Abstract
Duchenne muscular dystrophy (DMD) is the most common childhood myopathy, characterized by muscle loss and cardiorespiratory failure. While the genetic basis of DMD is well established, secondary mechanisms associated with dystrophic pathophysiology are not fully clarified yet. In order to obtain new insights into the molecular mechanisms of muscle dystrophy during earlier stages of the disease, we performed a comparative proteomic profile of the spared extraocular muscles (EOM) vs. affected diaphragm from the mdx mice, using a label based shotgun proteomic approach. Out of the 857 identified proteins, 42 to 62 proteins had differential abundance of peptide ions. The calcium-handling proteins sarcalumenin and calsequestrin-1 were increased in control EOM compared with control DIA, reinforcing the view that constitutional properties of EOM are important for their protection against myonecrosis. The finding that galectin-1 (muscle regeneration), annexin A1 (anti-inflammatory) and HSP 47 (fibrosis) were increased in dystrophic diaphragm provides novel insights into the mechanisms through which mdx affected muscles are able to counteract dystrophy, during the early stage of the disease. Overall, the shotgun technique proved to be suitable to perform quantitative comparisons between distinct dystrophic muscles and allowed the suggestion of new potential biomarkers and drug targets for dystrophinopaties.
Introduction
Duchenne muscular dystrophy (DMD) is the most common and devastating of the human muscular dystrophies. It is characterized by progressive muscle weakness and death from cardiorespiratory compromise around the second or third decade of life [1]–[3]. In DMD and in the mdx mice model of DMD [4], [5] the genetic abnormality is in the X chromosome, in which the nucleotide sequence responsible for the expression of the protein dystrophin is mutated. In the absence of dystrophin, instability of sarcolemma leads to progressive myonecrosis, followed by intense inflammation and fibrosis [1], [6].
Numerous proteomics analysis of dystrophic muscles in mdx and in DMD have been performed with the aim to unravel the molecular pathogenesis of muscular dystrophy [7]–[13]. Previous proteomic studies included the use of the differential gel electrophoresis (DIGE), which provided important data related to the nature of the dystrophic proteome and showed a great number of proteins that were differentially expressed in distinct dystrophic muscles and ages [7], [10]–[15].
The multidimensional protein identification technology (MudPIT) method is a gel free alternative [16]–[18] to conventional gel-based methods, which has revolutionized the proteomic field. Basically, complex proteins mixtures are digested to peptides, fractionated according to its different chemical properties and subsequently analyzed by tandem mass spectrometry (MS/MS) resulting in protein identification. By using MudPIT it is possible to overcome several drawbacks associated with two-dimensional gel electrophoresis (2D-PAGE), especially under-representation of extreme acid/basic proteins [19] and the poor sensitivity for lowly expressed proteins. Moreover the MudPIT method simplifies sample handling, avoids sample loss in gel matrix and increases throughput and data acquisition [20], [21].
Protein quantification is fundamental for any comparative proteomic study of biological systems. In a proteomic analysis, the number of extracted proteins in a sample is higher than the number of identified proteins, which in turn is higher than the total number of quantified proteins [22]. In MS-based proteomics, two basic possibilities of quantification exist: (i) a relative quantification of proteins in compared samples (e.g. control vs. disease state) or (ii) an absolute quantification [23]. Isobaric mass tagging reagents, as tandem mass tags (TMT), allow multiple and independent measures of protein abundance in the same experiment, enabling statistical estimates of protein quantification and comparisons between different samples [24].
In order to obtain new insights into the molecular mechanisms of muscle dystrophy, we here used a label based shotgun proteomic approach, combining TMT labels and MudPIT method to analyze the diaphragm of 2-month-old mdx mice. We considered the age of 2 months as an early phase of dystrophy, similar to other proteomic studies [12], given the worsening of disease overtime. Signs of necrosis have been reported to occur at about 5 weeks of age in the mdx diaphragm [25]. At 2 months of age, diaphragm shows active muscle damage, regeneration and inflammation. However, significant histopathological signs of the disease, such as extensive fibrosis, are not seen at this age, suggesting an ability to compensate for muscle degeneration with cycles of muscle regeneration, as reported for limb muscles [26], [27]. Fibrosis will be present later in mdx diaphragm, increasing progressively from 6 months to 1 year of age and onwards, a timepoint when the muscle mostly resembles DMD myopathy [13], [26], [28]. In our proteomic analysis we have also searched for compensatory mechanisms involved in dystrophic muscle protection against myonecrosis, by comparing the proteomic profiles from dystrophic mdx diaphragm against the one from mdx extraocular muscles, which do not show muscle degeneration [29].
Materials and Methods
Animals
Males and females, 2-month-old mdx (C57BL/10ScSn-mdx/J) mice (n = 15) and age-match wild-type mice (C57BL/10SnJ, n = 15) were obtained from Jackson Laboratory and maintained in the animal facilities of Biomedical Center (Lund) according to the animal care guidelines. All mouse experimentation was approved by the Malmö/Lund (Sweden) ethical committee for animal research (permit numbers M62-09 and M122-10).
TMT labelling of mouse muscle samples
Protein extraction and preparing of the samples
Mice were sacrificed by cervical dislocation and the diaphragm (DIA; Figure 1) muscle and extraocular muscles (EOM; Figure 1) were dissected out, frozen in liquid nitrogen and reduced to powder using a mortar. Three different pools for each group (mdx mice and wild-type mice) were made, each composed of five animals. The muscles were lysed in assay lysis buffer (10 mM NaHCO3, 5% SDS) containing freshly added protease and phosphatase inhibitors (Roche - Indianapolis, IN, USA). The samples were centrifuged for 10 min at 15,682 g, and the soluble fraction was removed. The protein concentration was determined using BCA Protein Assay Kit (Pierce).
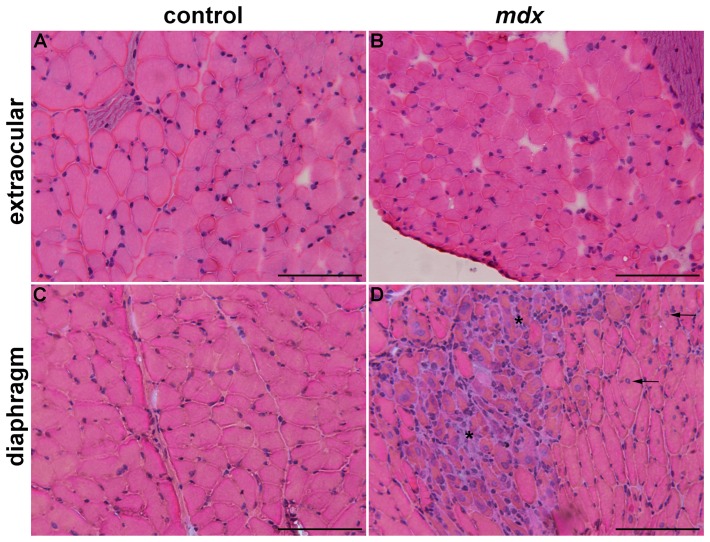
Figure 1. Histological differences between extraocular and diaphragm muscles.
Extraocular (A, B) and diaphragm (C, D) muscles of control (A, C) and mdx (B, D) mice. In controls (A, C), muscle fibers show peripheral nuclei. In the spared dystrophic EOM (B), fibers with peripheral nuclei indicate lack of muscle degeneration-regeneration. In the mdx DIA (D), fibers with central nuclei (arrows in D; regenerated fibers), and areas containing muscle fibers in regeneration surrounded by inflammatory cells (asterisk). Scale bar: 100 µm.
The samples were processed according to the instructions of the TMT isobaric Mass Tagging Kits and Reagents. In brief, 100 µg of protein per condition were mixed in six volumes of pre-chilled (−20°C) acetone and precipitated overnight. After centrifugation at 8,000 g for 10 minutes at 4°C, the pellet was dried. For protein digestion, 5 µl of 2% SDS, 45 µl of 200 mM TEAB were added to the sample and the final volume was adjusted to 100 µl with ultrapure water. Five microliter of 200 mM TCEP were added to the sample and incubated at 55°C for 1 hour. Then, 5 µl of the 375 mM iodoacetamide (with TEAB) were added and incubated for 30 min protected from light. To digest proteins, 2.5 µg of trypsin were added and kept overnight at 30–37°C. For protein labeling, 41 µl of the TMT Label Reagent were added to each sample and incubated for 1 h at room temperature. Eight microliter of 5% hydroxylamine were added and incubated for 15 min. Our labeling design allowed a label swap, in order to avoid possible bias due to technical errors (Table 1).
Table 1. Experimental setup for 2 muscles (EOM - extraocular and DIA - diaphragm) under 2 conditions (control and mdx) and with 3 biological replicates (Pool 1, 2 and 3).
| Groups | TMT label | |
| Pool 1 (n = 5) | EOM control | 127 |
| DIA control | 128 | |
| EOM mdx | 129 | |
| DIA mdx | 130 | |
| Internal standard | 126 | |
| Pool 2 (n = 5) | EOM control | 130 |
| DIA control | 129 | |
| EOM mdx | 128 | |
| DIA mdx | 127 | |
| Internal standard | 126 | |
| Pool 3 (n = 5) | EOM control | 129 |
| DIA control | 130 | |
| EOM mdx | 127 | |
| DIA mdx | 128 | |
| Internal standard | 126 |
The internal standard is a mixture of all samples.
SCX fractionation of the pooled TMT labelled samples
The pooled TMT-labelled samples were fractionated by strong cation-exchange (SCX, Applied Biosystems) using 500 µl of buffer A with 30, 60, 90, 120, 240, 300, 420, 500 mM KCl, respectively and collect as fractions 1–8, respectively. The fractions were cleaned on Ultra Microspin C18 columns (The Nest Group, Southboro, MA, USA), dried and resuspended in 30 µl 0.1% formic acid.
LC-MS/MS Analysis on LTQ-OrbitrapXL
The fractions were analyzed on an LTQ-OrbitrapXL (Thermo Fisher Scientific) interfaced with an in-house constructed nano-LC column. Two-microliter sample injections were made with an HTC-PAL autosampler (CTC Analytics AG) connected to an Agilent 1200 binary pump (Agilent Technologies). The peptides were trapped on a pre-column (40×0.075 mm i.d.) and separated on a reversed phase column, 200×0.05 mm. Both columns were packed in-house with 3 µm Reprosil-Pur C18-AQ particles. The flow through the analytical column was reduced by a split to approximately 100 nl/min and the gradient was as followed; 0–6 min 0.1% formic acid, 6–76 min 7–35% acetonitrile, 0.1% formic acid, 76–79 min 40–80% acetonitrile 0.1% formic acid.
LTQ-OrbitrapXL settings were: spray voltage 1.4 kV, 1 microscan for MS1 scans at 60 000 resolutions (m/z 400), full MS mass range m/z 400–2000. The LTQ-Orbitrap XL was operated in a data-dependent mode with one MS1 FTMS scan of precursor ions followed by CID (collision induced dissociation) and HCD (high energy collision dissociation), MS2 scans of the three most abundant doubly, triply and quadruply protonated ions in each FTMS scan. The settings for the MS2 were as follows: 1 microscans for HCD-MS2 at 7500 resolution (at m/z 400), mass range m/z 100–2000 with a collision energy of 50%, 1 microscans for CID-MS2 with a collision energy of 30%. Dynamic exclusion of a precursor selected for MS2 was used for 120 s after one repeat, enabling most of the co-eluting precursors to be selected for MS2.
Database Search and TMT Quantification
MS raw data files from all 8 SCX fractions per one TMT set and 3 MS runs were merged for relative quantification and identification using Proteome Discoverer version 1.3 (Thermo Fisher Scientific), respectively. Database search was performed by Mascot search engine using the following critera: Mus musculus in Swissprot protein database from April 2012, MS peptide tolerance as 10 ppm, MS/MS tolerance as 0.5 Da, trypsin digestion allowing 1 missed cleavages with variable modifications; methionine oxidation, cysteine methylthiol, and fixed modifications; N-terminal TMT6-plex label, lysine TMT6-plex label. The detected protein threshold in the software was set to a confidence using the FDR 1% method and identified proteins were grouped by sharing the same sequences to minimize redundancy.
For quantification, the ratios of TMT reporter ion intensities in MS/MS spectra (m/z 126.12, 127.13, 128.13, 129.14, 130.14) from raw data sets were used to calculate fold changes between samples via the relative ratio to the reference pool. Only peptides unique for a given protein were considered for relative quantitation, excluding those common to other isoforms or proteins of the same family. Only peptides with a score >10 and below the Mascot significance threshold filter of p = 0.05 were included. Single peptide identifications required a score equal to or above the Mascot identity threshold. Normalisation on protein median was used. The median of peptides was used for protein ratio and the resulting ratios were then exported into Excel for manual data interpretation.
Statistical analysis was performed by Student's t-test, with p values ≤0.05, with protein ratios smaller than −1.25 or greater than 1.25 and a coefficient of variation of less than 20% considered significantly different. For correction of false-positive values the FDR (false discovery rate) with estimated q-values was used [30].
Western blot analysis
Verification of the proteomic findings was carried out by comparative Western blot analysis. The levels of galectin-1 (Gal), annexin A5 (ANXA 5), β-dystroglycan (β-DG), calmodulin I (CaM) and calsequestrin-1 (CSQ) were quantified in DIA and EO muscles of control (n = 6) and mdx (n = 6) mice. The method was previously described [28], [31]. Muscles were lysed in assay lysis buffer containing freshly added protease and phosphatase inhibitors (1% Triton, 10 mM sodium pyrophosphate, 100 mM NaF, 10 µg/ml aprotinin, 1 mM PMSF, and 0.25 mM Na3VO4). The samples were centrifuged for 20 min at 12,581 g, and the soluble fraction was resuspended in 50 µl Laemmli loading buffer (2% SDS, 20% glycerol, 0.04 mg/ml bromophenol blue, 0.12 M Tris-HCl, pH 6.8, and 0.28 M ß-mercaptoethanol). An amount of 60 µg of total protein homogenate was loaded onto 8%–15% SDS-polyacrylamide gels. Proteins were transferred from the gels to a nitrocellulose membrane using a submersion electrotransfer apparatus (Bio-Rad Laboratories, Hercules, California, USA). Membranes were blocked for 2 h at room temperature with 5% skim milk/Tris-HCl buffered saline-Tween buffer (TBST; 10 mM Tris-HCl, pH 8, 150 mM NaCl, and 0.05% Tween 20). The membranes were incubated with the primary antibodies overnight at 4°C, washed in TBST, incubated with the peroxidase-conjugated secondary antibodies for 2 h at room temperature, and developed using the SuperSignal West Pico Chemiluminescent Substrate kit (Pierce Biotechnology, Rockford, Illinois, USA). To control for protein loading, Western blot transfer and nonspecific changes in protein levels, the blots were stripped and re-probed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The signal from western blotting bands was captured (G:Box iChemi camera; Syngene, Cambridge, UK) and quantified using the software Gene Tools Version 4.01, Syngene, Cambridge, UK.
The following primary antibodies were used: annexin A5, galectin-1, calmodulin I and GAPDH (Santa Cruz Biotechnology, Santa Cruz, California, USA); β-dystroglycan (Novocastra Laboratories Ltd., Benton Lane, Newcastle Upon Tyne, UK); and calsequestrin-1 (Affinity Bioreagents, Golden, Colorado, USA). The corresponding secondary antibody was peroxidase-labeled affinity-purified mouse or rabbit IgG antibody (H+L) (KPL, Gaithersburg, Maryland, USA).
Results
Shotgun proteomic analysis of DIA and EOM
By using the shotgun with MudPIT and TMT methodology, a total of 857 proteins were identified (DIA plus EOM). From this, about 48% (415 out of 857; Table S1) were common to the muscles studied, i.e., they were detected in both EOM and DIA, from both conditions (control and mdx) and in the three biological replicates. The criteria to identify proteins as being significantly changed between samples were proteins ratios with p-value ≤0.05 (Student's t-test), q-value ≤0.025 and protein ratios smaller than −1.25 or greater than 1.25. About 10% (42 out of 415; Table 2) of the proteins showed an altered expression pattern in the dystrophic DIA compared with the control DIA. We did not observe any proteins that had differential abundance of peptide ions in the mdx EOM (compared to control EOM), according to our established criteria. Overall, the proteins identified could be grouped into several biological processes such as immune system processes, energy and metabolism, sarcomeric and cytoskeletal proteins.
Table 2. The identified proteins that exhibit change in abundance in mdx diaphragm in comparison to control diaphragm.
| Accession | Description | MW [kDa] | ΣCoverage | Σ# Peptides | Fold Change |
| Q5SX39 | Myosin-4 | 222.7 | 57.9 | 133 | −1.49 |
| P13707 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic | 37.5 | 15.5 | 5 | −1.45 |
| Q8BW75 | Amine oxidase [flavin-containing] B | 58.5 | 2.7 | 2 | −1.42 |
| Q80XN0 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial | 38.3 | 8.8 | 3 | −1.35 |
| P19157 | Glutathione S-transferase P 1 | 23.6 | 12.9 | 2 | −1.33 |
| P10649 | Glutathione S-transferase Mu 1 | 26.0 | 28.9 | 6 | −1.32 |
| P28665 | Murinoglobulin-1 | 165.2 | 3.1 | 5 | −1.30 |
| P68134 | Actin, alpha skeletal muscle | 42.0 | 58.1 | 22 | −1.30 |
| P09542 | Myosin light chain 3 | 22.4 | 45.1 | 8 | −1.30 |
| Q924D0 | Reticulon-4-interacting protein 1, mitochondrial | 43.3 | 3.3 | 1 | −1.30 |
| P07310 | Creatine kinase M-type | 43.0 | 55.4 | 21 | −1.29 |
| Q9D0F9 | Phosphoglucomutase-1 | 61.5 | 10.0 | 5 | −1.29 |
| O09165 | Calsequestrin-1 | 45.6 | 23.8 | 10 | −1.29 |
| Q91V92 | ATP-citrate synthase | 119.7 | 3.2 | 3 | −1.29 |
| Q9CQ62 | 2,4-dienoyl-CoA reductase, mitochondrial | 36.2 | 27.2 | 8 | −1.28 |
| P20801 | Troponin C, skeletal muscle | 18.1 | 37.5 | 5 | −1.27 |
| P05064 | Fructose-bisphosphate aldolase A | 39.3 | 44.5 | 18 | −1.26 |
| O70250 | Phosphoglycerate mutase 2 | 28.8 | 24.5 | 8 | −1.26 |
| P14152 | Malate dehydrogenase, cytoplasmic | 36.5 | 28.1 | 7 | −1.25 |
| Q9CQN1 | Heat shock protein 75 kDa, mitochondrial | 80.2 | 4.4 | 2 | 1.26 |
| P19253 | 60S ribosomal protein L13a | 23.4 | 3.9 | 1 | 1.28 |
| P20029 | 78 kDa glucose-regulated protein | 72.4 | 10.2 | 5 | 1.28 |
| P35980 | 60S ribosomal protein L18 | 21.6 | 6.9 | 1 | 1.31 |
| P09103 | Protein disulfide-isomerase | 57.1 | 12.8 | 7 | 1.32 |
| Q8VDD5 | Myosin-9 | 226.2 | 6.3 | 11 | 1.33 |
| P47915 | 60S ribosomal protein L29 | 17.6 | 5.0 | 1 | 1.41 |
| Q9Z1N5 | Spliceosome RNA helicase Bat1 | 49.0 | 1.4 | 1 | 1.41 |
| Q9CZX8 | 40S ribosomal protein S19 | 16.1 | 18.6 | 3 | 1.42 |
| Q9D1R9 | 60S ribosomal protein L34 | 13.3 | 6.0 | 1 | 1.45 |
| Q91VR5 | ATP-dependent RNA helicase DDX1 | 82.4 | 1.8 | 1 | 1.47 |
| P47955 | 60S acidic ribosomal protein P1 | 11.5 | 14.0 | 1 | 1.54 |
| P14148 | 60S ribosomal protein L7 | 31.4 | 12.2 | 3 | 1.59 |
| Q8CGP6 | Histone H2A type 1-H | 13.9 | 27.3 | 3 | 1.61 |
| P19324 | Serpin H1 | 46.6 | 10.8 | 3 | 1.62 |
| P48036 | Annexin A5 | 35.7 | 23.2 | 8 | 1.65 |
| P17742 | Peptidyl-prolyl cis-trans isomerase A | 18.0 | 35.4 | 6 | 1.71 |
| Q8CI43 | Myosin light chain 6B | 22.7 | 34.3 | 7 | 1.75 |
| P20152 | Vimentin | 53.7 | 43.4 | 18 | 1.77 |
| P51881 | ADP/ATP translocase 2 | 32.9 | 38.9 | 12 | 1.95 |
| P16045 | Galectin-1 | 14.9 | 5.9 | 1 | 2.02 |
| Q61171 | Peroxiredoxin-2 | 21.8 | 14.7 | 2 | 2.03 |
| P10107 | Annexin A1 | 38.7 | 10.4 | 3 | 2.19 |
Comparative proteomic profiling: mdx DIA versus control DIA
By comparing dystrophic DIA with control DIA, we found that most of the proteins detected were present in the sarcoplasm or in the cytoskeleton. Mitochondrion was the organelle that displayed the highest percentage of proteins with altered levels, followed by the nucleus and the sarcoplasmic reticulum (Table 3). The majority of the identified proteins belonged to the class of metabolic proteins or to the class of immune system processes. All 8 protein metabolism-related ribosomal proteins had increased expression in the mdx DIA in comparison to control DIA. Regarding the immune system process, 2 proteins involved in responses to toxins (glutathione S-transferase P1 and glutathione S-transferase Mu1) were decreased in the mdx DIA compared to control DIA while 7 proteins were increased in the mdx DIA compared with control DIA, including proteins involved in responses to stress (heat shock protein 75 kDa, mitochondrial and 78 kDa glucose-regulated protein), induction of apoptosis (galectin-1), oxidative (ROS) processes (peroxiredoxin-2) and finally serine-type endopeptidase inhibitors (serpin H1 and murinoglobulin-1). Proteins involved in cellular respiration were also decreased in the mdx DIA, with 3 engaged in glycolysis (phosphoglucomutase-1, fructose-bisphosphate aldolase A and phosphoglucerate mutase 2), 2 in tricarboxylic acid cycle (ATP-citrate synthase and malate dehydrogenase) and 1 from the respiratory electron transport chain (amine oxidase [flavin-containing] B).
Table 3. Localization of shotgun identified proteins with altered expression in the comparisons: mdx diaphragm (DIA)×control (ct) DIA; ct DIA×ct extraocular (EO); mdx DIA×mdx EO.
| mdx DIA×ct DIA | ct DIA×ct EO | mdx DIA×mdx EO | |
| Sarcoplasm | 20.9% | 19.4% | 30.0% |
| Cytoskeleton | 30.2% | 21.0% | 14.3% |
| Extracellular Matrix | 7.0% | 3.2% | 10.0% |
| Mitochondrion | 18.6% | 29.0% | 17.1% |
| Nucleus | 14.0% | 9.7% | 4.3% |
| Sarcolemma | 2.3% | 8.1% | 15.7% |
| Sarcoplasmic reticulum | 4.7% | 6.5% | 8.6% |
| Peroxisome | 2.3% | 3.2% | 0.0% |
Comparative proteomic profiling: DIA versus EOM
About 13% (54 out of 415; Table 4) of the proteins showed differential abundance of peptide ions when comparing control DIA with control EOM and 15% (62 out of 415; Table 5) increased or decreased in the mdx DIA compared with the mdx EOM. By performing a further double comparison (i.e., control DIA×control EOM with mdx DIA×mdx EOM), 21 proteins were found in common, and may represent constitutive proteins related to embryological, morphological or functional differences between DIA and EOM muscles, rather than related to the pathogenesis of dystrophy per se. Among these 21 proteins, only annexin A1 showed a distinct pattern of change (increased or decreased) depending on the comparison (39% decreased in control DIA vs. control EOM and 51% increased in mdx DIA vs. mdx EOM). The remaining 20 proteins, most [11] were decreased in DIA vs. EOM (collagen alpha-1 and alpha-2; myosin 3, 4 and 11; SERCA 1; calsequestrin 1; sarcalumenin; aspartate aminotransferase; tropomyosin alpha -3 and mitochondrial 2-oxoglutarate/malate carrier protein), with a few [9] proteins increased in DIA×EOM (voltage-dependent anion selective channel protein 1; myosin 1, regulatory light chain 2 and light chain 3; uncharacterized protein C1orf93 homolog; isocitrate dehydrogenase (NADP) mitochondrial; L-lactate dehydrogenase B chain; C-X-C chemokine receptor type 1, and SERCA 2).
Table 4. The identified proteins that exhibit change in abundance in control diaphragm in comparison to control extraocular muscle.
| Accession | Description | MW [kDa] | ΣCoverage | Σ# Peptides | Fold Change |
| P08121 | Collagen alpha-1(III) chain | 138.9 | 0.8 | 1 | −4.12 |
| P13541 | Myosin-3 | 223.7 | 31.8 | 66 | −3.42 |
| O08638 | Myosin-11 | 226.9 | 4.6 | 8 | −2.44 |
| Q8CI43 | Myosin light chain 6B | 22.7 | 34.3 | 7 | −2.31 |
| Q01149 | Collagen alpha-2(I) chain | 129.5 | 4.7 | 6 | −2.28 |
| Q7TQ48 | Sarcalumenin | 99.1 | 27.8 | 18 | −2.18 |
| Q9CR62 | Mitochondrial 2-oxoglutarate/malate carrier protein | 34.1 | 12.4 | 4 | −2.04 |
| Q5SX39 | Myosin-4 | 222.7 | 57.9 | 133 | −1.87 |
| Q8R429 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | 109.4 | 31.7 | 31 | −1.76 |
| Q6PIE5 | Sodium/potassium-transporting ATPase subunit alpha-2 | 112.1 | 10.9 | 11 | −1.63 |
| P05202 | Aspartate aminotransferase, mitochondrial | 47.4 | 22.8 | 10 | −1.51 |
| P10922 | Histone H1.0 | 20.8 | 20.6 | 4 | −1.43 |
| P10107 | Annexin A1 | 38.7 | 10.4 | 3 | −1.39 |
| Q8CGP6 | Histone H2A type 1-H | 13.9 | 27.3 | 3 | −1.36 |
| P21107 | Tropomyosin alpha-3 chain | 32.8 | 49.3 | 21 | −1.35 |
| O09165 | Calsequestrin-1 | 45.6 | 23.8 | 10 | −1.33 |
| Q00896 | Alpha-1-antitrypsin 1–3 | 45.8 | 22.3 | 11 | −1.28 |
| Q6PB66 | Leucine-rich PPR motif-containing protein, mitochondrial | 156.5 | 3.1 | 5 | −1.28 |
| Q60714 | Long-chain fatty acid transport protein 1 | 77.9 | 19.9 | 12 | −1.25 |
| O88346 | Troponin T, slow skeletal muscle | 31.3 | 7.6 | 2 | 1.25 |
| P97447 | Four and a half LIM domains protein 1 | 31.9 | 16.1 | 5 | 1.26 |
| Q8BMS1 | Trifunctional enzyme subunit alpha, mitochondrial | 51.4 | 29.5 | 14 | 1.27 |
| P50544 | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | 70.8 | 13.0 | 7 | 1.27 |
| P51174 | Long-chain specific acyl-CoA dehydrogenase, mitochondrial | 47.9 | 25.8 | 12 | 1.28 |
| Q07417 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial | 44.9 | 10.2 | 3 | 1.28 |
| Q99JY0 | Trifunctional enzyme subunit beta, mitochondrial | 82.6 | 25.7 | 19 | 1.32 |
| P10649 | Glutathione S-transferase Mu 1 | 25.9 | 28.9 | 6 | 1.32 |
| P58771 | Tropomyosin alpha-1 chain | 32.7 | 72.2 | 34 | 1.34 |
| Q9CQ62 | 2,4-dienoyl-CoA reductase, mitochondrial | 36.2 | 27.2 | 8 | 1.35 |
| Q99LX0 | Protein DJ-1 | 20.0 | 8.0 | 2 | 1.35 |
| P15626 | Glutathione S-transferase Mu 2 | 25.7 | 16.1 | 4 | 1.35 |
| Q60932 | Voltage-dependent anion-selective channel protein 1 | 32.3 | 41.6 | 10 | 1.37 |
| Q9DB60 | Uncharacterized protein C1orf93 homolog | 21.7 | 4.0 | 1 | 1.39 |
| Q91WC3 | Long-chain-fatty-acid–CoA ligase 6 | 78.0 | 2.7 | 2 | 1.39 |
| P07310 | Creatine kinase M-type | 43.0 | 55.4 | 21 | 1.40 |
| Q924X2 | Carnitine O-palmitoyltransferase 1, muscle isoform | 88.2 | 4.3 | 3 | 1.40 |
| Q99LC5 | Electron transfer flavoprotein subunit alpha, mitochondrial | 35.0 | 19.8 | 5 | 1.42 |
| Q9DCW4 | Electron transfer flavoprotein subunit beta | 27.6 | 31.4 | 9 | 1.43 |
| P35550 | rRNA 2′-O-methyltransferase fibrillarin | 34.3 | 9.2 | 2 | 1.43 |
| Q921G7 | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial | 68.0 | 2.4 | 2 | 1.44 |
| P41216 | Long-chain-fatty-acid–CoA ligase 1 | 77.9 | 19.9 | 12 | 1.51 |
| P50247 | Adenosylhomocysteinase | 47.7 | 9.5 | 4 | 1.59 |
| Q8BW75 | Amine oxidase [flavin-containing] B | 58.5 | 2.7 | 2 | 1.66 |
| Q8CI51 | PDZ and LIM domain protein 5 | 18.0 | 35.4 | 6 | 1.67 |
| Q91Z83 | Myosin-7 | 222.7 | 36.5 | 81 | 1.73 |
| P19157 | Glutathione S-transferase P 1 | 23.6 | 12.9 | 2 | 1.78 |
| P16125 | L-lactate dehydrogenase B chain | 36.5 | 31.1 | 12 | 2.07 |
| Q5SX40 | Myosin-1 | 223.2 | 59.9 | 148 | 2.15 |
| P54071 | Isocitrate dehydrogenase [NADP], mitochondrial | 50.9 | 30.7 | 13 | 2.25 |
| Q8R0Y6 | 10-formyltetrahydrofolate dehydrogenase | 98.6 | 0.9 | 1 | 2.57 |
| P51667 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | 18.9 | 22.9 | 3 | 2.67 |
| O55143 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 114.8 | 17.5 | 19 | 2.73 |
| P09542 | Myosin light chain 3 | 22.4 | 45.1 | 8 | 3.28 |
| Q810W6 | C-X-C chemokine receptor type 1 | 40.0 | 1.71 | 1 | 3.78 |
Table 5. The identified proteins that exhibit change in abundance in mdx diaphragm in comparison to mdx extraocular muscle.
| Accession | Description | MW [kDa] | ΣCoverage | Σ# Peptides | Fold Change |
| P08121 | Collagen alpha-1(III) chain | 138.9 | 0.8 | 1 | −3.64 |
| Q5SX39 | Myosin-4 | 222.7 | 57.9 | 133 | −3.46 |
| O08638 | Myosin-11 | 226.9 | 4.6 | 8 | −3.36 |
| P32848 | Parvalbumin alpha | 11.9 | 74.6 | 15 | −2.86 |
| P13541 | Myosin-3 | 223.7 | 31.7 | 66 | −2.73 |
| Q8R429 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | 109.4 | 31.7 | 31 | −2.03 |
| Q01149 | Collagen alpha-2(I) chain | 129.5 | 4.7 | 6 | −1.99 |
| P11087 | Collagen alpha-1(I) chain | 137.9 | 4.9 | 6 | −1.88 |
| O09165 | Calsequestrin-1 | 45.6 | 23.8 | 10 | −1.78 |
| Q91V92 | ATP-citrate synthase | 119.7 | 3.2 | 3 | −1.75 |
| P21550 | Beta-enolase | 47.0 | 28.6 | 14 | −1.72 |
| Q9Z1E4 | Glycogen [starch] synthase, muscle | 83.9 | 7.6 | 5 | −1.63 |
| Q7TQ48 | Sarcalumenin | 99.1 | 27.8 | 18 | −1.63 |
| P13707 | Glycerol-3-phosphate dehydrogenase [NAD+], cytoplasmic | 37.5 | 15.5 | 5 | −1.62 |
| P05202 | Aspartate aminotransferase, mitochondrial | 47.4 | 22.8 | 10 | −1.62 |
| P21107 | Tropomyosin alpha-3 chain | 32.8 | 49.3 | 21 | −1.62 |
| P22599 | Alpha-1-antitrypsin 1–2 | 45.9 | 24.5 | 10 | −1.59 |
| P19096 | Fatty acid synthase | 272.3 | 3.2 | 7 | −1.53 |
| Q00898 | Alpha-1-antitrypsin 1–5 | 45.9 | 22.3 | 10 | −1.52 |
| Q5EBG6 | Heat shock protein beta-6 | 17.5 | 21.0 | 3 | −1.46 |
| P46412 | Glutathione peroxidase 3 | 25.4 | 15.9 | 3 | −1.44 |
| Q3UV70 | [Pyruvate dehydrogenase [acetyl-transferring]]-phosphatase 1, mitochondrial | 61.1 | 1.9 | 1 | −1.42 |
| Q6P8J7 | Creatine kinase S-type, mitochondrial | 47.4 | 29.1 | 13 | −1.42 |
| P07759 | Serine protease inhibitor A3K | 46.8 | 17.5 | 6 | −1.42 |
| P05064 | Fructose-bisphosphate aldolase A | 39.3 | 44.5 | 18 | −1.41 |
| Q8CI43 | Myosin light chain 6B | 22.7 | 34.3 | 7 | −1.37 |
| Q9WUB3 | Glycogen phosphorylase, muscle form | 97.2 | 34.6 | 30 | −1.32 |
| Q9CRB8 | Mitochondrial fission process protein 1 | 18.3 | 14.5 | 2 | −1.29 |
| Q61147 | Ceruloplasmin | 121.1 | 1.5 | 1 | −1.28 |
| Q9CR62 | Mitochondrial 2-oxoglutarate/malate carrier protein | 34.1 | 12.4 | 4 | −1.28 |
| Q8BH59 | Calcium-binding mitochondrial carrier protein Aralar1 | 74.5 | 31.2 | 15 | −1.25 |
| Q924D0 | Reticulon-4-interacting protein 1, mitochondrial | 43.3 | 3.3 | 1 | −1.25 |
| P11499 | Heat shock protein HSP 90-beta | 83.3 | 24.6 | 17 | 1.25 |
| P09103 | Protein disulfide-isomerase | 57.1 | 12.8 | 7 | 1.26 |
| Q60932 | Voltage-dependent anion-selective channel protein 1 | 32.3 | 41.6 | 10 | 1.29 |
| Q9CZX8 | 40S ribosomal protein S19 | 16.1 | 18.6 | 3 | 1.30 |
| Q62009 | Periostin | 93.1 | 1.7 | 1 | 1.33 |
| Q9CXT8 | Mitochondrial-processing peptidase subunit beta | 54.6 | 3.7 | 2 | 1.34 |
| P14602 | Heat shock protein beta-1 | 23.0 | 27.3 | 5 | 1.35 |
| Q8VDD5 | Myosin-9 | 226.2 | 6.3 | 11 | 1.36 |
| P15864 | Histone H1.2 | 21.3 | 33.0 | 9 | 1.40 |
| P17742 | Peptidyl-prolyl cis-trans isomerase A | 18.0 | 35.4 | 6 | 1.43 |
| P14148 | 60S ribosomal protein L7 | 31.4 | 12.2 | 3 | 1.44 |
| O09161 | Calsequestrin-2 | 48.2 | 12.1 | 5 | 1.45 |
| Q5SX40 | Myosin-1 | 223.2 | 59.9 | 148 | 1.46 |
| Q9D1R9 | 60S ribosomal protein L34 | 13.3 | 6.0 | 1 | 1.47 |
| P47955 | 60S acidic ribosomal protein P1 | 11.5 | 14.0 | 1 | 1.49 |
| Q9D0K2 | Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mitochondrial | 56.0 | 8.3 | 3 | 1.50 |
| P10107 | Annexin A1 | 38.7 | 10.4 | 3 | 1.51 |
| P19324 | Serpin H1 | 46.6 | 10.8 | 3 | 1.52 |
| P47915 | 60S ribosomal protein | 17.6 | 5.0 | 1 | 1.52 |
| Q9D8N0 | Elongation factor 1-gamma | 50.0 | 7.8 | 4 | 1.56 |
| P16045 | Galectin-1 | 14.9 | 5.9 | 1 | 1.63 |
| Q9DB60 | Uncharacterized protein C1orf93 homolog | 21.7 | 4.0 | 1 | 1.71 |
| P51881 | ADP/ATP translocase 2 | 32.9 | 38.9 | 12 | 1.81 |
| P54071 | Isocitrate dehydrogenase [NADP], mitochondrial | 50.9 | 30.7 | 13 | 1.82 |
| P16125 | L-lactate dehydrogenase B chain | 36.5 | 32.6 | 11 | 1.87 |
| P51667 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | 18.9 | 75.7 | 15 | 2.07 |
| P11404 | Fatty acid-binding protein, heart | 14.8 | 48.9 | 7 | 2.19 |
| P09542 | Myosin light chain 3 | 22.4 | 45.1 | 8 | 2.25 |
| Q810W6 | C-X-C chemokine receptor type 1 | 40.0 | 1.7 | 1 | 3.27 |
| O55143 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 114.8 | 17.5 | 19 | 3.91 |
Some proteins [41] were found exclusively in the comparison of the mdx DIA with the mdx EOM and therefore are more likely to be directly involved in the processes of muscle degeneration-regeneration or to the protection against myonecrosis. Regarding their biological processes classifications, most of them (37.2%) were related to carbohydrate, lipid or protein metabolism. Some were related to the immune system processes: in the dystrophic DIA, 3 proteins (HSP beta-6, glutathione peroxidase 3 and ceruloplasmin) were decreased and 5 proteins (HSP 90-beta, HSP beta-1, peptidyl-prolyl cis-trans isomerase A, elongation factor 1-gamma and galectin-1) were increased.
To select proteins that could be directly involved in dystrophic muscle degeneration we made a further double comparison (mdx DIA vs. control DIA with mdx DIA vs. mdx EOM) and found 19 proteins in common. Among these proteins, the majority (11 proteins; protein disulfide isomerase, 40S ribosomal protein S19; peptidyl-prolyl cis-trans isomerase; 60S ribosomal protein L7, L29 and L34; 60S acidic ribosomal protein P1; annexin A1; serpin H1; galectin-1 and ADP/ATP translocase 2) were increased in the mdx DIA (which presents muscle degeneration-regeneration in comparison to mdx EOM). Fewer (6 proteins; myosin-4, calsequestrin 1, ATP-citrate synthase, glycerol-3-phosphate dehydrogenase, fructose bisphosphate aldolase A, reticulon-4-interacting protein 1) were increased in control DIA and in the mdx EOM (which do not show muscle degeneration). Two myosins were increased or decreased depending on the comparison: myosin light chain 3 was 30% decreased in mdx DIA vs. control DIA and 124% increased in mdx DIA vs. mdx EOM and myosin light chain 6B was 76% increased in mdx DIA vs. control DIA and 37% decreased in mdx DIA vs. mdx EOM.
Western blot analysis
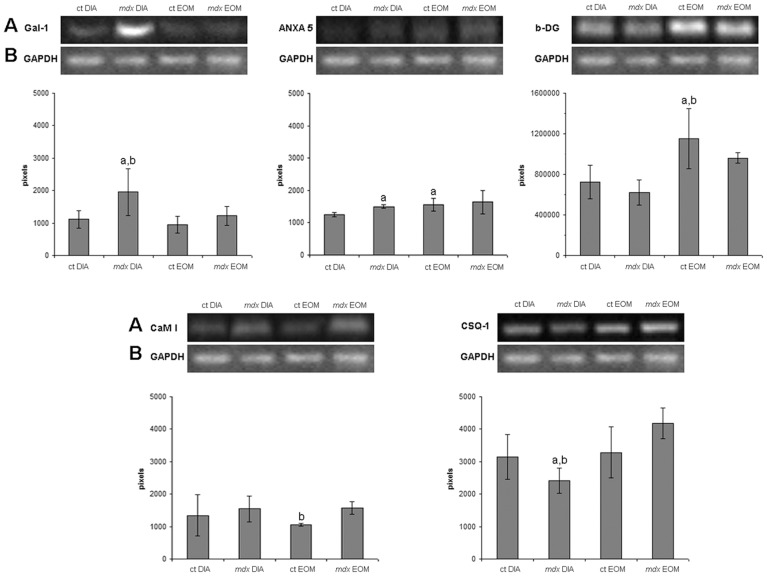
Western blot was performed in order to independently validate the identification and quantification of some proteins in DIA and EOM muscles of control and mdx mice (Figure 2). Western blot analysis indicated that galectin-1 levels were significantly higher in mdx DIA compared to control DIA (75% increase) and to mdx EOM (60% increase; p≤0.05, ANOVA; Figure 2). This is in agreement with the proteomic analysis showing a significant increase of galectin-1 in mdx DIA compared with control DIA (fold change of 2.02, Table 2) and with mdx EOM (fold change of 1.63, Table 5).
Figure 2. Western blot analysis of some proteins revealed by proteomics.
Quantification of galectin-1 (Gal-1), annexin A5 (ANXA5), β-dystroglycan (b-DG), calmodulin I (CaM I) and calsequestrin-1 (CSQ-1) by Western blot analysis in crude extracts of diaphragm (DIA) and extraocular (EO) muscles from control (ct) and dystrophic (mdx) mice. In A, Western blot of proteins. In B, the same blot reprobed for GAPDH as a loading control. Graphs represent the level of proteins expressed in pixels. Bars represent standard deviation. a Significantly different from ct DIA (p≤0.05, ANOVA). b Significantly different from mdx EOM (p≤0.05, ANOVA).
Annexin A5 also presented similar changes as those detected with MudPIT-TMT for most of the comparisons, mainly the increased (20%) levels of this protein in mdx DIA vs. control DIA (Table 2) and no differences in mdx EOM vs. control EOM (Figure 2). While Western blot analysis showed lower (20%) levels of annexin A5 in control DIA in relation to control EOM, proteomics showed comparable levels of annexin A5 between these muscles (Figure 2, Table 4).
The proteins related to calcium homeostasis, calmodulin I and calsequestrin-1, also displayed similar expression changes using both Western blot and proteomics analyses. Calmodulin levels were similar among the groups (blot and proteome), with a tendency to be increased in the mdx EO compared to control EO (significance only detected by Western blot analysis) (Figure 2), in agreement with our previous work [31]. Calsequestrin-1 was reduced in mdx DIA compared with control DIA (23% reduction) and with mdx EO (42% reduction; Figure 2) in line with the proteomic finding showing a significant decrease of calsequestrin-1 in mdx DIA compared with control DIA (fold change of −1,29, Table 2) and with mdx EO (fold change of −1.78, Table 5).
Western blot analysis of β-dystroglycan expression confirmed previous studies [32] demonstrating a persistent expression of this protein in the mdx EO concomitant with a significant decrease in the affected muscle (mdx DIA; Figure 2C). We also found that dystroglycan (α and β) expression was decreased in mdx DIA compared with mdx EO (−1.12) and control EO (−1.11) and similar results were described in a recent proteomic study [33].
Discussion
Shotgun proteomic analysis
By using the shotgun proteomic analysis, we have here identified a total of 857 proteins in the DIA and EOM from control and dystrophic mdx mice at 2 months of age. Out of the 857 proteins, 42 had differential abundance of peptide ions in the DIA of the mdx mice. Previous studies using the DIGE proteomic analysis detected 2398 2D spots of which 19 [7] or 35 [8] proteins showed a differential abundance of spots in mdx DIA compared to control DIA. By using the 2D gel-based proteomic technique, one protein made from one gene may exhibit an average of 10–15 different spots [34] due to pos-translational modifications and protein degradation [35], [36]. Additionally, spots could appear in the gels due to disulfide bridges because the current DIGE protocol does not require alkylation during the isoelectric focusing step [34]. Therefore, the fact that multiple spots correspond to one protein may explain the difference between the number of spots detected by DIGE and the number of proteins detected in the present study. Nevertheless, the present study demonstrates that the shotgun methodology allowed the identification and quantification of most proteins present in a small amount of muscle (100 µg) and in a short period of time (one run in the mass spectrometer).
In the dystrophic DIA, we observed that some proteins already displayed abnormal levels at this early stage of the disease (2 months of age). Some of the changed proteins detected in the DIA had not been described before in other proteomic studies, such as galectin-1, annexin, serpin H1 and periostin. We also found proteins that had been described by other proteomic techniques. For instance, by using DIGE analysis, malate dehydrogenase, myosin light chain 3, myosin light chain 6B, myosin-4, myosin-9 and vimentin were found to be altered in the mdx diaphragm at 9 weeks of age [7], [8]; phosphoglucomutase-1 and phosphoglycerate mutase 2 were changed in the 4–7 week old mdx gastrocnemius [12] and 2,4-dienoyl-CoA reductase mitochondrial, myosin light chain 3 and peroxiredoxin were affected in the 9 months of age mdx heart [10]; troponin T slow skeletal muscle and four and a half LIM domains protein 1 expression was also affected in mdx muscles when comparing the proteomic profile of dystroglycan-interacting proteins [33]. Therefore, the shotgun technique proves to be effective in demonstrating new altered proteins as well as proteins already described by the seminal DIGE studies [7]–[10], [12], [13], [37]. Furthermore, given that the analysis of different samples is performed at one run in the mass spectrometer, we were able to compare the proteomic profile of two different muscles: the affected DIA and the non-affected EOM.
The MudPIT technique has never been used before to investigate the molecular aspects of dystrophin absence in the mdx mice. In the present study, we demonstrate that this technique contributes to new insights to the pathophysiology of dystrophy. However, it is important to note that, as with any other technique, the shotgun proteomics has its weaknesses. For improved proteome coverage, optimization of fractionation processes and enrichment of purified organelles could allow a more comprehensive view of the molecular aspects of the disease [20], [38], [39]. For instance, in a recent study of mdx limb muscles, immunoprecipitation coupled with shotgun proteomics allowed proteomic analyzes of the dystrophin-associated protein complex per se and identification of new dystroglycan-associated proteins [33]. In addition, the combination of different proteomic approaches could lead to a more complete coverage of the proteomic profile [38].
Proteins related to protection against myonecrosis
The comparison of EOM with DIA muscle revealed altered expression of cytoskeletal proteins (myosin and troponin) and of extracellular matrix components, such as collagen. Furthermore, proteins related to calcium homeostasis and ion channels also exhibited different expression levels. The differential levels of calequestrin1, SERCA1 and SERCA2 observed here, possibly related to fiber type, suggest a better calcium homeostasis, and consequent protection against myonecrosis, in the EOM compared with DIA. This finding is in agreement with previous observations in the mdx EOM using Western blotting analysis [31]. Other mdx spared muscles, such as the intrinsic laryngeals, also show higher levels of SERCA1 in comparison to normal ILM muscles [40]. Moreover, we observed that the calcium buffering proteins sarcalumenin and calsequestrin 1 [41], [42] were increased in EOM compared to DIA, even in the control group. Therefore, the present results support previous observations (with Western blotting, ELISA and immunocytochemistry techniques) that constitutional properties of the EOM compensate for the lack of dystrophin, allowing a better response against myonecrosis [11], [43].
Proteins related to degeneration and regeneration
Oxidative stress and fibrosis
Proteins involved in the oxidative stress response (HSP 75 kDa, 78 kDa glucose-related protein, serpin H1 or HSP 47 and peroxiredoxin-2) were increased in the mdx DIA compared to control DIA. While peroxiredoxins are antioxidant enzymes that control peroxide levels induced by cytokines [44], the HSPs have chaperone functions [45]. The higher levels of these proteins may reflect an attempt of the dystrophic DIA to control oxidative stress at this stage of the disease [46], [47]. However, HSP 47 is also related to increased collagen production and fibrosis [48], which will be morphologically prominent in the mdx DIA but only at later stages of the disease [28], as demonstrated by proteomics [13]. Interestingly, several proteins related to fibrosis that were demonstrated to be increased in the old mdx DIA muscle compared with younger mdx DIA, such as collagen α1 (VI) chain, minecan and actinin-α2 [13] had no differential abundance of peptide ions at the age studied here (comparing mdx DIA×control DIA in the present study). This suggests that time-related changes of the proteomic profile occur and this may be of relevance for future studies of drug therapy for DMD.
Inflammation, apoptosis and regeneration
We detected an increased level of the annexins A1 and A5 in mdx DIA. The annexins bind to negative charged phospholipids in a calcium-dependent manner and participate in many physiological processes, such as cell shape changes, transport and organization of vesicles, exocytosis and endocytosis [49]. Annexin A1 prevents muscle degeneration due to sarcolemma resealing repair [50], [51]. Extracellular annexins act in fibrinolysis, coagulation and apoptosis [52], [53]. The annexins A1 and A2 are overexpressed in different forms of muscular dystrophies, possibly related to their anti-inflammatory activity [54]. Annexin A1 is a glucocorticoid-inducible protein [55], [56] able to mimic the anti-inflammatory effects of glucocorticoids in several experimental models of inflammation both in vivo and in vitro [57]. Besides participating in apoptotic processes, annexin A5 presents anti-inflammatory properties by inhibiting phospholipase A2 and phosphadylserine-catalyzed inflammation [58], [59]. Therefore, the increased level of annexins early in the mdx DIA may suggest a potential ability of the dystrophic DIA to modulate inflammation.
Reticulon-4-interacting protein 1 is a mitochondrial protein that reduces the anti-apoptotic activity of Bcl-2 and Bcl-XL [60], [61] and was decreased in the mdx DIA compared to control DIA. This finding indicates that apoptosis may be involved in mdx pathology [62]–[66], at least during the early phase, although apoptotic fibers have not been consistently detected in the mdx, at later stages [67], [68]. Galectin-1 was also overexpressed in mdx DIA. This protein is produced by myoblasts and other cell types [69], [70], and participates in muscle regeneration [71]–[76], and may be involved in the regenerative ability of the dystrophic DIA during this period, since at later stages fibrosis is a hallmark of diaphragm dystrophy. Galectin-1 also has a protective effect on skeletal muscle by reducing inflammation [77].
Conclusions
In the present study we demonstrated that the shotgun proteomics approach adds to the former proteomic techniques [7]–[10], [12], [13], [37] as a suitable alternative to track possible changes in the levels of proteins in dystrophic muscles, during the early phase of the disease. We would like to highlight some advantages of the technique that include the small amount of sample required, the relatively short time to accomplish the analysis and the possibility to perform qualitative and quantitative comparisons between distinct muscles and experimental groups.
The current proteomics study of the dystrophic DIA, in the phase prior to more advanced disease [13], [26], demonstrates an increase in proteins involved in muscle regeneration (galectin-1 [76], [77]), calcium handling (calsequestrin 1 [31]), inflammation (annexin A1 [50], [51], [54]) and fibrosis (HSP 47 [48]), making them valuable candidates for being potential drug targets and exploratory biomarkers.
Supporting Information
Proteins identified in extraocular and diaphragm muscles of control and mdx mice in three biological replicates.
(XLS)
Acknowledgments
We acknowledge the help with the protein analysis from the SCIBLU Proteomics Resource Centre at Lund University (especially Karin Hansson) and from the Proteomics Core Facility at Gothenburg University (especially Carina Sihlbom), and the Wallenberg Stiftelse (KAW).
Funding Statement
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, grants 11/51697-6; 08/58491-1) and the Swedish Research Council. MJM is recipient of fellowships from Conselho Nacional de Pesquisas (301306/2010-9). CYM was recipient of a fellowship from Fundação de Amparo à Pesquisa do Estado de São Paulo (grant 08/54775-5) and from CAPES (grant 2014-10-6). MS data were obtained from the Proteomics Core Facility, Gothenburg University, on equipment obtained by a grant from Knut and Alice Wallenberg (KAW grant 2007.0118). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Engel AG, Yamamoto M, Fischbeck KH (1994) Dystrophinopathies. In: Engel AG, Franzini-Armstrong C, editors. Myology. 2 ed. New York: McGraw-Hill; pp. 1133–1187.
- 2. Petrof BJ (1998) The molecular basis of activity-induced muscle injury in Duchenne muscular dystrophy. Molec Cel Biochem 179 1–2: 111–123. [DOI] [PubMed] [Google Scholar]
- 3. Biggar DW (2006) Duchenne Muscular Dystrophy. Pediatr Rev 401: 83–88. [DOI] [PubMed] [Google Scholar]
- 4. Bulfield G, Siller WG, Wight PAL, Moore KJ (1984) X chromosome-liked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA 81: 1189–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Durbeej M, Campbell KP (2002) Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr Opin Genet Dev 12 3: 349–361. [DOI] [PubMed] [Google Scholar]
- 6. Grounds MD, Sorokin L, White J (2005) Strength at the extracellular matrix-muscle interface. Scand J Med Sci Sports 15: 381–391. [DOI] [PubMed] [Google Scholar]
- 7. Doran P, Dowling P, Donoghue P, Buffini M, Ohlendieck K (2006) Reduced expression of regucalcin in young and aged mdx diaphragm indicates abnormal cytosolic calcium handling in dystrophin-deficient muscle. Biochim Biophys Acta 1764 4: 773–785. [DOI] [PubMed] [Google Scholar]
- 8. Doran P, Martin G, Dowling P, Jockusch H, Ohlendieck K (2006) Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics 6 16: 4610–4621. [DOI] [PubMed] [Google Scholar]
- 9. Doran P, Donoghue P, O'Connell K, Gannon J, Ohlendieck K (2009) Proteomics of skeletal muscle aging. Proteomics 9 4: 989–1003. [DOI] [PubMed] [Google Scholar]
- 10. Lewis C, Jockusch H, Ohlendieck K (2010) Proteomic profiling of the dystrophin-deficient mdx heart reveals drastically altered levels of key metabolic and contractile proteins. J Biomed Biotech 648501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lewis C, Ohlendieck K (2010) Proteomic profiling of naturally protected extraocular muscles from the dystrophin-deficient mdx mouse. Biochem Biophys Res Commun 396 4: 1024–1029. [DOI] [PubMed] [Google Scholar]
- 12. Gardan-Salmon D, Dixon JM, Lonergan SM, Selsby JT (2011) Proteomic assessment of the acute phase of dystrophin deficiency in mdx mice. Eur J Appl Physiol 111 11: 2763–2773. [DOI] [PubMed] [Google Scholar]
- 13. Carberry S, Zweyer M, Swandulla D, Ohlendieck K (2012) Proteomics reveals drastic increase of extracellular matrix proteins collagen and dermatopontin in the aged mdx diaphragm model of Duchenne muscular dystrophy. Int J Mol Med 30 2: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge Y, Molloy MP, Chamberlain JS, Andrews PC (2003) Proteomic analysis of mdx skeletal muscle: Great reduction of adenylate kinase 1 expression and enzymatic activity. Proteomics 3 10: 1895–1903. [DOI] [PubMed] [Google Scholar]
- 15. Doran P, Gannon J, O'Connell K, Ohlendieck K (2007) Proteomic profiling of animal models mimicking skeletal muscle disorders. Proteomics Clin Appl 1: 1169–84. [DOI] [PubMed] [Google Scholar]
- 16. Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, et al. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem 75 8: 1895–904. [DOI] [PubMed] [Google Scholar]
- 17. Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–69. [DOI] [PubMed] [Google Scholar]
- 18. Aggarwal K, Choe LH, Lee KH (2005) Quantitative analysis of protein expression using amine-specific isobaric tags in Escherichia coli cells expressing rhsA elements. Proteomics 5 9: 2297–2308. [DOI] [PubMed] [Google Scholar]
- 19. Martins de Souza D, Oliveira BM, Castro-Dias E, Winck FV, Horiuchi RS, et al. (2008) The untiring search for the most complete proteome representation: reviewing the methods. Brief Funct Genomic Proteomic 7 4: 312–321. [DOI] [PubMed] [Google Scholar]
- 20. Geiser L, Vaezzadeh AR, Deshusses JM, Hochstrasser DF (2011) Shotgun proteomics: a qualitative approach applying isoelectric focusing on immobilized pH gradient and LC-MS/MS. Methods Mol Biol 681: 449–58. [DOI] [PubMed] [Google Scholar]
- 21. Lewis C, Doran P, Ohlendieck K (2012) Proteomic analysis of dystrophic muscle. Methods Mol Biol 798: 357–69. [DOI] [PubMed] [Google Scholar]
- 22. Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389 4: 1017–1031. [DOI] [PubMed] [Google Scholar]
- 23. Yates JR, Ruse CI, Nakorchevsky A (2009) Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng 11: 49–79. [DOI] [PubMed] [Google Scholar]
- 24. Aggarwal K, Choe LH, Lee KH (2006) Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic 5 2: 112–120. [DOI] [PubMed] [Google Scholar]
- 25. Reutenauer-Patte J, Boittin FX, Patthey-Vuadens O, Ruegg UT, Dorchies OM (2012) Urocortins improve dystrophic skeletal muscle structure and function through both PKA- and Epac-dependent pathways. Am J Pathol 180 2: 749–762. [DOI] [PubMed] [Google Scholar]
- 26. Stedman HH, Sweeney HL, Shrager JB, Maguire HC, Panettieri RA, et al. (1991) The mdx mouse diaphragm reproduces the degenerative changes of Duchenne muscular dystrophy. Nature 352 6335: 536–539. [DOI] [PubMed] [Google Scholar]
- 27. Grounds MD, Radley HG, Lynch GS, Nagaraju K, De Luca A (2008) Towards developing standard operating procedures for pre-clinical testing in the mdx mouse model of Duchenne muscular dystrophy. Neurobiol Dis 31: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Taniguti AP, Pertille A, Matsumura CY, Santo Neto H, Marques MJ (2011) Prevention of muscle fibrosis and myonecrosis in mdx mice by suramin, a TGF-β1 blocker. Muscle Nerve 43 1: 82–87. [DOI] [PubMed] [Google Scholar]
- 29. Andrade FH, Porter JD, Kaminski HJ (2004) Eye muscle sparing by the muscular dystrophies: lessons to be learned? Micros Res Tec 48: 192–203. [DOI] [PubMed] [Google Scholar]
- 30. Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100 16: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pertille A, Tonizza de Carvalho CL, Matsumura CY, Santo Neto H, Marques MJ (2010) Calcium-binding proteins in skeletal muscles of the mdx mice: potential role in the pathogenesis of Duchenne muscular dystrophy. Int J Exp Pathol 91 1: 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dowling P, Lohan J, Ohlendieck K (2003) Comparative analysis of Dp427-deficient mdx tissues shows that the milder dystrophic phenotype of extraocular and toe muscle fibres is associated with a persistent expression of beta-dystroglycan. Eur J Cell Biol 82 5: 222–230. [DOI] [PubMed] [Google Scholar]
- 33. Yoon JH, Johnson E, Xu R, Martin LT, Martin PT, et al. (2012) Comparative proteomic profiling of dystroglycan-associated proteins in wild type, mdx, and Galgt2 transgenic mouse skeletal muscle. J Proteome Res 11 9: 4413–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu WW, Wang G, Baek SJ, Shen RF (2006) Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res 5 3: 651–658. [DOI] [PubMed] [Google Scholar]
- 35. Fountoulakis M, Juranville JF, Berndt P, Langen H, Suter L (2000) Two-dimensional database of mouse liver proteins. An update. Electrophoresis 22 9: 1747–1763. [DOI] [PubMed] [Google Scholar]
- 36. Fountoulakis M, Berndt P, Langen H, Suter L (2002) The rat liver mitochondrial proteins. Electrophoresis 23 2: 311–328. [DOI] [PubMed] [Google Scholar]
- 37. Doran P, Dowling P, Lohan J, McDonnell K, Poetsch S, et al. (2004) Subproteomics analysis of Ca2+-binding proteins demonstrates decreased calsequestrin expression in dystrophic mouse skeletal muscle. Eur J Biochem 271 19: 3943–3952. [DOI] [PubMed] [Google Scholar]
- 38. Kubota K, Kosaka T, Ichikawa K (2005) Combination of two-dimensional electrophoresis and shotgun peptide sequencing in comparative proteomics. J Chromatogr B Analyt Technol Biomed Life Sci 815 1–2: 3–9. [DOI] [PubMed] [Google Scholar]
- 39. Malmström J, Lee H, Aebersold R (2007) Advances in proteomic workflows for systems biology. Curr Opin Biotechnol 18 4: 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ferretti R, Marques MJ, Pertille A, Santo Neto H (2009) Sarcoplasmic-endoplasmic-reticulum Ca2+-ATPase and calsequestrin are overexpressed in spared intrinsic laryngeal muscles of dystrophin-deficient mdx mice. Muscle Nerve 39 5: 609–615. [DOI] [PubMed] [Google Scholar]
- 41. Dowling P, Doran P, Ohlendieck K (2004) Drastic reduction of sarcalumenin in Dp427-deficient fibres indicates that abnormal calcium handling plays a key role in muscular dystrophy. Biochem J 379: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rossi AE, Dirksen RT (2006) Sarcoplasmatic reticulum: the dynamic calcium governor of muscle. Muscle Nerve 33: 715–731. [DOI] [PubMed] [Google Scholar]
- 43. Porter JD, Merriam AP, Khanna S, Andrade AH, Richmonds CR, et al. (2003) Constitutive properties, not molecular adaptations, mediate extraocular muscle sparing in dystrophic mdx mice. FASEB J 17: 893–895. [DOI] [PubMed] [Google Scholar]
- 44. Rhee SG (2005) Chae HZ, Kim K (2005) Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic Biol Med 38 12: 1543–52. [DOI] [PubMed] [Google Scholar]
- 45. Craig EA, Gambill BD, Nelson RJ (1993) Heat-shock proteins: molecular chaperones of protein biogenesis. Mol Cell Biol 13: 5637–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Banfi G, Dolci A, Verna R, Corsi MM (2004) Exercise raises serum heat-shock protein 70 (Hsp70) levels. Clin Chem Lab Med 42: 1445–1446. [DOI] [PubMed] [Google Scholar]
- 47. Paepe BD, Creus KK, Weis J, Bleecker JL (2012) Heat shock protein families 70 and 90 in Duchenne muscular dystrophy and inflammatory myopathy: balancing muscle protection and destruction. Neuromuscul Disord 22 1: 26–33. [DOI] [PubMed] [Google Scholar]
- 48. Razzaque1 MS, Taguchi T (1999) The possible role of colligin/HSP47, a collagen-binding protein, in the pathogenesis of human and experimental fibrotic diseases. Histol Histopathol 14: 1199–1212. [DOI] [PubMed] [Google Scholar]
- 49. Gerke V, Creutz CE, Moss SE (2005) Annexins: linking Ca2+ signalling to membrane dynamics. Nat Rev Mol Cell Biol 6 6: 449–461. [DOI] [PubMed] [Google Scholar]
- 50. McNeil AK, Rescher U, Gerke V, McNeil PL (2006) Requirement for annexin A1 in plasma membrane repair. J Biol Chem 281 46: 35202–35207. [DOI] [PubMed] [Google Scholar]
- 51. Waddell LB, Lemckert FA, Zheng XF, Tran J, Evesson FJ, et al. (2011) Dysferlin, annexin A1, and mitsugumin 53 are upregulated in muscular dystrophy and localize to longitudinal tubules of the T-system with stretch. J Neuropathol Exp Neurol 70 4: 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gerke V, Moss S (2002) Annexins: from structure to function. Physiol Rev 82 2: 331–371. [DOI] [PubMed] [Google Scholar]
- 53. van Genderen HO, Kenis H, Hofstra L, Narula J, Reutelingsperger CP (2008) Extracellular annexin A5: functions of phosphatidylserine-binding and two-dimensional crystallization. Biochim Biophys Acta 1783 6: 953–963. [DOI] [PubMed] [Google Scholar]
- 54. Probst-Cousin S, Berghoff C, Neundörfer B, Heuss D (2004) Annexin expression in inflammatory myopathies. Muscle Nerve 30 1: 102–110. [DOI] [PubMed] [Google Scholar]
- 55. Flower RJ (1988) Lipocortin and the mechanisms of action of the glucocorticoids. Br J Pharmacol 94: 987–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goulding NJ, Guyre PM (1993) Glucocorticoids, lipocortins and the immune response. Curr Opin Immunol 5: 108–113. [DOI] [PubMed] [Google Scholar]
- 57. Parente L, Solito E (2004) Annexin 1: more than an anti-phospholipase protein. Inflamm Res 53: 125–132. [DOI] [PubMed] [Google Scholar]
- 58. Reutelingsperger CP, van Heerde WL (1997) Annexin V, the regulator of phosphatidylserine-catalyzed inflammation and coagulation during apoptosis. Cell Mol Life Sci 53: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buckland AG, Wilton DC (1998) Inhibition of human cytosolic phospholipase A2 by human annexin V. Biochem J 329: 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li Q, Qi B, Oka K, Shimakage M, Yoshioka N, et al. (2001) Link of a new type of apoptosis-inducing gene ASY/Nogo-B to human cancer. Oncogene 20: 3929–3936. [DOI] [PubMed] [Google Scholar]
- 61. Tagami S, Eguchi Y, Kinoshita M, Takeda M, Tsujimoto Y (2000) A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene 19: 5736–5746. [DOI] [PubMed] [Google Scholar]
- 62. Tidball JG, Albrecht DE, Lokensgard BE (1995) Spencer, M. J. Apoptosis precedes necrosis of dystrophin-deficient muscle. J Cell Sci 108: 2197–204. [DOI] [PubMed] [Google Scholar]
- 63. Matsuda RA, Nishikawa A, Tanaka H (1995) Vizualization of dystrophic muscle fibers in mdx mouse by vital staining with Evans blue: evidence of apoptosis in dystrophin-deficient muscle. J Bioch 118: 959–964. [DOI] [PubMed] [Google Scholar]
- 64. Sandri M, Podhorska-Okolow M, Geromel V, Rizzi C, Arslan P, et al. (1997) Exercise induces myonuclear ubiquitination and apoptosis in dystrophin-deficient muscle of mice. J Neuropathol Exp Neurol 56: 45–57. [DOI] [PubMed] [Google Scholar]
- 65. Tews DS, Goebel HH (1977) DNA-fragmentation and expression of apoptosis-related proteins in muscular dystrophies. Neuropathol Appl Neurobiol 23: 331–338. [PubMed] [Google Scholar]
- 66. Pereira JA, Taniguti AP, Matsumura C, Marques MJ, Neto HS (2012) Doxycycline ameliorates the dystrophic phenotype of skeletal and cardiac muscles in mdx mice. Muscle Nerve 46 3: 400–406. [DOI] [PubMed] [Google Scholar]
- 67. Abmayr S, Crawford RW, Chamberlain JS (2004) Characterization of ARC, apoptosis repressor interacting with CARD, in normal and dystrophin-deficient skeletal muscle. Hum Mol Genet 13 2: 213–221. [DOI] [PubMed] [Google Scholar]
- 68. Dominov JA, Kravetz AJ, Ardelt M, Kostek CA, Beermann ML, et al. (2005) Muscle-specific BCL2 expression ameliorates muscle disease in laminin α2-deficient, but not in dystrophin-deficient, mice. Hum Mol Genet 14 8: 1029–1040. [DOI] [PubMed] [Google Scholar]
- 69. Goldring K, Jones GE, Thiagarajah R, Watt DJ (2002) The effect of galectin-1 on the differentiation of fibroblasts and myoblasts in vitro. J Cell Sci 115 2: 355–366. [DOI] [PubMed] [Google Scholar]
- 70. Stowell SR, Arthur CM, Mehta P, Slanina KA, Blixt O, et al. (2008) Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J Biol Chem 283: 10109–10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nowak TP, Haywood PL, Barondes SH (1976) Developmentally regulated lectin in embryonic chick muscle and a myogenic cell line. Biochem Biophys Res Commun 68: 650–657. [DOI] [PubMed] [Google Scholar]
- 72. Barondes SH, Haywood-Reid PL (1981) Externalization of an endogenous chicken muscle lectin with in vivo development. J Cell Biol 91: 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cooper DN, Barondes SH (1990) Evidence for export of a muscle lectin from cytosol to extracellular matrix and for a novel secretory mechanism. J Cell Biol 110 5: 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Harrison FL, Wilson TJ (1992) The 14 kDa beta-galactoside binding lectin in myoblast and myotube cultures: Localization by confocal microscopy. J Cell Sci 101: 635–646. [DOI] [PubMed] [Google Scholar]
- 75. Poirier F, Timmons PM, Chan CT, Guénet JL, Rigby PW (1992) Expression of the L14 lectin during mouse embryogenesis suggests multiple roles during pre- and post-implantation development. Development 115: 143–155. [DOI] [PubMed] [Google Scholar]
- 76. Watt DJ, Jones GE, Goldring K (2004) The involvement of galectin-1 in skeletal muscle determination, differentiation and regeneration. Glycoconj J 19: 615–619. [DOI] [PubMed] [Google Scholar]
- 77. Rabinovich GA, Sotomayor CE, Riera CM, Bianco I, Correa SG (2000) Evidence of a role for galectin-1 in acute inflammation. Eur J Immunol 30: 1331–1339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins identified in extraocular and diaphragm muscles of control and mdx mice in three biological replicates.
(XLS)