Abstract
Background
Clostridium botulinum and related clostridia express extremely potent toxins known as botulinum neurotoxins (BoNTs) that cause severe, potentially lethal intoxications in humans. These BoNT-producing bacteria are categorized in seven major toxinotypes (A through G) and several subtypes. The high diversity in nucleotide sequence and genetic organization of the gene cluster encoding the BoNT components poses a great challenge for the screening and characterization of BoNT-producing strains.
Methodology/Principal Findings
In the present study, we designed and evaluated the performances of a resequencing microarray (RMA), the PathogenId v2.0, combined with an automated data approach for the simultaneous detection and characterization of BoNT-producing clostridia. The unique design of the PathogenID v2.0 array allows the simultaneous detection and characterization of 48 sequences targeting the BoNT gene cluster components.
This approach allowed successful identification and typing of representative strains of the different toxinotypes and subtypes, as well as the neurotoxin-producing C. botulinum strain in a naturally contaminated food sample. Moreover, the method allowed fine characterization of the different neurotoxin gene cluster components of all studied strains, including genomic regions exhibiting up to 24.65% divergence with the sequences tiled on the arrays.
Conclusions/Significance
The severity of the disease demands rapid and accurate means for performing risk assessments of BoNT-producing clostridia and for tracing potentials sources of contamination in outbreak situations. The RMA approach constitutes an essential higher echelon component in a diagnostics and surveillance pipeline. In addition, it is an important asset to characterise potential outbreak related strains, but also environment isolates, in order to obtain a better picture of the molecular epidemiology of BoNT-producing clostridia.
Introduction
Botulism is a severe neuroparalytic disease caused by botulinum toxin, and characterized by acute descending flaccid paralysis. The disease is caused by consumption of food contaminated with pre-formed toxin (foodborne botulism), or by absorption of toxin produced in situ in wounds (wound botulism) or colonized intestinal tracts (infant/intestinal adult botulism) [1]. Botulinum neurotoxins (BoNT) are the most potent toxins known and are considered as one of the six highest risk threat agents of bioterrorism [2],[3].
BoNT are produced by six physiologically and genetically distinct bacteria, namely Clostridium botulinum Groups I to IV, and occasionally strains of C. butyricum and C. barati [4], [5]. These neurotoxin-producing bacteria can be further categorized in seven major toxinotypes (A, B, C, D, E, F and G) based on the antigenic properties of the toxins they produce [6],[7]. Toxinotypes A, B, E and more rarely F are responsible for human botulism cases, while infections by C and D toxinotypes are observed mainly in animals. Although most strains produce only one toxin, bivalent strains producing two different toxins (Ab, Af, Ba, and Bf) have also been reported [8]–[11]. In addition, C. botulinum strains producing a single toxin but carrying a silent gene for another (A(B)) [10], and strains producing a chimeric neurotoxin (C/D or D/C) have been described [12].
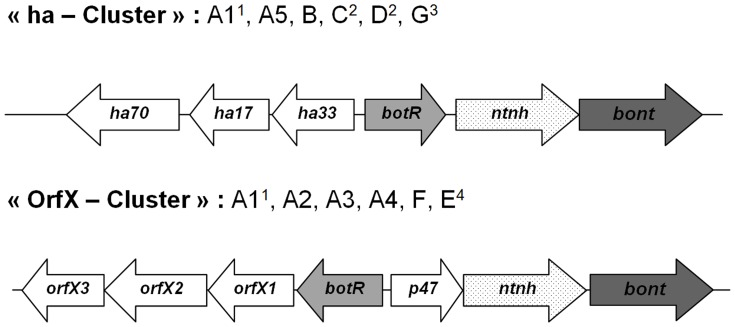
The genes encoding the neurotoxins (bont) can be found on the chromosome, a plasmid or a bacteriophage, and are located in a cluster with other genes encoding associated non-toxic proteins (ANTPs) forming the botulinum neurotoxin complexes [13]. The recent sequencing of the bont genes has revealed significant sequence variations within the toxinotypes and has led to the recognition of several subtypes, with differences ranging between 2–32% at the amino acid level [14]. In addition, the BoNT gene cluster varies in structure and organization [5], leading to the differentiation of two different conserved cluster types, termed the “ha cluster” and the “orf-X cluster” (Figure 1). The “ha cluster” consists of a set of hemaglutinin (HA) genes while the “orf-X cluster” consists of a set of genes of unknown function called “orf-Xs” associated to the bont gene. The diversity in nucleotide sequence and genetic cluster organization can be explained by the occurrence of many recombination and insertion events [15], as well as bont gene transfer between some toxinotypes of C. botulinum, and with non-botulinum species [7].
Figure 1. Neurotoxin gene cluster organization.
In the gene cluster encoding the botulinum neurotoxin complex, the “ha cluster” appears to be associated with type A1, A5, B, C, D and G bont genes, and the “orf-X cluster” with type A1, A2, A3, A4, F and E bont genes. 1Although the type A1 bont gene is mostly found in a ha cluster, it has been found associated with an orf-X cluster in some single toxin strains and in all bivalent strains [18],[17].2The botR gene is in front of the ha genes in all toxinotype C and D strains.3The Ha33 gene is absent in all toxinotype G strains.4The botR gene is absent in all toxinotype E strains
This variability not only has implications for antibody neutralization strategies [14], but also poses a great challenge for screening and characterization of BoNT-producing strains. Sensitive laboratory detection techniques are necessary for establishing monitoring programs to track C. botulinum contamination in animals, food and environmental samples. In addition, characterization methods are essential for tracing potentials sources of contamination in outbreak situations or forensic investigations, and for performing molecular risk assessments of BoNT-producing clostridia. Moreover, characterization of genetic differences allows the development of improved diagnostics and therapeutic agents for the treatment of botulism [14]. In consequence, efforts have been centered on the development of molecular techniques allowing simultaneous detection and typing of C. botulinum toxinotypes, such as multiplex real time PCR and focused DNA microarray [16], [17] [17]–[19]. These assays are highly sensitive but rely on hybridization of sequence-specific primers for sufficient DNA amplification and/or on a limited number of probes for detection of target genomic regions, and may therefore be suboptimal when bacterial sequence divergence is high. In addition, although some multiplex assays allow simultaneous detection and differentiation of toxinotype A subtypes and strains [16], [20], they do not allow typing further than the toxin type or in-depth genetic characterization of strains belonging to other known toxinotypes. Several genotyping methods such as pulse-field gel electrophoresis, randomly amplified polymorphic DNA, multilocus variable number tandem repeat analysis and comparative genomic hybridization microarray have been applied to further differentiate strains within a toxinotype. However, such methods are often technically demanding, difficult to standardize between laboratories and sometimes produce poorly interpretable results, as subtype and strain discrimination relies on differences in observed electrophoretic or hybridization patterns [18], [19], [21]–[24]. In addition, these approaches often require high amounts of genomic DNA obtained from pure bacterial cultures. The use of high-yield random DNA amplification methods, combined to sequence-based technologies targeting multiple molecular markers, is indispensable to overcome these limitations and obtain a better overall resolution.
To overcome the problem of limiting amount of available DNA in biological samples, whole genome amplification (WGA) methods have been developed and successfully applied for a number of genotyping assays [25]–[29]. In addition, high-density resequencing microarrays (RMA) have emerged as a rapid detection and molecular characterization tool for a broad range of bacterial and viral agents [30],[31]. This technology has demonstrated reliable detection of sequences differing up to 10–15% from the prototype sequences on the array, enhancing the spectrum of detection [31],[32]. In order to determine a target nucleotide sequence, resequencing microarrays use closely overlapping (“tiled”) probe sets of 25 mers, which contain one perfectly matched and three mismatched probes per base for both strands of the target genes [33]. In the present study, we evaluated the PathogenID v2.0 resequencing microarray, containing probes for the detection and characterization of neurotoxin-producing Clostridium species.
Results
Design of PathogenID microarray for Clostridium detection and characterization
Detailed description of prototype sequences selected for Clostridium detection and characterization is shown in table 1. The array contains 4 tiled sequences targeting housekeeping genes of Clostridium species: the complete rrs gene of C. botulinum toxinotype A and partial rpoB genes of C. difficile, C. perfringens and C. botulinum toxinotype A. In addition, the RMA includes 48 tiled sequences ranging from 149 to 200 nt in length, targeting representative botulinum neurotoxin gene cluster components (bont, ntnh, botR, ha17, ha70, ha33, orf-X1, orf-X2, orf-X3 and p47) of toxinotypes A through G, as well as three toxinotype A subtypes (A1, A2 and A3). Finally, it contains 19 tiled regions of 200 nt in length for the detection and characterization of other specific toxins produced by C. tetani, C. difficile, C. perfringens, C. sordellii, C. septicum and C. oedematiens.
Table 1. Description of the sequences tiled on the PathogenID v2.0 microarray for the detection and characterization of BoNT-producing clostridia.
| Tiled sequence description | Species | Type | Strain | Genbank accession no. | Position (nt) | length (nt) | |
| Housekeeping genes | |||||||
| rrs | 16S rRNA | C. botulinum | A1 | DSM1734 | X73442.1 | 7..1513 | 1506 |
| rpoB | RNA polymerase (subunit β) | C. botulinum | A1 | NCTC7272 | Y16466.1 | 1249..1755 | 506 |
| rpoB | RNA polymerase (subunit β) | C. perfringens | ATCC13124 | CP000246.1 | 2986532..2986026 | 506 | |
| rpoB | RNA polymerase (subunit β) | C. difficile | 630 | AM180355.1 | 91744..92253 | 509 | |
| Neurotoxin complex genes | |||||||
| bont/A1 | Botulinum neurotoxin type A1 | C. botulinum | A1 | Hall | AF461540.1 | 11250..11450 | 200 |
| bont/A2 | Botulinum neurotoxin type A2 | C. botulinum | A2 | Kyoto | X73423.1 | 1771..1971 | 200 |
| bont/B1 | Botulinum neurotoxin type B1 | C. botulinum | B1 | Okra | AB232927.1 | 14210..14410 | 200 |
| bont/Bnp | Botulinum neurotoxin type Bnp | C. botulinum | Bnp | ATCC25765 | X71343.1 | 3586..3786 | 200 |
| bont/C1 | Botulinum neurotoxin type C1 | C. botulinum | C1 | 468 | X53751.1 | 1876..2076 | 200 |
| bont/D | Botulinum neurotoxin type D | C. botulinum | D | 1873 | X54254.1 | 1697..1897 | 200 |
| bont/E | Botulinum neurotoxin type E | C. botulinum | E1 | Beluga | X62089.1 | 1213..1413 | 200 |
| bont/F | Botulinum neurotoxin type F | C. botulinum | F6 | 202 | M92906.1 | 606..806 | 200 |
| bont/G | Botulinum neurotoxin type G | C. botulinum | G | NCFB3012 | X74162.1 | 1651..1851 | 200 |
| ntnh/A1 | Non-toxigenic non-hemagglutinin | C. botulinum | A1 | Hall | AF461540 | 8501..8695 | 194 |
| ntnh/A2 | Non-toxigenic non-hemagglutinin | C. botulinum | A2 | Kyoto | X87974 | 2453..2647 | 194 |
| ntnh/A3 | Non-toxigenic non-hemagglutinin | C. botulinum | A3 | Mascarpone | DQ310546 | 5690..5884 | 194 |
| ntnh/Bp | Non-toxigenic non-hemagglutinin | C. botulinum | B1 | Okra | AB232927 | 9649..9843 | 194 |
| ntnh/C | Non-toxigenic non-hemagglutinin | C. botulinum | C1 | 468 | X72793 | 4757..4966 | 209 |
| ntnh/D | Non-toxigenic non-hemagglutinin | C. botulinum | D | 1873 | AB012112 | 4483..4692 | 209 |
| ntnh/E | Non-toxigenic non-hemagglutinin | C. botulinum | E | Mashike | D12697 | 2644..2838 | 194 |
| ntnh/Ebut | Non-toxigenic non-hemagglutinin | C. butyricum | E4 | BL6340 | D12739 | 2644..2838 | 194 |
| ntnh/F | Non-toxigenic non-hemagglutinin | C. botulinum | F6 | 202 | S73676 | 2664..2858 | 194 |
| ntnh/G | Non-toxigenic non-hemagglutinin | C. botulinum | G | ATCC27322 | X87972 | 3390..3599 | 209 |
| ha17/A | Hemagglutinin 17 | C. botulinum | A1 | Hall | AF461540 | 4022..3822 | 200 |
| ha17/Bp | Hemagglutinin 17 | C. botulinum | B1 | Okra | AB232927 | 5170..4970 | 200 |
| ha17/C | Hemagglutinin 17 | C. botulinum | C1 | 468 | X72793 | 3181..2981 | 200 |
| ha17/D | Hemagglutinin 17 | C. botulinum | D | 1873 | AB012112 | 2907..2707 | 200 |
| ha17/G | Hemagglutinin 17 | C. botulinum | G | ATCC27322 | X87972 | 1749..1600 | 149 |
| ha70/A | Hemagglutinin 70 | C. botulinum | A | Hall | AF461540 | 3166..3005 | 161 |
| ha70/Bp | Hemagglutinin 70 | C. botulinum | B1 | Okra | AB232927 | 4314..4153 | 161 |
| ha70/C/D | Hemagglutinin 70 | C. botulinum | C1 | 468 | X72793 | 2337..2173 | 164 |
| ha70/G | Hemagglutinin 70 | C. botulinum | G | ATCC27322 | X87972 | 945..787 | 158 |
| ha33/A | Hemagglutinin 33 | C. botulinum | A1 | Hall | AF461540 | 4603..4403 | 200 |
| ha33/Bp | Hemagglutinin 33 | C. botulinum | B1 | Okra | AB232927 | 5748..5548 | 200 |
| ha33/C/D | Hemagglutinin 33 | C. botulinum | C1 | 468 | X72793 | 3750..3550 | 200 |
| orfX1/A2 | Orf-X component 1 | C. botulinum | A2 | Kyoto | AB004778 | 524..315 | 209 |
| orfX1/A3 | Orf-X component 1 | C. botulinum | A3 | Mascarpone | DQ310546 | 1001..792 | 209 |
| orfX1/E | Orf-X component 1 | C. botulinum | E | Iwanai | D88418 | 1046..837 | 209 |
| orfX2/A2 | Orf-X component 2 | C. botulinum | A2 | Kyoto | AY497358 | 4619..4373 | 246 |
| orfX2/E | Orf-X component 2 | C. botulinum | E | Iwanai | D88418 | 256..10 | 246 |
| orfX3/A2 | Orf-X component 3 | C. botulinum | A2 | Kyoto | AY497358 | 2445..2236 | 209 |
| botR/A1 | Transcriptionnal regulator BotR | C. botulinum | A1 | Hall | AF461540 | 5502..5702 | 200 |
| botR/A2 | Transcriptionnal regulator BotR | C. botulinum | A2 | Kyoto | X96493 | 229..29 | 200 |
| botR/A3 | Transcriptionnal regulator BotR | C. botulinum | A3 | Mascarpone | DQ310546 | 1438..1238 | 200 |
| botR/B | Transcriptionnal regulator BotR | C. botulinum | B1 | Okra | AB232927 | 6653..6853 | 200 |
| botR/C | Transcriptionnal regulator BotR | C. botulinum | C1 | 468 | X72793 | 512..712 | 200 |
| botR/D | Transcriptionnal regulator BotR | C. botulinum | D | 1873 | AB012112 | 238..438 | 200 |
| botR/G | Transcriptionnal regulator BotR | C. botulinum | G | ATCC27322 | X87972 | 2595..2795 | 200 |
| p47/A2 | P 47 | C. botulinum | A2 | Kyoto | X96493 | 779..937 | 158 |
| p47/A3 | P 47 | C. botulinum | A3 | Mascarpone | DQ310546 | 1976..2134 | 158 |
| p47/E | P 47 | C. botulinum | E | Iwanai | D88418 | 1881..2081 | 200 |
| p47/F | P 47 | C. botulinum | F6 | 202 | Y10770 | 946..1116 | 170 |
| Clostridial toxin genes | |||||||
| C2 | C2 toxin (component 1) | C. botulinum | (C)-203U28 | D88982.1 | 838..1038 | 200 | |
| C2 | C2 toxin (component 2) | C. botulinum | (C)-203U28 | D88982.1 | 2245..2445 | 200 | |
| tent | Tetanus neurotoxin | C. tetani | Massachusetts | X04436.1 | 2026..2226 | 200 | |
| toxB | Toxin B | C. difficile | VPI10463 | X53138.1 | 1518..1718 | 200 | |
| toxB | Toxin BF | C. difficile | 1470 | Z23277.1 | 1183..1383 | 200 | |
| toxA | Toxin A | C. difficile | VPI10463 | X51797.1 | 2521..2721 | 200 | |
| cpa | α-toxin | C. perfringens | S13 | L43546.1 | 1317..1517 | 200 | |
| cpb1 | β-toxin | C. perfringens | NCTC 8533 | L13198.1 | 510..710 | 200 | |
| cpb2 | β2-toxin | C. perfringens | Porcine | CWC245 | L77965.1 | 418..618 | 200 |
| cpb2 | β2-toxin | C. perfringens | Equine | D21/98 | AJ537535.1 | 199..399 | 200 |
| cpb2 | β2-toxin | C. perfringens | Bovine | JGS4147 | AY609175 | 151..351 | 200 |
| etxD | ε-toxin D | C. perfringens | NCTC 8346 | M95206.1 | 714..914 | 200 | |
| pfoR | τ-toxin (perfringolysin) | C. perfringens | lambda gt10 | M81080.1 | 3529..3729 | 200 | |
| enterotoxin | Enterotoxin | C. perfringens | NCTC8239 | M98037.1 | 787..987 | 200 | |
| toxin-iota-a | ι-toxin (component Ia) | C. perfringens | NCIB10748 | X73562.1 | 2254..2454 | 200 | |
| toxin-iota-b | ι-toxin (component Ib) | C. perfringens | NCIB10748 | X73562.1 | 3515..3715 | 200 | |
| toxin-LT82 | Lethal toxin 82 | C. sordellii | 82 | X82638.1 | 1595..1795 | 200 | |
| α-toxin | α-toxin | C. septicum | NCTC547 | D17668.1 | 1062..1262 | 200 | |
| α-toxin | α-novyi toxin | C. oedematiens | ATCC19402 | Z48636.1 | 1286..1486 | 200 | |
| Quinolone-resistance determining regions | |||||||
| gyrA-qrdr | Gyrase (subunit A) | C. perfringens | ATCC13124 | CP000246.1 | 7269..7466 | 197 | |
| gyrA-qrdr | Gyrase (subunit A) | C. difficile | 630 | AM180355.1 | 6231..6428 | 197 | |
| parC-qrdr | Topoisomerase (subunit IV) | C. perfringens | SM101 | CP000312.1 | 2254972..2255160 | 188 | |
Resequencing of Clostridium with the PathogenID v2.0 microarray
The ability of the PathogenID v2.0 prototype sequences to detect and subtype neurotoxin-producing C. botulinum strains was assessed by performing a blind analysis of genomic DNA from eleven well-characterized strains representative of the main toxinotypes and subtypes. The observed call-rate for each tiled sequence is detailed in table 2. The call-rates marked in bold indicate sequences retained after filtering using the defined threshold. The average base call rate was 91.6% (range 52.5–100%) for strains for which genome sequence was identical to the tiled sequences. The analysis of more distant genomic sequences showed that these were successfully retrieved by RMA (Figure 2), and retained after filtering when they presented up to 24.7% divergence. The mean resequencing accuracy (correctly re-sequenced bases in comparison to reference sequencing results or database sequences) was 98.9% (range 87.1–100.0%). For all strains analysed by the RMA and the automated filtering approach, at least one sequence targeting a housekeeping gene and one sequence targeting a neurotoxin gene cluster component were successfully retrieved. For all bivalent strains analysed by RMA, two distinct bont gene prototype sequences could be retrieved: bont/B and/F sequences for strain 168.08 (toxinotype Bf2), and bont/A and/B sequences for strains NCTC 2916 [toxinotype A1(B)] and 1430-11 [toxinotype A5(B′)]. Additionally, sequences of two complete sets of ha and orf-X cluster components were retrieved for strains 168.08 and NCTC 2916, but as expected only sequences of ha cluster components were recovered for strain 1430-11. Finally, the RMA did not only allow the retrieval of sequences specifically targeting BoNT gene cluster components antp genes from the toxinotype of the strain analysed, but also sequences targeting antp genes from other toxinotypes belonging to the same taxonomic group. In particular, toxinotype B prototype sequences (e.g. botR/B, ha17/B) were successfully retrieved for strain Hall (toxinotype A), although these sequences show up to 11% divergence with the corresponding regions of the Hall strain genome. Similarly, prototype sequences targeting toxinotype A1 antp genes were retrieved for the BL6 strain (toxinotype B), and prototype sequences targeting toxinotypes A2 and A3 antp genes were retrieved for the NCIB10658 strain (toxinotype F). This was also observed for strains belonging to the C. botulinum taxonomic group III strains, as both toxinotype C and toxinotype D antp sequences were retrieved for strains 468 and 1873.
Table 2. Call-rates observed for the different C. botulinum sequences retrieved by the PathogenID V2.0 microarray for all tested bacterial strains.
| Tiled C. botulinum sequence | Clostridium botulinum toxin subtype and strain | ||||||||||
| A1 | A5(B′) | A1(B) | A3 | B2 | Bf2 | F6 | E1 | C1 | D | G | |
| Hall | 1430.11 | 2916 | Loch Maree | BL6 | 168.08 | NCIB 10658 | K16 | 468 | 1873 | NCIB 10714 | |
| Housekeeping genes | |||||||||||
| rrs A1 | 88,1 | 83,5 | 83,9 | 80,6 | 90,7 | 21,5 | 85,1 | 69,8 | 71,7 | 61,7 | 69,2 |
| RpoB A1 | 96,5 | 96,3 | 94,6 | 91,1 | 90,7 | 90,7 | 95,2 | 16,4 | 23,6 | 15,7 | 16,4 |
| Neurotoxin genes | |||||||||||
| bont/A1 | 97,2 | 87,0 | 97,7 | 23,7 | 6,2 | 8,5 | 11,9 | 7,3 | 16,4 | 11,3 | 7,9 |
| bont/A2 | 18,1 | 12,4 | 21,5 | 88,7 | 8,5 | 9,0 | 16,4 | 7,9 | 7,9 | 2,3 | 7,9 |
| bont/B1 | 14,7 | 18,1 | 78,5 | 11,3 | 19,8 | 88,7 | 11,3 | 10,2 | 13,6 | 1,7 | 10,7 |
| bont/Bnp | 10,2 | 27,1 | 35,6 | 10,7 | 20,3 | 35,6 | 10,2 | 3,4 | 8,5 | 2,8 | 11,3 |
| bont/C1 | 14,1 | 8,5 | 10,7 | 14,1 | 4,5 | 7,3 | 16,9 | 10,7 | 99,4 | 1,1 | 11,9 |
| bont/D | 13,6 | 2,8 | 12,4 | 21,5 | 4,5 | 5,7 | 19,2 | 15,3 | 13,6 | 84,7 | 12,4 |
| bont/E | 6,8 | 1,7 | 2,8 | 9,6 | 4,5 | 1,1 | 6,2 | 67,2 | 5,6 | 0,6 | 2,3 |
| bont/F | 16,4 | 4,0 | 16,9 | 23,7 | 11,9 | 49,2 | 53,1 | 11,3 | 13,6 | 3,4 | 8,5 |
| bont/G | 11,9 | 7,9 | 11,9 | 11,3 | 5,6 | 9,0 | 11,3 | 8,5 | 10,2 | 3,4 | 76,8 |
| Non-toxin non-hemagglutinin gene | |||||||||||
| ntnh/A1 | 99,4 | 81,9 | 90,6 | 75,4 | 10,5 | 78,4 | 55,6 | 9,4 | 8,2 | 1,2 | 6,4 |
| ntnh/A2 | 86,0 | 78,4 | 87,7 | 60,8 | 15,8 | 76,6 | 80,7 | 13,5 | 7,6 | 1,2 | 8,2 |
| ntnh/A3 | 70,2 | 73,7 | 81,3 | 69,6 | 8,8 | 81,3 | 48,5 | 8,2 | 7,0 | 0,6 | 7,0 |
| ntnh/Bp | 11,1 | 2,3 | 14,6 | 12,3 | 97,1 | 98,2 | 19,9 | 8,8 | 20,5 | 4,1 | 29,2 |
| ntnh/C | 26,9 | 11,8 | 17,2 | 17,7 | 16,1 | 9,7 | 13,4 | 14,5 | 96,2 | 72,0 | 9,7 |
| ntnh/D | 32,8 | 21,0 | 24,2 | 19,4 | 22,0 | 17,2 | 13,4 | 14,5 | 96,2 | 79,6 | 9,1 |
| ntnh/E | 21,1 | 12,3 | 24,0 | 26,3 | 7,6 | 9,9 | 15,8 | 87,1 | 15,8 | 2,3 | 11,1 |
| ntnh/Ebut | 21,6 | 11,7 | 24,0 | 26,3 | 6,4 | 9,9 | 15,2 | 86,5 | 15,2 | 1,8 | 11,1 |
| ntnh/F | 45,0 | 38,6 | 39,8 | 39,8 | 9,4 | 43,3 | 33,3 | 22,8 | 14,0 | 1,8 | 9,4 |
| ntnh/G | 27,4 | 14,5 | 22,6 | 22,0 | 16,1 | 15,1 | 14,0 | 7,0 | 24,2 | 5,9 | 64,5 |
| ha-cluster components | |||||||||||
| ha70/A | 94,9 | 63,8 | 91,3 | 13,0 | 76,8 | 96,4 | 9,4 | 3,6 | 9,4 | 0,0 | 7,2 |
| ha70/Bp | 88,4 | 77,5 | 85,5 | 13,8 | 84,8 | 92,0 | 6,5 | 3,6 | 8,0 | 0,7 | 8,0 |
| ha70/C/D | 7,8 | 4,3 | 7,1 | 13,5 | 5,7 | 7,1 | 5,7 | 5,7 | 100,0 | 79,4 | 6,4 |
| ha70/G | 14,8 | 23,0 | 15,6 | 10,4 | 23,0 | 11,9 | 12,6 | 5,2 | 25,2 | 5,2 | 45,9 |
| ha17/A | 98,9 | 80,8 | 97,2 | 15,8 | 86,4 | 96,6 | 15,8 | 11,3 | 19,8 | 8,5 | 11,9 |
| ha17/Bp | 84,7 | 84,2 | 82,5 | 15,8 | 84,2 | 84,2 | 14,7 | 13,0 | 19,2 | 9,6 | 13,0 |
| ha17/C | 10,7 | 7,9 | 5,6 | 18,1 | 8,5 | 7,3 | 10,2 | 7,3 | 100,0 | 89,3 | 7,9 |
| ha17/D | 11,3 | 8,5 | 4,5 | 16,4 | 8,5 | 6,8 | 7,9 | 6,2 | 100,0 | 84,7 | 9,0 |
| ha17/G | 11,1 | 3,2 | 4,8 | 9,5 | 3,2 | 5,6 | 5,6 | 6,3 | 9,5 | 0,0 | 57,9 |
| ha33/A | 96,0 | 45,2 | 78,5 | 14,1 | 49,2 | 85,3 | 8,5 | 6,2 | 13,6 | 4,0 | 9,0 |
| ha33/Bp | 30,5 | 30,5 | 33,3 | 7,3 | 55,4 | 41,8 | 6,8 | 8,5 | 6,8 | 2,3 | 4,5 |
| ha33/C/D | 19,2 | 5,1 | 15,3 | 10,7 | 9,0 | 4,0 | 10,7 | 7,9 | 99,4 | 0,6 | 9,0 |
| botR/A1 | 93,2 | 59,3 | 74,6 | 18,1 | 83,6 | 89,3 | 13,6 | 5,1 | 16,4 | 0,6 | 10,7 |
| botR/Bp | 81,9 | 68,4 | 88,1 | 20,3 | 67,2 | 93,2 | 13,0 | 7,3 | 17,5 | 0,6 | 10,7 |
| botR/C | 7,9 | 5,1 | 14,1 | 11,3 | 9,6 | 13,6 | 14,1 | 7,3 | 98,9 | 45,2 | 7,3 |
| botR/D | 9,0 | 6,8 | 13,6 | 11,3 | 8,5 | 11,9 | 15,3 | 7,3 | 98,9 | 52,5 | 8,5 |
| botR/G | 15,8 | 4,5 | 11,9 | 11,3 | 9,0 | 2,3 | 7,9 | 4,5 | 13,0 | 0,6 | 46,9 |
| orfX -cluster components | |||||||||||
| orfX1-A2 | 5,4 | 1,1 | 57,5 | 90,3 | 6,5 | 59,7 | 73,7 | 3,8 | 7,5 | 3,8 | 6,5 |
| orfX2-A2 | 14,8 | 4,5 | 90,1 | 88,8 | 9,0 | 68,2 | 83,9 | 11,2 | 10,8 | 6,7 | 12,6 |
| orfX3-A2 | 9,7 | 2,2 | 87,1 | 97,8 | 10,8 | 96,8 | 86,6 | 14,0 | 20,4 | 7,5 | 9,7 |
| orfX1-A3 | 8,6 | 5,4 | 78,0 | 61,3 | 7,5 | 41,9 | 54,3 | 5,4 | 7,5 | 1,6 | 5,9 |
| orfX1-E | 6,5 | 5,9 | 9,1 | 11,3 | 3,2 | 9,1 | 7,5 | 68,3 | 8,1 | 7,5 | 6,5 |
| orfX2-E | 9,9 | 5,4 | 15,2 | 22,4 | 6,3 | 7,2 | 17,0 | 71,3 | 10,3 | 1,8 | 7,2 |
| botR/A2 | 9,6 | 1,1 | 39,0 | 74,0 | 8,5 | 62,1 | 42,9 | 2,8 | 10,2 | 2,3 | 4,5 |
| botR/A3 | 8,5 | 0,6 | 68,4 | 41,8 | 2,8 | 69,5 | 39,5 | 5,1 | 10,2 | 1,7 | 6,2 |
| p47/A2 | 9,6 | 3,7 | 40,7 | 31,1 | 6,7 | 36,3 | 36,3 | 6,7 | 14,1 | 1,5 | 8,1 |
| p47/A3 | 10,4 | 1,5 | 90,4 | 61,5 | 6,7 | 69,6 | 72,6 | 5,9 | 15,6 | 1,5 | 7,4 |
| p47/E | 5,1 | 1,1 | 21,5 | 33,3 | 2,3 | 23,2 | 19,8 | 80,8 | 4,0 | 0,0 | 2,3 |
| p47/F | 8,8 | 0,7 | 8,8 | 6,1 | 4,8 | 95,9 | 19,0 | 6,1 | 8,8 | 1,4 | 12,9 |
Sequences retained after filtering using the defined threshold are designated in bold.
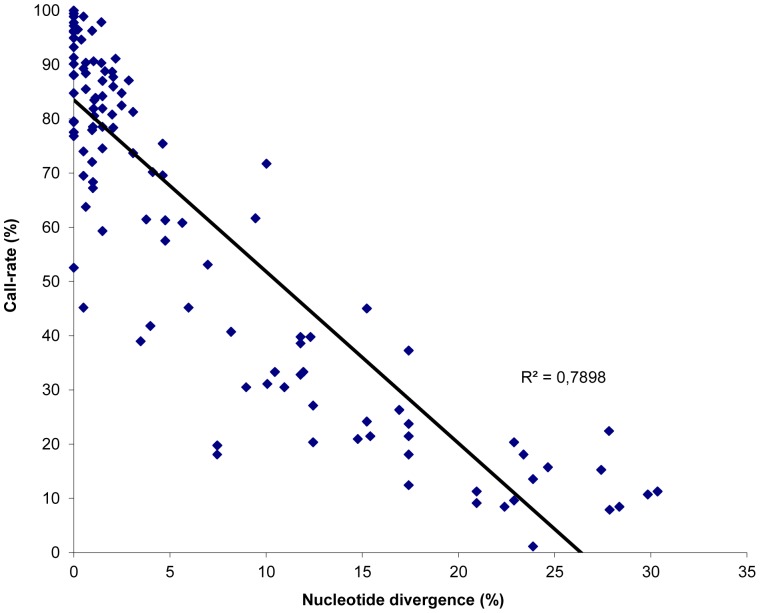
Figure 2. Detection spectrum of the PathogenID_v2.0 according to the nucleotide diversity of the tested BoNT-producing strains.
The spectrum of detection of BoNT-producing clostridia by the PathogenID v2.0 was assessed using 9 well-characterized strains representative of all toxinotypes and several toxin A subtypes. Each sequence obtained by the RMA for which the percentage of nucleotide divergence compared to the corresponding tiled sequence is known (n = 118), is indicated by a blue diamond, and presented according to the percentage of nucleotide bases determined by the RMA (call rate). The linear association between these two parameters is shown, and demonstrates a good correlation (correlation coefficient R2 of 0.79).
Automated filtering and taxonomic identification of resequencing results
Automated filtering by TaxFinder using the defined parameters retained between 7 and 27 C. botulinum sequences for each strain (Table 2). Of the retained sequences, 98.8% (168/170) allowed retrieval of at least one BLAST hit, and 99,4 % (167/168) of the retrieved BLAST hits lead to correct taxonomic identification at least to the toxinotype level. Moreover, blasting of all remaining unfiltered C. botulinum prototype sequences (n = 358) did not return any BLAST hit, indicating that no useful sequence was excluded. For all reference strains the systematic BLAST strategy of retained sequences obtained by the RMA allowed successful identification of the clostridial species and taxonomic group using housekeeping genes prototype sequences (Table 3). The strategy also permitted in-depth characterization of all tested strains using the BoNT gene cluster prototype sequences (Table 3). Moreover, BLAST analysis allowed simultaneous typing of the neurotoxin up to the subtype level, as for each strain a unique best hit or multiple best bacterial hits were retrieved and corresponded to the correct subtype. Finally, the turnaround time of this automated approach was inferior to 2 hours, extensively decreasing the time needed for filtering of sequences, blasting of retained sequences and result analysis.
Table 3. Identification and characterization of C. botulinum strains based on the sequences retrieved by the PathogenID v2.0 microarray and the BLAST analysis results.
| Strain | Toxin subtype | PathogenID v2.0 RMA results | ||
| Taxonomic identification | Neurotoxin subtype | BoNT gene cluster characterization | ||
| Hall | A1 | C. botulinum group I | A1 | ha+/OrfX− |
| 1430.11 | A5(B′) | C. botulinum group I | A5, B | ha+/OrfX− |
| 2916 | A1(B) | C. botulinum group I | A1, bivalent B | ha+/OrfX+ |
| Loch Maree | A3 | C. botulinum group I | A3 | ha-/OrfX+ |
| BL6 | B2 | C. botulinum group I | proteolytic B | ha+/OrfX− |
| 168.08 | Bf2 | C. botulinum group I | bivalent B, F2 | ha+/OrfX+ |
| NCIB10658 | F6 | C. botulinum group I | proteolytic F | ha-/OrfX+ |
| K16 | E1 | C. botulinum group II | E1 | ha-/OrfX+ |
| 468 | C1 | C. botulinum group III | C1 | ha+/OrfX− |
| 1873 | D | C. botulinum group III | D | ha+/OrfX− |
| NCIB10714 | G | C. botulinum group IV | G | ha+/OrfX− |
Identification, typing and characterization of a foodborne botulism outbreak strain
To demonstrate the capacity of the RMA approach to detect C. botulinum target sequences in the context of a foodborne outbreak, DNA extracts from naturally contaminated food samples were analysed. Food specimens were collected during the investigation of a C. botulinum family outbreak in Corsica [34]. RMA was performed on the DNA extract from a contaminated salad sample, as well as the enrichment culture of the salad sample. For DNA obtained directly from the salad specimen, a total of 286 bacterial prototype sequences were retained, including 3 sequences targeting C. botulinum genes (rrs, rpoB and ha33). However, none led to correct taxonomic identification of the C. botulinum infecting strain. For the DNA extract obtained from the enrichment culture of the contaminated salad, a total of 15 C. botulinum prototype sequences were retained after filtering, with call-rates ranging between 46.7 to 97.7%. In addition, sequences targeting partial C. perfringens-specific housekeeping genes (rpoB, gyrA, and parC) and toxin genes (cpa, cpb2 and pfoR) were retrieved (call-rates ranging between 70.1 and 98.3%). All filtered sequences led to the retrieval of at least one BLAST hit and corresponding taxonomic identity. The highest scoring BLAST results obtained for C.perfringens prototype sequences retrieved by RMA all designated C. perfringens strains as taxonomic hit (data not shown). The results of the highest scoring BLAST alignment for each retained C. botulinum sequence (total score, coverage and taxonomic identity) are presented in figure 3. Strains belonging to the toxin subtype A2 were recognized as best hit for most of the retrieved sequences of the neurotoxin gene cluster components. In addition, a gene cluster type ha-/orfX+ was identified. Phylogenetic analysis of the retrieved neurotoxin sequence with the corresponding sequences of five strains representative of the different toxin A subtypes (A1 to A5), demonstrated an overall identity of 100% with the neurotoxin sequence of the toxin subtype A2 reference strain. The only mismatches in the multiple alignment were due to unidentified bases (Figure 4).
Figure 3. C. botulinum sequences retrieved by the PathogenID_v2.0 in contaminated food and BLAST analysis results.
A schematic representation of the BoNT gene cluster sequences obtained by the RMA for the enrichment culture of the contaminated salad specimen is shown. The orientation and arrangement of these components were reproduced after Franciosa et al [59]. For each component, the highest call-rate observed and the results of the best retrieved BLAST hit(s) (defined as the BLAST hit demonstrating the highest total score) are given i.e. the number of best hits retrieved, as well as the corresponding query sequence coverage, total score, and taxonomy.
Figure 4. Phylogenetic tree of the bont/A sequence retrieved in contaminated food and several toxinotype A strains.
A neighbor-joining tree was constructed based on the bont/A sequence retrieved by the RMA for the enrichment culture of the C. botulinum strain in a naturally contaminated salad specimen, and the corresponding sequences of 6 strains representative of 5 known C. botulinum toxinotype A subtypes: strain Hall (A1), NCTC 2916 (A[B]), Kyoto (A2), Loch Maree (A3), 657 (Ba4) and A661222 (A5). Bootstrap values and genetic distance (bar) are shown. The Genbank accession number of each strain is indicated in parentheses. The bont sequence obtained from the contaminated salad specimen is marked in bold.
Discussion
Although botulism is quite rare in developed countries, recent outbreaks have demonstrated the potential severity of this hazard [35]–[40]. In such a context, rapid identification and characterization of BoNT-producing clostridia in home-made or manufactured food is a priority to stop the spread of botulism infections and trace back the source of contamination [41]–[43]. While identification of BoNT-encoding genes, using rapid nucleic-acid detection tools, is usually satisfactory for molecular risk assessment of BoNT-producing clostridia in food and environmental samples, it is not sufficient to discriminate strains within a toxinotype for source-tracing and molecular epidemiology [16]. For this reason, recent recommendations have stated that new assays should be able to detect variants for all toxinotypes, should be type-specific to determine proper treatment, and should be sensitive to perform risk assessment (NIAID Expert Panel on Botulism Diagnostics, Bethesda Maryland, May 2003). The development of such universal genetic characterization tools is hampered by the great variability of the genomic background, the sequence of the bont gene and the arrangement of the BoNT gene cluster of C. botulinum strains, and therefore only available for a limited number of strains.
In the present study, we designed and evaluated the performances of a resequencing microarray combined with an automated data approach for the simultaneous detection and characterization of BoNT-producing clostridia. The described RMA, designated PathogenID v2.0, and similar arrays have been proven successful for the characterization of numerous bacteria including Neisseria meningitidis, Streptococcus pyogenes, Streptococcus pneumoniae, Bacillus anthracis, and several species of the Enterobacteriaceae family [30], [44]–[46]. For the purpose of this study, prototype sequences of housekeeping genes rrs and rpoB from clostridia pathogenic for humans and most frequently associated with outbreaks were incorporated in the RMA for identification of clostridial species and lineages [47]–[49]. In addition, sequences from the BoNT gene cluster were included for their involvement in toxicity and usefulness for studying genetic variation [18], [50], [51].
The method was validated with DNA extracts from a representative panel of reference C. botulinum strains. The obtained results demonstrated that this approach has the potential to detect neurotoxin-producing C. botulinum strains, but also phenotypically related strains, by providing high quality sequence data. The RMA approach reliably detected and characterized the neurotoxin complex genetic determinants for all tested toxino-(sub)types, including bivalent strains. Further analysis of resequencing results showed that targeted sequences were successfully retrieved by the RMA when they presented up to 24.7% divergence with the tiled probes. This data supports the broadness of detection of the RMA, as the divergence of the tiled probes compared to the respective bont gene regions from all known toxin A, B, C, D, E and G subtypes, and most toxin F subtypes, was calculated to be inferior to 21%. Although the tiled bont/F sequence demonstrated respectively 70.65% and 55.72% with the corresponding gene region of the rare toxin subtypes F5 and F7, the high homology of the additional housekeeping gene and antp gene sequences targeted by the RMA should allow detection and correct identification of these strains.
In addition to sequence divergence, differences in DNA extract quality, genomic copy number variation, location of targeted genomic regions and possible secondary structures, as well as the G and C content of the tiled probes and the presence of nonspecific nucleotide stretches could have affected genome amplification and hybridization efficacy, possibly causing reduced base-call rates. Nevertheless, this did not hamper the correct identification and characterization of the analyzed strains.
Subsequently, the pathogenID v2.0 was used to identify C. botulinum strains in contaminated food specimens. The analysis of the DNA extract obtained directly from these specimens by the RMA did not permit quality resequencing of the infecting C. botulinum strain sequences. These results are due to the presence of numerous cultivable and non-cultivable environmental bacteria naturally present in food, competing with the C. botulinum strain during the whole genome amplification step, and subsequently increasing the noise level observed at the time of the analysis. Nevertheless, a 48 h enrichment culture of the contaminated food samples was sufficient to successfully detect and characterize the infecting C. botulinum strain as well as a co-cultured C. perfringens strain. The results obtained by BLASTing and phylogenetic analysis of the retrieved bont sequence, as well as the characterization of the BoNT gene cluster components, classified the strain as a C. botulinum toxinotype A2. Moreover, the examination of the taxonomic BLAST hits indicated a close relatedness to strain Mascarpone. As the analysis was performed with a partial sequence of the bont gene (200 nt), containing 4% unidentified bases, misclassification of the toxin subtype may have occurred. However, this was unlikely as the retained sequence contained specific determinants found only in bont/A2 genes, and unidentified residues were located in regions that are not discriminant for subtype differentiation. The C. botulinum strain detected by RMA could be isolated from the food sample, and confirmed as subtype A2 by sequencing the specifically PCR-amplified bont gene (genbank accession number JQ954970).
The many advantages of using DNA microarrays for characterization of bacteria have been discussed in the past and can be applied to this assay. First, the genetic characterization results can be used to support epidemiological associations in outbreaks involving a large number of samples or those from multiple geographical locations[17]. Secondly, they can be used to trace or confirm the source of an outbreak or to perform environmental risk assessments [52], [53]. In the specific context of botulism, the results provided by DNA microarray can further serve to design specific primers for rapid nucleic acid detection methods, and allow a preliminary differentiation of the strains before applying more laborious genotyping methods such as PFGE [17].
In addition to these advantages, the specific design and properties of high-density RMAs allow the detection of a broad range of strains, including potentially emerging variants [44], [54]. Firstly, the use of closely overlapping 25 nt probe sets in the RMA approach instead of unique probes to cover a target region of 100–500 nt increases the chance for hybridization of divergent target DNA to the array. Indeed, studies using focused or comparative genomic hybridization microarrays have reported a maximum value of 15–18% nucleotide sequence divergence of probe regions for positive microarray detection of C. botulinum genomic DNA, as compared to 24.7% for the RMA described here [17]–[19], [24]. In addition, the relatedness between strains can be quantified more precisely by using a sequence-based approach such as the RMA, rather than by analysis of differences in hybridization patterns provided by classic microarrays. This is particularly important in the case of botulism, as the frequent occurrence of gene recombination, insertion and transfer observed between Clostridium strains indicate that such events are likely to occur again. In such a case, the sequence information provided by the RMA will be most valuable for the characterization of the emerging variants and the development of improved rapid detection tools. This also emphasizes the need for bacterial culture isolates as well as well-characterized microbial strain collections, in order to monitor the molecular evolution of the strains.
The unique design of the PathogenID v2.0 array allows the simultaneous detection and characterization of 48 sequences targeting the BoNT gene cluster components. These sequences could easily be used in the design of specific RMAs with reduced probe densities by decreasing the number of sequences, thereby diminishing production costs and computing time. Using standard methods to obtain the same level of resolution would imply the use of specific PCRs followed by Sanger sequencing. Although these assays are cheaper as standalone tests, running enough of them to cover the same number of genes with a single sample quickly surpasses the RMA in cost, time and logistical complexity of running the analysis. Moreover these methods rely on sequence-specific primer hybridization for target DNA amplification instead of random hexamers for unbiased whole genome amplification, and may therefore fail to amplify and detect emerging variants. Other technologies with universal coverage could be used, such as high-throughput sequencing, however they are still much more laborious and expensive. Finally, the use of an automated approach allows an unbiased, exhaustive and rapid retrieval of taxonomic results.
In summary, the RMA allowed successful detection and fine characterization of neurotoxin-producing clostridia in both pure and polymicrobial cultures. The RMA approach, combined with the automated filtering and retrieval of taxonomic identities, allowed efficient and accurate subtyping of the neurotoxin and detailed characterization of the BoNT gene cluster components. This assay will be used as a higher echelon component in a diagnostics and surveillance pipeline. Importantly, it will be used in further studies to characterise potential outbreak related strains, but also environment isolates, in order to obtain a better picture of the molecular epidemiology of BoNT-producing clostridia.
Materials and Methods
Design of the Pathogen ID v2.0 resequencing microarray (RMA)
The second generation of a broad-spectrum resequencing microarray (RMA) was used in this study: the PathogenID v2.0 array, able to detect 124 bacteria, 126 viruses and 673 genes involved in toxin production, pathogenicity or antibiotic resistance [31]. This array included specific housekeeping genes, namely rrs and rpoB, for detection and characterization of clostridial species. In addition, it included prototype sequences of each component of the neurotoxin gene cluster from all known toxinotypes, as well as several subtypes. Tiled probes targeting these genes were selected using multiple sequence alignments of genome sequences from various toxin subtypes available in GenBank during the time of the RMA design, in order to select conserved regions within toxinotypes for optimal hybridization efficacy. Additional probes were added to improve the detection of specific subtypes, if strains demonstrated sequence variation within the selected probe region. In addition, the bont/C and bont/D sequences tiled on the array were specifically designed to maximize the probability of detection of mosaic C. botulinum toxinotype C/D or D/C strains, by targeting regions highly homologous to either mosaic bont/C/D or bont/D/C genes. Finally, 19 additional sequences were tiled on the array for detection of a large panel of clostridial toxins other than neurotoxins.
Bacterial strains and biological samples analysed
Eleven well-characterized C. botulinum strains belonging to toxinotypes A through G served as reference: strains Hall (type A1), 1430-11 [type A5(B′)], NCTC 2916 [type A1(B)], Loch Maree (type A3), BL6 (type B2), 161.08 (type Bf2), NCIB 10658 (type F6), K16 (type E1), 468 (type C1), 1873 (type D) and NCIB 10714 (type G). The characteristics and origin of most strains have been described elsewhere. The C. botulinum type A5(B) strain 1430-11 was isolated from contaminated commercial food (pasta). The C. botulinum strain 161.08 was isolated from a foodborne botulism case. Pure cultures of Clostridium spp. strains were performed as described previously [55]. Genome sequences from reference strains were retrieved from the genbank database. The bont sequences for strain 1430-11 (genbank accession numbers KC683799 and KC683800) and strain 161.08 (genbank accession numbers KC471328 and KC471329) were obtained by sanger sequencing of specifically PCR-amplified bont genes and deposited in genbank.
Biological samples included a naturally contaminated food specimen (salad) and a 48 h enrichment culture of this food sample in Fortified Cooked Meat Medium (FCMM) at 37°C in anaerobic conditions [56]. Presence of C.botulinum toxinotype A in the food samples was confirmed by SYBR green real-time PCR with primers P1646 (5′-TCTTACGCGAAATGGTTATGG-3′) and P1647 (5′-TGCCTGCACCTAAAAGAGGA-3′) for bont/A gene, P1648 (5′-CCTGGGCCAGTTTTAAATGA-3′) and P1649 (5′-GCGCCTTTGTTTTCTTGAAC-3′) for bont/B gene, P1650 (5′-GTGCCCGAAGGTGAAAATAA-3′) and P1651 (5′-TAATGCTGCTTGCACAGGTT-3′) for bont/E gene, P2107 (5′-TGCACAATGAATTTTCAAAACA-3′) and P2108 (5′-TCCAAAAGCATCCATTACTGC-3′) for bont/F gene. Real time PCR was performed in a total volume of 25 µl containing 12.5 µl of 2× concentration of iQ SYBR green Supermix (Biorad), 5 pmole of each primer, 5 µl of template DNA and 7.3 µl of DNase-RNase free ditilled water (Gibco). Amplifications were carried out on a CFX96 Real Time System (Biorad) using 96-well microwell plates and according to the following temperature profile: one cycle of 95°C for 10 minutes, 40 cycles of 95°C for 15 sec and 60°C for 30 sec with fluorescence signal capture at the end of each 60°C step, an extension phase of 1 cycle at 95°C for 60 sec, 60°C for 60 sec and 95°C for 60 sec, followed by a default melt (disassociation) stage.
Extraction and amplification of bacterial DNA
Total genomic DNA was isolated from C. botulinum cultures by lysozyme and proteinase K treatment as described previously [57]. DNA extraction from food samples was performed with Powerfood Microbial DNA isolation kit (MoBio Laboratories Inc) according to the manufacturer's recommendations. After extraction, DNA was amplified using the whole genome amplification (WGA) protocol (RepliG Midi kit, Qiagen) as described previously [54].
Resequencing microarray assay
The amplification products obtained from genomic DNA by WGA were quantified by Quantit BR (Invitrogen) according to the manufacturer's instructions. A recommended amount of target DNA was fragmented and labeled according to the GeneChip Resequencing Assay manual (Affymetrix). The obtained products were coded with unrelated numbers by a non-observer to ensure that the study was performed blindly. The microarray hybridization process was carried out according to the protocol recommended by the manufacturer (Affymetrix). The details and parameter settings for the data analysis (essentially conversion of raw image files obtained from scanning of the microarrays into FASTA files containing the sequences of called bases for each tiled region of the microarray) have been described previously [54]. The base call rate refers to the percentage of base calls generated from the full-length tiled sequence.
Automated data analysis
A Perl-based program, designated “TaxFinder”, was used for the automated analysis of re-sequencing data provided by PathogenID v2.0. The program read the FASTA file generated for each sample, which contains all the sequences read by the GSEQ software (Affymetrix) from the hybridization results. A filtering process, based on the one described by Malanoski et al. [58] was applied to these sequences. For each sequence, TaxFinder considers the Nb first bases of the sequence, Nb defined by the user in the beginning of the experiment and corresponding to the minimal length accepted for the sequence to keep. If the percentage of No-call in this fragment is inferior to an elongation threshold defined by the user, a base is added to the fragment and the percentage of No-call is recalculated. As soon as it is higher that the elongation threshold, the No-call at the ends of the fragment are removed. If the length of the nucleotide fragment is still higher than the minimal threshold established, then it is conserved. If the percentage is higher than the threshold, the fragment is skipped, and the program considers the fragment of size Nb and starting position one base upstream than the precedent fragment. This process is reiterated until the end of the sequence is reached. In this study, sequences that did not contain subsequences with a minimum nucleotide length of 20 nt and a maximum undetermined nucleotide content of 10% were discarded. Filtered sequences individually underwent a blastn analysis to search for sequence homologues in the NCBI nucleotide collection (nr/nt database) with adjusted settings to restrict the search to bacteria entries. Blast + application distributed by NCBI is used for the automated blast research (http://www.ncbi.nlm.nih.gov/books/NBK1763/). The following algorithm parameters were modified: the expected value cut-off was fixed to 100, the minimum word size was set to 7 and the upper limit of displayed descriptions of database sequences per query was decreased to 50. The best BLAST hits were classified according to their total score (the sum of the high-scoring segment pairs) and their corresponding taxonomies were retrieved from the NCBI Taxonomy database. When several hits obtained the highest total score, the script automatically retrieved the taxonomies of the 10 first BLAST hits.
Phylogenetic analysis
The resulting bont sequence obtained from the enrichment culture of the C. botulinum strain in a contaminated food specimen was compared with the corresponding bont sequences of reference strains from the 5 known C. botulinum toxinotype A subtypes (GenBank accessed 21/02/2013). Multiple sequence alignment was performed using the CLC Bio software and checked for accuracy by eye. A neighbor-joining tree of these sequences was constructed using the Jukes-Cantor method with the SeaView v4.2.1 software. The level of support for each node is provided by 100 bootstrap replications.
Funding Statement
This program was supported by grant UC1 AI062613 (to GC Kennedy) from the U.S. National Institute of Allergy and Infectious Diseases, National Institute of Health; by the “Programme Transversal de Recherche” (DEVA No. 246), financed by the Institut Pasteur (Paris, France), sponsorship by the Fondation Total-Institut Pasteur and the Elisabeth Taub award from the Académie Nationale de Medecine. The funders had no role in study design, data analysis or preparation of the manuscript.
References
- 1. Sobel J (2005) Botulism. Clin Infect Dis 41: 1167–1173 doi:10.1086/444507 [DOI] [PubMed] [Google Scholar]
- 2. Arnon SS, Schechter R, Inglesby TV, Henderson DA, Bartlett JG, et al. (2001) Botulinum Toxin as a Biological Weapon. JAMA 285: 1059–1070 doi:10.1001/jama.285.8.1059 [DOI] [PubMed] [Google Scholar]
- 3. Villar RG, Elliott SP, Davenport KM (2006) Botulism: The Many Faces of Botulinum Toxin and its Potential for Bioterrorism. Infect Dis Clin North Am 20: 313–327 doi:16/j.idc.2006.02.003 [DOI] [PubMed] [Google Scholar]
- 4.Peck MW (2009) Biology and Genomic Analysis of Clostridium botulinum. Academic Press, Vol. 55: . 183–265, 320. Available: http://www.sciencedirect.com/science/article/pii/S0065291109055039. Accessed 2011 Aug 3. [DOI] [PubMed] [Google Scholar]
- 5.Brüggemann H, Woller A, Mazuet C, Popoff MR (2011) Clostridium botulinum. Genomes of Foodborne and Waterborne Pathogens. Washington DC: Fratamico P., Liu Y., Kathariou S. 185–212.
- 6.Smith TJ, Hill KK, Foley BT, Detter JC, Munk AC, et al. (2007) Analysis of the Neurotoxin Complex Genes in Clostridium botulinum A1–A4 and B1 Strains: BoNT/A3,/Ba4 and/B1 Clusters Are Located within Plasmids. PLoS One. 2 .doi:10.1371/journal.pone.0001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hill KK, Smith TJ, Helma CH, Ticknor LO, Foley BT, et al. (2007) Genetic Diversity among Botulinum Neurotoxin-Producing Clostridial Strains. J Bacteriol 189: 818–832 doi:10.1128/JB.01180-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hutson RA, Zhou Y, Collins MD, Johnson EA, Hatheway CL, et al. (1996) Genetic Characterization of Clostridium botulinum Type A Containing Silent Type B Neurotoxin Gene Sequences. J Biol Chem 271: 10786–10792 doi:10.1074/jbc.271.18.10786 [DOI] [PubMed] [Google Scholar]
- 9. Santos-Buelga JA, Collins MD, East AK (1998) Characterization of the Genes Encoding the Botulinum Neurotoxin Complex in a Strain of Clostridium botulinum Producing Type B and F Neurotoxins. Curr Microbiol 37: 312–318 doi:10.1007/s002849900384 [DOI] [PubMed] [Google Scholar]
- 10. Kirma N, Ferreira JL, Baumstark BR (2004) Characterization of six type A strains of Clostridium botulinum that contain type B toxin gene sequences. FEMS Microbiol Lett 231: 159–164 doi:10.1016/S0378-1097(03)00911-X [DOI] [PubMed] [Google Scholar]
- 11. Lúquez C, Raphael BH, Maslanka SE (2009) Neurotoxin Gene Clusters in Clostridium botulinum Type Ab Strains. Appl Environ Microbiol 75: 6094–6101 doi:10.1128/AEM.01009-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moriishi K, Koura M, Abe N, Fujii N, Fujinaga Y, et al. (1996) Mosaic structures of neurotoxins produced from Clostridium botulinum types C and D organisms. Biochim Biophys Acta 1307: 123–126 doi:10.1016/0167-4781(96)00006-1 [DOI] [PubMed] [Google Scholar]
- 13. Brüggemann H (2005) Genomics of clostridial pathogens: implication of extrachromosomal elements in pathogenicity. Curr Opin Microbiol 8: 601–605 doi:16/j.mib.2005.08.006 [DOI] [PubMed] [Google Scholar]
- 14. Smith TJ, Lou J, Geren IN, Forsyth CM, Tsai R, et al. (2005) Sequence Variation within Botulinum Neurotoxin Serotypes Impacts Antibody Binding and Neutralization. Infect Immun 73: 5450–5457 doi:–10.1128/IAI.73.9.5450–5457.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hill KK, Xie G, Foley BT, Smith TJ, Munk AC, et al. (2009) Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol 7: 66–66 doi:10.1186/1741-7007-7-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fach P, Fenicia L, Knutsson R, Wielinga PR, Anniballi F, et al. (2011) An innovative molecular detection tool for tracking and tracing Clostridium botulinum types A, B, E, F and other botulinum neurotoxin producing Clostridia based on the GeneDisc cycler. Int J Food Microbiol 145: S145–S151 doi:16/j.ijfoodmicro.2010.04.006 [DOI] [PubMed] [Google Scholar]
- 17. Raphael BH, Joseph LA, McCroskey LM, Lúquez C, Maslanka SE (2010) Detection and differentiation of Clostridium botulinum type A strains using a focused DNA microarray. Mol Cell Probes 24: 146–153 doi:16/j.mcp.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 18. Carter AT, Paul CJ, Mason DR, Twine SM, Alston MJ, et al. (2009) Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum. BMC Genomics 10: 115–115 doi:10.1186/1471-2164-10-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MTG, et al. (2007) Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17: 1082–1092 doi:10.1101/gr.6282807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Umeda K, Seto Y, Kohda T, Mukamoto M, Kozaki S (2010) A novel multiplex PCR method for Clostridium botulinum neurotoxin type A gene cluster typing. Microbiol Immunol 54: 308–312 doi:10.1111/j.1348-0421.2010.00213.x [DOI] [PubMed] [Google Scholar]
- 21. Umeda K, Seto Y, Kohda T, Mukamoto M, Kozaki S (2009) Genetic Characterization of Clostridium botulinum Associated with Type B Infant Botulism in Japan. J Clin Microbiol 47: 2720–2728 doi:10.1128/JCM.00077-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leclair D, Pagotto F, Farber JM, Cadieux B, Austin JW (2006) Comparison of DNA Fingerprinting Methods for Use in Investigation of Type E Botulism Outbreaks in the Canadian Arctic. J Clin Microbiol 44: 1635–1644 doi:–10.1128/JCM.44.5.1635–1644.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fillo S, Giordani F, Anniballi F, Gorgé O, Ramisse V, et al. (2011) Clostridium botulinum Group I Strain Genotyping by 15-Locus Multilocus Variable-Number Tandem-Repeat Analysis. J ClinMicrobiol 49: 4252–4263 doi:10.1128/JCM.05396-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindström M, Hinderink K, Somervuo P, Kiviniemi K, Nevas M, et al. (2009) Comparative Genomic Hybridization Analysis of Two Predominant Nordic Group I (Proteolytic) Clostridium botulinum Type B Clusters. Appl Environ Microbiol 75: 2643–2651 doi:10.1128/AEM.02557-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han T, Chang CW, Kwekel JC, Chen Y, Ge Y, et al. (2012) Characterization of whole genome amplified (WGA) DNA for use in genotyping assay development. BMC Genomics 13: 217 doi:10.1186/1471-2164-13-217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Erlandsson L, Rosenstierne MW, McLoughlin K, Jaing C, Fomsgaard A (2011) The Microbial Detection Array Combined with Random Phi29-Amplification Used as a Diagnostic Tool for Virus Detection in Clinical Samples. PLoS One 6: e22631 doi:10.1371/journal.pone.0022631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bouzid M, Heavens D, Elwin K, Chalmers RM, Hadfield SJ, et al. (2010) Whole genome amplification (WGA) for archiving and genotyping of clinical isolates of Cryptosporidium species. Parasitology 137: 27–36 doi:10.1017/S0031182009991132 [DOI] [PubMed] [Google Scholar]
- 28. Gadkar V, Rillig MC (2005) Suitability of genomic DNA synthesized by strand displacement amplification (SDA) for AFLP analysis: genotyping single spores of arbuscular mycorrhizal (AM) fungi. J Microbiol Methods 63: 157–164 doi:10.1016/j.mimet.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 29. Berthet N, Reinhardt AK, Leclercq I, van Ooyen S, Batéjat C, et al. (2008) Phi29 polymerase based random amplification of viral RNA as an alternative to random RT-PCR. BMC Mol Biol 9: 77 doi:10.1186/1471-2199-9-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zwick ME, Mcafee F, Cutler DJ, Read TD, Ravel J, et al. (2005) Microarray-based resequencing of multiple Bacillus anthracis isolates. Genome Biol 6: R10–R10 doi:10.1186/gb-2004-6-1-r10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dacheux L, Berthet N, Dissard G, Holmes EC, Delmas O, et al. (2010) Application of Broad-Spectrum Resequencing Microarray for Genotyping Rhabdoviruses. J Virol 84: 9557–9574 doi:10.1128/JVI.00771-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Z, Malanoski AP, Lin B, Kidd C, Long NC, et al. (2008) Resequencing microarray probe design for typing genetically diverse viruses: human rhinoviruses and enteroviruses. BMC Genomics 9: 577–577 doi:10.1186/1471-2164-9-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin B, Malanoski AP (2009) Resequencing Arrays for Diagnostics of Respiratory Pathogens. In: Dufva M, DNA Microarrays for Biomedical Research. Totowa, NJ: Humana Press ,Vol. 529 . 231–257. Available: http://www.springerlink.com/content/q1102392j0782ql1/#section=55300&page=1. Accessed 2011 Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Oriot C, D'Aranda E, Castanier M, Glaizal M, Galy C, et al. (2011) One collective case of type A foodborne botulism in Corsica. Clin Toxicol 49: 752–754 doi:10.3109/15563650.2011.606222 [DOI] [PubMed] [Google Scholar]
- 35. Lai LS, Wang YM, Lin CH (2011) Foodborne botulinum type E intoxication associated with dried bean curd: first case report in Taiwan. Acta Neurol Taiwan 20: 138–141. [PubMed] [Google Scholar]
- 36.Jalava K, Selby K, Pihlajasaari A, Kolho E, Dahlsten E, et al. (2011) Two cases of food-borne botulism in Finland caused by conserved olives, October 2011. Euro Surveill. 16: : 20034. Available: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20034. Accessed 2011 Dec. 27. [DOI] [PubMed] [Google Scholar]
- 37.Browning LM, Prempeh H, Little C, Houston C, Grant K, et al. (2011) An outbreak of food-borne botulism in Scotland, United Kingdom, November 2011. Euro Surveill. 16: : 20036. Available: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20036. Accessed 2011 Dec 27. [DOI] [PubMed] [Google Scholar]
- 38.Pingeon JM, Vanbockstael C, Popoff MR, King LA, Deschamps B, et al. (2011) Two outbreaks of botulism associated with consumption of green olive paste, France, September 2011. Euro Surveill. 16: : 20035. Available: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20035. Accessed 2011 Dec 27. [DOI] [PubMed] [Google Scholar]
- 39. Daminelli P, De Nadai V, Bozzo G, Finazzi G, Oliverio E, et al. (2011) Two unlinked cases of foodborne botulism in Italy at the beginning of 2010. New Microbiol 34: 287–290. [PubMed] [Google Scholar]
- 40. Date K, Fagan R, Crossland S, MacEachern D, Pyper B, et al. (2011) Three Outbreaks of Foodborne Botulism Caused by Unsafe Home Canning of Vegetables Ohio and Washington, 2008 and 2009. J Food Prot 74: 2090–2096 doi:10.4315/0362-028X.JFP-11-128 [DOI] [PubMed] [Google Scholar]
- 41. Case definitions for infectious conditions under public health surveillance. Centers for Disease Control and Prevention (1997) MMWR Recomm Rep 46: 1–55. [PubMed] [Google Scholar]
- 42.Therre H. (1999) Botulism in the European Union. Euro Surveill.. 4: : 2–7. Available:http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=48. Accessed 28 December 2011. [DOI] [PubMed] [Google Scholar]
- 43. Lindström M, Korkeala H (2006) Laboratory Diagnostics of Botulism. Clin Microbiol Rev 19: 298–314 doi:–10.1128/CMR.19.2.298–314.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandya GA, McEllistrem MC, Venepally P, Holmes MH, Jarrahi B, et al. (2011) Monitoring the Long-Term Molecular Epidemiology of the Pneumococcus and Detection of Potential “Vaccine Escape” Strains. PLoS One 6 . doi:10.1371/journal.pone.0015950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corless CE, Kaczmarski E, Borrow R, Guiver M (2008) Molecular Characterization of Neisseria meningitidis Isolates Using a Resequencing DNA Microarray. J Mol Diagn 10: 265–271 doi:10.2353/jmoldx.2008.070152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davignon L, Walter EA, Mueller KM, Barrozo CP, Stenger DA, et al. (2005) Use of Resequencing Oligonucleotide Microarrays for Identification of Streptococcus pyogenes and Associated Antibiotic Resistance Determinants. J Clin Microbiol 43: 5690–5695 doi:–10.1128/JCM.43.11.5690–5695.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hutson RA, Thompson DE, Lawson PA, Schocken-Itturino RP, Böttger EC, et al. (1993) Genetic interrelationships of proteolytic Clostridium botulinum types A, B, and F and other members of the Clostridium botulinum complex as revealed by small-subunit rRNA gene sequences. Antonie Van Leeuwenhoek 64: 273–283. [DOI] [PubMed] [Google Scholar]
- 48. Pourshaban M, Franciosa G, Fenicia L, Aureli P (2002) Taxonomic identity of type E botulinum toxin-producing Clostridium butyricum strains by sequencing of a short 16S rDNA region. FEMS Microbiol Lett 214: 119–125. [DOI] [PubMed] [Google Scholar]
- 49. Sasaki Y, Takikawa N, Kojima A, Norimatsu M, Suzuki S, et al. (2001) Phylogenetic positions of Clostridium novyi and Clostridium haemolyticum based on 16S rDNA sequences. Int J Syst Evol Microbiol 51: 901–904. [DOI] [PubMed] [Google Scholar]
- 50. Franciosa G, Floridi F, Maugliani A, Aureli P (2004) Differentiation of the gene clusters encoding botulinum neurotoxin type A complexes in Clostridium botulinum type A, Ab, and A(B) strains. Appl Environ Microbiol 70: 7192–7199 doi:–10.1128/AEM.70.12.7192–7199.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Raphael BH, Luquez C, McCroskey LM, Joseph LA, Jacobson MJ, et al. (2008) Genetic Homogeneity of Clostridium botulinum Type A1 Strains with Unique Toxin Gene Clusters. Appl Environ Microbiol 74: 4390–4397 doi:10.1128/AEM.00260-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leski TA, Malanoski AP, Gregory MJ, Lin B, Stenger DA (2011) Application of a Broad-Range Resequencing Array for Detection of Pathogens in Desert Dust Samples from Kuwait and Iraq. Appl Environ Microbiol 77: 4285–4292 doi:10.1128/AEM.00021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leski TA, Lin B, Malanoski AP, Wang Z, Long NC, et al. (2009) Testing and Validation of High Density Resequencing Microarray for Broad Range Biothreat Agents Detection. PLoS One 4 . doi:10.1371/journal.pone.0006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berthet N, Dickinson P, Filliol I, Reinhardt AK, Batejat C, et al. (2008) Massively parallel pathogen identification using high-density microarrays. Microb Biotechnol 1: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fach P, Micheau P, Mazuet C, Perelle S, Popoff M (2009) Development of real-time PCR tests for detecting botulinum neurotoxins A, B, E, F producing Clostridium botulinum, Clostridium baratii and Clostridium butyricum. J Appl Microbiol 107: 465–473 doi:–10.1111/j.1365–2672.2009.04215.x [DOI] [PubMed] [Google Scholar]
- 56. Takeda M, Tsukamoto K, Kohda T, Matsui M, Mukamoto M, et al. (2005) Characterization of the neurotoxin produced by isolates associated with avian botulism. Avian Dis 49: 376–381. [DOI] [PubMed] [Google Scholar]
- 57. Dineen SS, Bradshaw M, Johnson EA (2003) Neurotoxin Gene Clusters in Clostridium botulinum Type A Strains: Sequence Comparison and Evolutionary Implications. Curr Microbiol 46: 345–352 doi:10.1007/s00284-002-3851-1 [DOI] [PubMed] [Google Scholar]
- 58. Malanoski AP, Lin B, Wang Z, Schnur JM, Stenger DA (2006) Automated identification of multiple micro-organisms from resequencing DNA microarrays. Nucleic Acids Res 34: 5300–5311 doi:10.1093/nar/gkl565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Franciosa G, Maugliani A, Floridi F, Aureli P (2006) A novel type A2 neurotoxin gene cluster in Clostridium botulinum strain Mascarpone. FEMS Microbiol Lett 261: 88–94 doi:10.1111/j.1574-6968.2006.00331.x [DOI] [PubMed] [Google Scholar]