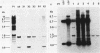
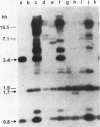
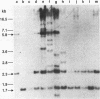
Abstract
We have constructed phage lambda and plasmid DNA substrates (lambda tk2 and ptk2) that contain two defective herpesvirus thymidine kinase (tk) genes that can be used to detect homologous recombination during the transfer of DNA into mouse L cells deficient in thymidine kinase activity. The recombination event reconstructs a wild-type tk gene and is scored because it converts Tk- cells to Tk+. Using this system, we have shown that (i) both intramolecular and intermolecular homologous recombination can be detected after gene transfer; (ii) the degree of recombination decreases with decreasing tk gene homology; and (iii) the efficiency of recombination can be stimulated 10- to 100-fold by cutting the tk2 DNA with restriction enzymes at appropriate sites relative to the recombining sequences. Based on the substrate requirements for these recombination events, we propose a model to explain how recombination might occur in mammalian cells. The essential features of the model are that the cut restriction site ends are substrates for cellular exonucleases that degrade DNA strands. This process exposes complementary strands of the two defective tk genes, which then pair. Removal of unpaired DNA at the junction between the paired and unpaired regions permits a gap repair process to reconstruct an intact gene.
Full text
PDF
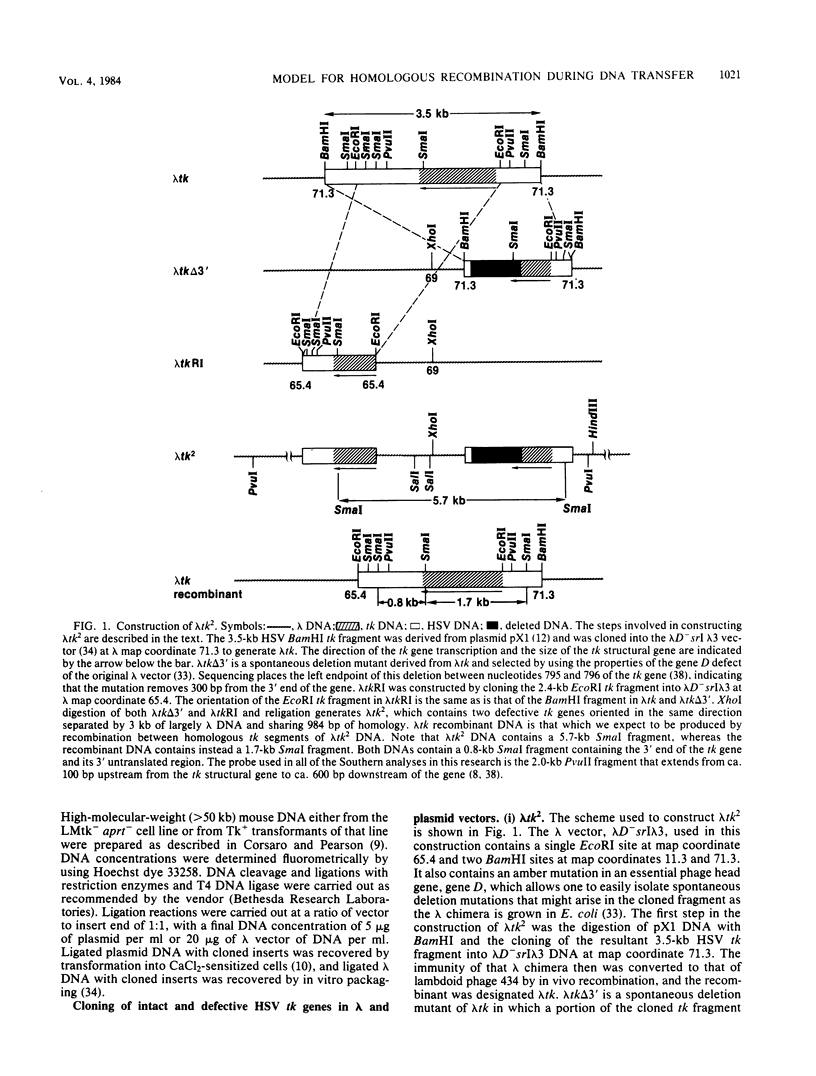





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckingham K. A plasmid cloning vector for Kpnl-cleaved DNA. Plasmid. 1980 Nov;4(3):354–356. doi: 10.1016/0147-619x(80)90074-8. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Broker T. R., Lehman I. R. Branched DNA molecules: intermediates in T4 recombination. J Mol Biol. 1971 Aug 28;60(1):131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- Campbell C. E., Worton R. G. Segregation of recessive phenotypes in somatic cell hybrids: role of mitotic recombination, gene inactivation, and chromosome nondisjunction. Mol Cell Biol. 1981 Apr;1(4):336–346. doi: 10.1128/mcb.1.4.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon J., Shenk T. E., Berg P. Biochemical procedure for production of small deletions in simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1392–1396. doi: 10.1073/pnas.72.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colbere-Garapin F., Chousterman S., Horodniceanu F., Kourilsky P., Garapin A. C. Cloning of the active thymidine kinase gene of herpes simplex virus type 1 in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3755–3759. doi: 10.1073/pnas.76.8.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro C. M., Pearson M. L. Enhancing the efficiency of DNA-mediated gene transfer in mammalian cells. Somatic Cell Genet. 1981 Sep;7(5):603–616. doi: 10.1007/BF01549662. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dasgupta U. B., Summers W. C. Genetic recombination of herpes simplex virus, the role of the host cell and UV-irradiation of the virus. Mol Gen Genet. 1980;178(3):617–623. doi: 10.1007/BF00337869. [DOI] [PubMed] [Google Scholar]
- Dubbs D. R., Rachmeler M., Kit S. Recombination between temperature-sensitive mutants of simian virus 40. Virology. 1974 Jan;57(1):161–174. doi: 10.1016/0042-6822(74)90117-2. [DOI] [PubMed] [Google Scholar]
- Enquist L. W., Vande Woude G. F., Wagner M., Smiley J. R., Summers W. C. Construction and characterization of a recombinant plasmid encoding the gene for the thymidine kinase of Herpes simplex type 1 virus. Gene. 1979 Nov;7(3-4):335–342. doi: 10.1016/0378-1119(79)90052-0. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Wong E. A., Wahl G., Capecchi M. R. Patterns of integration of DNA microinjected into cultured mammalian cells: evidence for homologous recombination between injected plasmid DNA molecules. Mol Cell Biol. 1982 Nov;2(11):1372–1387. doi: 10.1128/mcb.2.11.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Kafatos F. C., Jones C. W., Efstratiadis A. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 1979 Nov 24;7(6):1541–1552. doi: 10.1093/nar/7.6.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer P. J., Bowman A. H., Huberman M. H., Sanders-Haigh L., Killos L., Anderson W. F. Recovery of recombinant bacterial plasmids from E. coli transformed with DNA from microinjected mouse cells. Nucleic Acids Res. 1981 Nov 25;9(22):6199–6217. doi: 10.1093/nar/9.22.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird C. D. Chromatid structure: relationship between DNA content and nucleotide sequence diversity. Chromosoma. 1971 Mar 16;32(4):378–406. doi: 10.1007/BF00285251. [DOI] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Rosenstraus M. J., Chasin L. A. Separation of linked markers in Chinese hamster cell hybrids: mitotic recombination is not involved. Genetics. 1978 Dec;90(4):735–760. doi: 10.1093/genetics/90.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Shapira G., Stachelek J. L., Letsou A., Soodak L. K., Liskay R. M. Novel use of synthetic oligonucleotide insertion mutants for the study of homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4827–4831. doi: 10.1073/pnas.80.15.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J., Scangos G. Recombination during gene transfer into mouse cells can restore the function of deleted genes. Science. 1983 Jan 14;219(4581):174–176. doi: 10.1126/science.6294829. [DOI] [PubMed] [Google Scholar]
- Soberon X., Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. IV. Deletion derivatives of pBR322 and pBR325. Gene. 1980 May;9(3-4):287–305. doi: 10.1016/0378-1119(90)90328-o. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sternberg N. A characterization of bacteriophage P1 DNA fragments cloned in a lambda vector. Virology. 1979 Jul 15;96(1):129–142. doi: 10.1016/0042-6822(79)90179-x. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D., Enquist L., Weisberg R. A simple technique for the isolation of deletion mutants of phage lambda. Gene. 1979 Dec;8(1):35–51. doi: 10.1016/0378-1119(79)90006-4. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Tiemeier D., Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977 May;1(3-4):255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Vogel T. Recombination between endogenous and exogenous simian virus 40 genes. III. Rescue of SV40 tsA and tsBC mutants by passage in permissive transformed monkey lines. Virology. 1980 Jul 15;104(1):73–83. doi: 10.1016/0042-6822(80)90366-9. [DOI] [PubMed] [Google Scholar]
- Wagner M. J., Sharp J. A., Summers W. C. Nucleotide sequence of the thymidine kinase gene of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M. J., Sharp J. A., Summers W. C. Nucleotide sequence of the thymidine kinase gene of herpes simplex virus type 1. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Defined oligomeric SV40 DNA: a sensitive probe of general recombination in somatic cells. Cell. 1980 Aug;21(1):141–148. doi: 10.1016/0092-8674(80)90121-x. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Wilson J. H. Simian virus 40 recombinants are produced at high frequency during infection with genetically mixed oligomeric DNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2876–2880. doi: 10.1073/pnas.76.6.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C. S., Silverstein S. J. The kinetics of adenovirus recombination in homotypic and heterotypic genetic crosses. Virology. 1980 Mar;101(2):503–515. doi: 10.1016/0042-6822(80)90464-x. [DOI] [PubMed] [Google Scholar]
- de Saint Vincent B. R., Wahl G. M. Homologous recombination in mammalian cells mediates formation of a functional gene from two overlapping gene fragments. Proc Natl Acad Sci U S A. 1983 Apr;80(7):2002–2006. doi: 10.1073/pnas.80.7.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]