Abstract
In this work we build the first BI-RADS parser for Portuguese free texts, modeled after existing approaches to extract BI-RADS features from English medical records. Our concept finder uses a semantic grammar based on the BIRADS lexicon and on iterative transferred expert knowledge. We compare the performance of our algorithm to manual annotation by a specialist in mammography. Our results show that our parser’s performance is comparable to the manual method.
Keywords: feature extraction, breast cancer, BI-RADS descriptors
I. Introduction
Breast cancer is the most frequent type of cancer among women, and one of the main causes of death in western countries [1]. The main tool for early detection of breast cancer is mammography, or X-ray of the breast. Although more advanced techniques exist, mammography is considered the cheapest and most efficient way of detecting breast cancer at an early stage [2]. A routine asymptomatic mammography exam is called a screening mammography, while a more detailed exam following symptoms or a higher risk is called a diagnostic mammography.
Mammography findings are described by a radiologist according to the Breast Imaging Reporting and Data System (BI-RADS), created by the American College of Radiology [3]. The BI-RADS annotation system specifies a lexicon that forms a common language used by specialists in the area of breast cancer. It is composed of 43 descriptors, organized in a hierarchy. More advanced hospital database systems allow the annotation of BI-RADS features in a standard database format [4]. Nevertheless, most databases either do not support a standard format, or complement it with free-text reports. Hence the need for automated BI-RADS feature extraction from free-text.
Many researchers used natural language processing techniques to extract relevant concepts from medical texts. For example, MedLEE [5] is capable of extracting complex concepts from medical reports written in English. The National Library of Medicine’s Unified Medical Language System (UMLS) compiled a large number of medical dictionaries as well as a vocabulary that specifies a great number of biomedical concepts [6]. Other meta-thesauri exist too, like caTIES and SNOMED CT. None of these extracting tools and thesauri incorporate BI-RADS concepts.
In fact, few researchers tackled the problem of BI-RADS feature extraction from mammography free-text reports. Burnside et al. mapped frequent words in medical reports to BI-RADS terms using Linear Least Squares Fit [7]. Even though this approach is language independent, it performs poorly. Nassif et al. used a simple and effective parser, based on regular grammar expressions, to extract BI-RADS terms from English free-text documents [8]. This parser was recently extended to extract breast composition [9].
In this work, we construct a parser to extract Portuguese BI-RADS features, based on [8]. We refine the parser using input from a radiology specialist. We validate our method on a dataset of 153 patients, comparing the algorithm’s performance to manual annotation. Our parser achieves a performance comparable to the manual method performed by a specialist. According to our knowledge, this is the first work on extracting BI-RADS features from medical reports written in Portuguese.
II. MATERIALS AND METHODS
We used data collected for women undergoing mammography, between 2008 and 2009, at Centro Hospitalar São João in Porto, one of the largest hospitals in Portugal. Data were properly de-identified before the experiments. In total, we had medical evaluations for 622 women and 1,129 mammography reports. After removing redundancies, preprocessing the data, and linking together reports for the same patient, we ended up with 153 instances. Each resulting instance has both kinds of mammography reports: the basic screening and the detailed diagnostic. To best validate our method, we constructed a non-skewed data composed of both types. These 153 dual-instances were also given to a specialist that manually extracted the BI-RADS features after reading each text report.
In order to build our parser, our first step was to translate the BI-RADS lexicon to Portuguese. This was done with the help of a specialist. We then built a dictionary of synonyms for every BI-RADS term. Using an iterative process, we supplemented this list using expert knowledge to differentiate between different uses of the same word, to gauge the proximity of the words of a multi-word concept, and to capture medical wording practices and idiosyncrasies [8]. To illustrate some of the terms, the sentence “lesão da pele” (skin lesion) is captured by the presence of both words: lesão (lesion) and pele (skin), within a relative short distance. Thus we established an order for the combination of terms and a degree of proximity. We perform stemming and group words in the same concept if they are synonyms or typos. For example, “adenomegalia”, “axila positiva” and “gãnglio axilar” are all associated with the same concept: “Adenopatia Axilar”.
After detecting a concept, we proceed to the treatment of negations. Fortunately, medical texts tend to have a less complex semantic structure, a limited purpose, and are lexically less ambiguous than unrestricted documents [10]. Clinical negations tend to be much more direct and straightforward, especially in radiology reports [11]. A very small set of negation words accounts for the large majority of clinical negations. Following [12], we identify a set of negation triggers: “não” (not) when not preceded by “onde” (where), “sem” (without), and “nem” (nor). We found that negation triggers usually precede, but sometimes fall within or succeed, the concept they negate.
We implemented our algorithm in Perl. The evaluation of the parser was done in 3 phases. In the first phase, we only used the translated terms to extract the features. After reviewing the results with the specialist, we augmented our parser with synonyms and fine-tuned the word proximity for multi-words concepts. We performed this process of iterative expert knowledge incorporation over two iterations, constituting phases 2 and 3 of our analysis. The algorithmic performance also prompted the radiologist to update her own classification, since the parser was discovering BI-RADS features that she overlooked in her manual annotation.
In all phases, we generated a binary matrix of 153 rows by 43 columns, corresponding to the presence (1) or absence (0) of each feature for a given patient. In order to compare the performance of the parser with the performance of the manual annotation, we counted the number of agreements between both, as well as the number of disagreements related to each. We further examined these disagreement cases to determine their correct classification.
III. RESULTS
Tables I and II show the total number of features extracted by the parser and the radiologist during the 3 different phases, for both the screening and the diagnostic mammograms. We group the extracted features according to the BIRADS hierarchy.
Table I.
Number of attributes extracted from the screening mammograms, grouped by category
| Concept | 1st phase | 2nd phase | 3rd phase | ||
|---|---|---|---|---|---|
| Radiologist | Parser | Parser | Radiologist | Parser | |
| Shape (Forma) | 8 | 11 | 16 | 8 | 14 |
| Margin (Margem) | 15 | 12 | 26 | 15 | 20 |
| Density (densidade) | 0 | 4 | 2 | 0 | 1 |
| Calc. Morphology | 5 | 4 | 6 | 4 | 6 |
| Calc. Distribution | 8 | 2 | 8 | 8 | 9 |
| Special Cases | 9 | 7 | 8 | 7 | 7 |
| Associated Findings | 21 | 4 | 21 | 21 | 22 |
| Total | 66 | 44 | 87 | 63 | 80 |
Table II.
Number of attributes extracted from the diagnostic mammograms, grouped by category
| Concept | 1st phase | 2nd phase | 3rd phase | ||
|---|---|---|---|---|---|
| Radiologist | Parser | Parser | Radiologist | Parser | |
| Shape (Forma) | 3 | 1 | 4 | 3 | 3 |
| Margin (Margem) | 22 | 18 | 24 | 22 | 24 |
| Density (densidade) | 21 | 4 | 21 | 21 | 22 |
| Calc. Morphology | 9 | 9 | 13 | 10 | 14 |
| Calc. Distribution | 13 | 30 | 14 | 13 | 12 |
| Special Cases | 11 | 1 | 18 | 15 | 15 |
| Associated Findings | 43 | 8 | 35 | 43 | 36 |
| Total | 122 | 71 | 129 | 127 | 126 |
During the first phase of evaluation, and using the screening mammogram reports, the parser extracted 44 features while the radiologist extracted 66 (Table I). Out of 92 distinct extracted features, both methods had 18 features in common (20%), and disagreed on the remaining 74. Using the diagnostic mammography reports, the parser returned 71 features, and the manual method 122 (Table II). Out of 160 distinct extracted features, both methods agreed on 33 (21%), and disagreed on the remaining 127. This was a double-blind experiment, where the parser and radiologist were not influenced by each other.
We discussed the first set of results with the radiologist, reviewing the parser’s vocabulary. We refined its internal rules accordingly, and parsed the texts again. On the screening reports, the parser now returns 87 features. Out of 99 distinct extracted features, the parser and radiologist had 54 cases in common (54%), a substantial improvement from the first phase. For the diagnostic reports, the parser extracts 129 features. From a total of 146 distinct extracted features, 107 are agreements (73%) while 37 are disagreements, a significant improvement related to the first phase.
After phase 2, we performed a second round of parser fine-tuning. The radiologist too revised her annotations, removing 3 features from the screening matrix and adding 5 to the diagnostic matrix. Clearly the first manual extraction did not constitute ground truth. In fact, correctly labeling a text corpus is complicated enough that even experts need several passes to reduce labeling errors [13]. We can not assert what is ground truth, nor the actual number of features truly present in the text. Hence, we assume that the cases that both computational and manual methods agree upon are correctly classified, and we focus our attention on analyzing and re-labeling the disputed cases.
In the last phase, considering the screening reports, the parser returns 80 features and the radiologist 63. The two methods agreed on 59 extracted features, and differed on 25. Re-labeling the latter cases, the parser correctly classifies 14, versus 11 for the manual method. For the diagnostic reports, the parser and the radiologist respectively extract 126 and 127 features, forming 115 agreements and 23 disagreements. Our program correctly classified 11 of the disputed cases, while the radiologist got 12.
Combining both data subsets together, we can see that our method extracted 206 features, 174 of which are in accordance with manual extraction (84.5%). It extracted 32 features that the expert didn’t, while the radiologist had 16 extra features. Out of these 48 disputed cases, the parser edges the radiologist by correctly classifying 25 (52.1%). The parser is thus able to discover features missed or missclassified by the radiologist, and exhibits a similar performance.
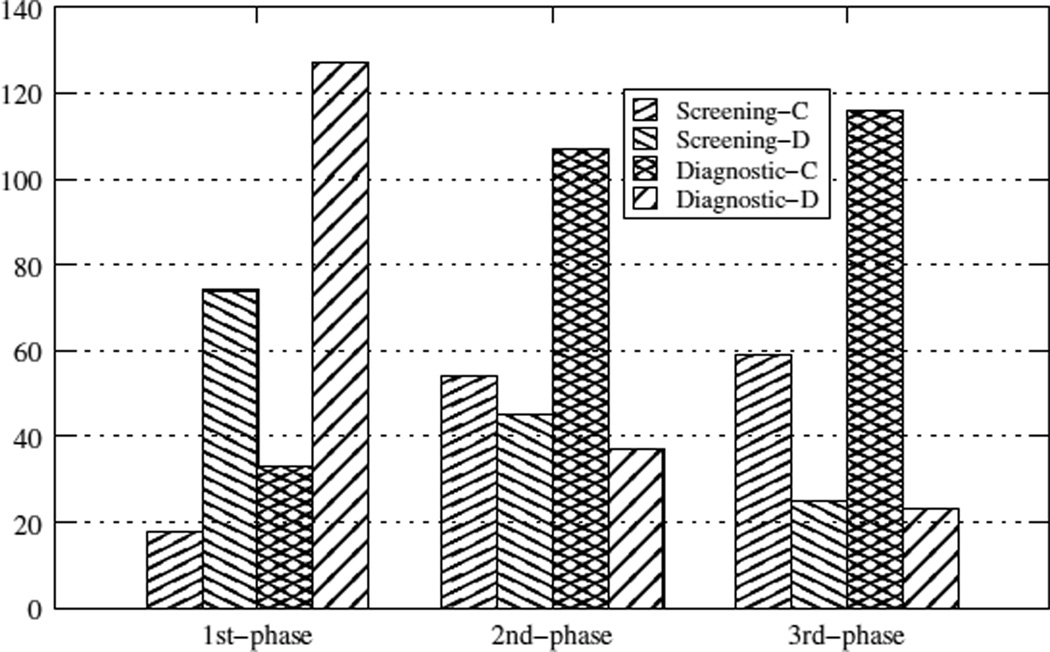
Figure 1 summarizes the improvements of the parser during the three phases of the experiment, in terms of concordant and discordant extracted features. Each phase is represented by four bars. The first two bars correspond to the screening reports while the next two correspond to the diagnostic reports. Taken in pairs, the left bar (Screening-C and Diagnostic-C) reports the number of concordances between the parser and the radiologist, while the right bar (Screening-D and Diagnostic-D) reports the discordances, features that were either extracted by the parser or by the radiologist but not by both. For the diagnostic reports, we observe a drastic improvement between the first and second phases, and an additional slight improvement by the third phase. For the screening reports, the improvement is not so pronounced, because this type of reports is less thorough and detects less BI-RADS features. In both cases, we managed to achieve a high level of concordance while reducing the number of discordances.
Figure 1.
Number of concordant and discordant extracted features by the parser and the manual methods, over the three phases and both data subsets
The number of features found in the screening mammography reports is much smaller than in its diagnostic counterpart, as expected. However, the parser manages to return approximately the same number of features extracted by the radiologist. The differences in the labeling of features related to shape and margin are explained by sentences containing ambiguous texts about “irregular shape” and “indistinct” or “ill defined” margin. As the expressions “irregular”, “indistinct” and “ill defined” can be used to categorize both shape and margin, the parser ended up extracting more features than needed for shape and margin. In such situations a human inspection of the text can be more effective than the parser.
According to Table II, there is a clear difference in extraction for the concept “Associated Findings”. The parser did not manage to extract all the information available in the medical reports. This difference is due to sentences related to “distorção arquitetural do estroma” (architectural stromal distortion). The different ways of defining the same concept were not well captured by the parser, but were captured by the radiologist. Our parser still has room for improvement. On the other hand, the parser extracted features related to “popcorn” calcifications, while the radiologist missed them.
IV. CONCLUSION AND FUTURE WORK
Feature extraction from free-text medical reports is still a challenge. In this work, we introduce the first BI-RADS parser for the Portuguese language. We use a simple approach, based on regular expressions, to capture most of the BI-RADS features expressed by radiologists in Portuguese free-text medical reports. We applied our technique to screening and diagnostic mammography reports. Our method is comparable to manual annotation. The parser was in accordance with the manual method over 84.5% of its extracted features, and correctly classified 52.1% of the disputed cases.
Our parser may be used as an automated double reader, or as an assessment tool of radiologist’s labeling of mammography reports. But most importantly, it is an automated method for extracting BI-RADS features from free-text reports and populating structural databases. Breast cancer models and classifiers are built using structured databases [14], and our parser is a necessary step to integrate free-text datasets to such models.
Our next step is to apply our parser to other medical reports not used in this study. We are also working on integrating this parser to a medical system that transcribes audio speech into text. We can thus perform feature extraction on-line, and emit alerts to the speaker in case of errors or ambiguities. Finally, we plan on using the features extracted by the parser to build classifiers that can distinguish between malignant and benign cancer findings in Portuguese medical records.
Acknowledgments
This work has been supported by the projects DigiScope (PTDC/EIA-CCO/100844/2008), HORUS (PTDC/EIA-EIA/ 100897/2008), the Fundação para a Ciência e Tecnologia (FCT/Portugal), and the US National Institute of Health grant R01CA127379-01.
References
- 1.American Cancer Society, Global Cancer Facts & Figures. 2nd ed. Atlanta, GA: American Cancer Society, Inc.; 2011. [Google Scholar]
- 2.Boyle P, Levin B, editors. World Cancer Report 2008. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 3.American College of Radiology, Breast Imaging Atlas: Breast Imaging Reporting and Data System. 4th ed. American College of Radiology, Inc.; 2003. [Google Scholar]
- 4.National Mammography Database. American College of Radiology; 2001. [Google Scholar]
- 5.Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J. Am. Med. Inform. Assn. 2004;vol. 11:392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Method. Inform. Med. 1993;vol. 32:281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnside B, Strasberg H, Rubin D. Automated indexing of mammography reports using linear least squares fit. 14th International Congress and Exhibition on Computer Assisted Radiology and Surgery. 2000:449–454. [Google Scholar]
- 8.Nassif H, Woods R, Burnside E, Ayvaci M, Shavlik J, Page D. Information extraction for clinical data mining: A mammography case study. 2009 IEEE International Conference on Data Mining Workshops. 2009:37–42. doi: 10.1109/icdmw.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by bi-rads breast tissue composition class. J. Am. Med. Inform. Assn. 2012 doi: 10.1136/amiajnl-2011-000607. Published Online First. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruch P, Baud R, Geissbuhler A, Rassinoux AM. Proc. of the 10th World Congress on Medical Informatics. Pt 1. vol. 10. London, UK: 2001. Comparing general and medical texts for information retrieval based on natural language processing: an inquiry into lexical disambiguation; pp. 261–265. [PubMed] [Google Scholar]
- 11.Mutalik PG, Deshpande A, Nadkarni PM. Use of general-purpose negation detection to augment concept indexing of medical documents: A quantitative study using the UMLS. J. Am. Med. Inform. Assn. 2001;vol. 8(no. 6):598–609. doi: 10.1136/jamia.2001.0080598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gindl S, Kaiser K, Miksch S. Syntactical negation detection in clinical practice guidelines. Proc. of the 21st International Congress of the European Federation for Medical Informatics; Göteborg, Sweden. 2008. pp. 187–192. [PMC free article] [PubMed] [Google Scholar]
- 13.Eskin E. Detecting errors within a corpus using anomaly detection. Proc. of the 1st North American chapter of the Association for Computational Linguistics Conference; San Francisco, CA. 2000. pp. 148–153. [Google Scholar]
- 14.Davis J, Burnside E, Dutra I, Page D, Ramakrishnan R, Costa VS, Shavlik J. View learning for statistical relational learning: With an application to mammography. Proc. of the 19th International Joint Conference on Artificial Intelligence; Edinburgh, Scotland. 2005. pp. 677–683. [Google Scholar]