 doi: 10.1023/A:1006830131173 [DOI] [PubMed] [Google Scholar]
- 73. Mehta KIS, Sharma NL, Kanga AK, Mahajan VK, Ranjan N (2007) Isolation of Sporothrix schenckii from the environmental sources of cutaneous sporotrichosis patients in Himachal Pradesh, India: results of a pilot study. Mycoses 50: 496–501 doi:10.1111/j.1439-0507.2007.014 [DOI] [PubMed] [Google Scholar]
- 74. O'Reilly LC, Altman SA (2006) Macrorestriction analysis of clinical and environmental isolates of Sporothrix schenckii . J Clin Microbiol 44: 2547–2552 doi: 10.1128/JCM.00078-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Song Y, Li SS, Zhong SX, Liu YY, Yao L, et al. (2013) Report of 457 sporotrichosis cases from Jilin province, northeast China, a serious endemic region. J Eur Acad Dermatol Venereol 27: 313–318 doi: 10.1111/j.1468-3083.2011.04389.x [DOI] [PubMed] [Google Scholar]
- 76. Kusuhara M, Hachisuka H, Sasai Y (1988) Statistical survey of 150 cases with sporotrichosis. Mycopathologia 102: 129–133 doi: 10.1007/BF00437450 [DOI] [PubMed] [Google Scholar]
- 77. Le Clec'h W, Braquart-Varnier C, Raimond M, Ferdy J-B, Bouchon D, et al. (2012) High virulence of Wolbachia after host switching: When autophagy hurts. PLoS Pathog 8: e1002844 doi:10.1371/journal.ppat.1002844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mu J, Joy DA, Duan J, Huang Y, Carlton J, et al. (2005) Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol 22: 1686–1693 doi: 10.1093/molbev/msi160 [DOI] [PubMed] [Google Scholar]
- 79. Nelson MI, Viboud C, Simonsen L, Bennett RT, Griesemer SB, et al. (2008) Multiple reassortment events in the evolutionary history of H1N1 Influenza A Virus since 1918. PLoS Pathog 4: e1000012 doi:10.1371/journal.ppat.1000012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Johnson L, Johnson R, Akamatsu H, Salamiah A, Otani H, et al. (2001) Spontaneous loss of a conditionally dispensable chromosome from the Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr Genet 40: 65–72 doi: 10.1007/s002940100233 [DOI] [PubMed] [Google Scholar]
- 81. Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, et al. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature 464: 367–373 doi:10.1038/nature08850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Park RF, Wellings CR (2012) Somatic hybridization in the Uredinales. Annu Rev Phytopathol 50: 219–239 doi: 10.1146/annurev-phyto-072910-095405 [DOI] [PubMed] [Google Scholar]
- 83. Forche A, Alby K, Schaefer D, Johnson AD, Berman J, et al. (2008) The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol 6: e110 doi:10.1371/journal.pbio.0060121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Schoustra SE, Debets AJM, Slakhorst M, Hoekstra RF (2007) Mitotic recombination accelerates adaptation in the fungus Aspergillus nidulans . PLoS Genet 3: e68 doi:10.1371/journal.pgen.0030068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Inami K, Yoshioka-Akiyama C, Morita Y, Yamasaki M, Teraoka T, et al. (2012) A genetic mechanism for emergence of races in Fusarium oxysporum f. sp. lycopersici: Inactivation of avirulence gene AVR1 by transposon insertion. PLoS ONE 7: e44101 doi:10.1371/journal.pone.0044101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Goss EM, Cardenas ME, Myers K, Forbes GA, Fry WE, et al. (2011) The plant pathogen Phytophthora andina emerged via hybridization of an unknown Phytophthora species and the Irish potato famine pathogen, P. infestans . PLoS ONE 6: e24543 doi:10.1371/journal.pone.0024543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH (2012) Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Natl Acad Sci U S A 109: 10954–10959 doi: 10.1073/pnas.1201403109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Farrer RA, Weinert LA, Bielby J, Garner TWJ, Balloux F, et al. (2011) Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc Natl Acad Sci U S A 108(46): 18732–18736 doi: 10.1073/pnas.1111915108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Brasier CM, Kirk SA (2010) Rapid emergence of hybrids between the two subspecies of Ophiostoma novo-ulmi with a high level of pathogenic fitness. Plant Pathol 59: 186–199 doi: 10.1111/j.1365-3059.2009.02157.x [Google Scholar]
- 90. Hilmer S, Algar D, Neck D, Schleucher E (2010) Remote sensing of physiological data: Impact of long term captivity on body temperature variation of the feral cat (Felis catus) in Australia, recorded via Thermochron iButtons. J Therm Biol 35: 205–210 doi: 10.1016/j.jtherbio.2010.05.002 [Google Scholar]
- 91. Casadevall A (2012) Fungi and the rise of mammals. PLoS Pathog 8: e1002808 doi:10.1371/journal.ppat.1002808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Casadevall A, Fang FC, Pirofski L-a (2011) Microbial virulence as an emergent property: Consequences and opportunities. PLoS Pathog 7: e1002136 doi:10.1371/journal.ppat.1002136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Robert VA, Casadevall A (2009) Vertebrate endothermy restricts most fungi as potential pathogens. J Infect Dis 200: 1623–1626 doi: 10.1086/644642 [DOI] [PubMed] [Google Scholar]
- 94. Chaves AR, de Campos MP, Barros MBL, do Carmo CN, Gremião IDF, et al. (2013) Treatment abandonment in feline sporotrichosis – Study of 147 cases. Zoonoses Public Health 60: 149–153 doi: 10.1111/j.1863-2378.2012.01506.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide diversity (%π) and haplotype diversity (1 – Σfi2) from Brazilian clinical isolates belonging to the Sporothrix schenckii complex.
(DOC)
Identification of the haplotypes in the Sporothrix species according to the calmodulin (CAL) or elongation factor (EF1-α) loci.
(DOC)