Abstract
The evolution of biological signalling systems and apparently altruistic or cooperative traits in diverse organisms has required selection against the subversive tendencies of self-interested biological entities. The bacterial signalling and response system known as quorum sensing or Acylated Homoserine Lactone (AHL) mediated gene expression is thought to have evolved through kin selection. In this in vitro study on the model quorum sensing bioluminescent marine symbiont Vibrio fischeri, competition and long-term sub culturing experiments suggest that selection for AHL synthesis (encoded by the AHL synthase gene luxI) is independent of the quorum sensing regulated phenotype (bioluminescence encoded by luxCDABE). Whilst results support the hypothesis that signal response (AHL binding and transcriptional activation encoded by the luxR gene) is maintained through indirect fitness benefits (kin selection), signal synthesis is maintained in the V. fischeri genome over evolutionary time through direct fitness benefits at the individual level from an unknown function.
Introduction
The behaviour of biological systems requiring forms of signalling and cooperation between individual partner organisms have long garnered interest from evolutionary biologists [1]. The interest generally stems from the challenge of explaining how cooperative traits resist corruption by the self-interest of partner individuals. So-called ‘cheating’ has received much attention in the scientific literature in diverse organisms including microbes and several possible strategies for microbial cheater control have been postulated [2]–[4].
Bacterial quorum sensing is a term used to describe the release of signalling molecules by bacterial cells into the extracellular environment and a response to the accumulation of the signalling molecule through changes in transcription and hence phenotype [5]. The phenotypes regulated by quorum sensing are typically ecologically relevant only when expressed in concert by large populations of cells [6], [7]. In this way both the concerted production of signalling molecules and the orchestrated expression of its regulated phenotypes are distinct cooperative traits that have survived the subversive tendencies of self-interested individuals over evolutionary time.
N-acyl-L-homoserine lactone (AHL) mediated gene expression is one such bacterial quorum sensing system that has received an enormous amount of attention primarily owing to the direct role of many of the regulated phenotypes to human and plant infection [5]. AHL mediated gene expression involves the production of AHLs by an AHL synthase enzyme most commonly encoded by a homologue of the luxI gene of the marine symbiont Vibrio fischeri. AHLs typically diffuse through cell membranes and increase in concentration in the local extracellular environment if there are many producing cells in one location [8] and there are barriers to diffusion preventing AHL loss from that location [9]. Once a threshold intracellular concentration is reached, AHLs interact directly with transcriptional regulators encoded by homologues of the luxR gene of V. fischeri resulting in a transcriptional response. In the case of V. fischeri, in which AHL mediated gene expression was discovered, the transcriptional response encodes for the production of light that forms the basis of symbiotic interactions with marine fish and the bobtail squid Euprymna scolopes [10], [11].
In recent years, experiments have been conducted to investigate the vulnerability of the two distinct cooperative activities constituting quorum sensing: 1) the production of a communal stimulatory pool of AHLs and 2) the orchestrated expression of a communally beneficial phenotype [4]. Specifically, signal synthesis and response mutants were cultured in vitro in media requiring the communally beneficial phenotype (elastase production) for growth. The results suggested that exploitative individuals of the opportunistic human pathogen Pseudomonas aeruginosa can avoid both the cost of contributing to the communal AHL pool or the cost of contributing to the communally beneficial phenotype. It was concluded that kin selection is responsible for maintenance of genes encoding AHL production and response.
Additionally, there has been recent interest in the evolution of the V. fischeri lux system. Bose and colleagues have shown in vitro that bioluminescence represents a cost [12] and that V. fischeri strains isolated from different environments have diverse bioluminescence output [13]. The variations in the light output were attributed to rapid evolution in the intergenic sequences between the divergently transcribed luxR and luxI genes controlling their transcription rather than mutations in the quorum sensing genes themselves.
In related work, Schuster and colleagues [14] developed an exquisite experimental evolution model in which the luminescent output of free-living (strain WH1) or fish associated (strain MJ11) V. fischeri isolates decreased over approximately 300 generations in association with E. scolopes. Whilst no sequencing was conducted to link the changes in light output to genetic mutation, an AHL bioassay and responses to exogenous AHL addition revealed, for the most part, that changes were not derived from changes in AHL synthesis or response [14]. The evolution of the bioluminescent phenotype in V. fischeri in the absence of selection for it in vitro has not been investigated.
Differences in the specific hypotheses tested and approaches used in studies on the evolution of AHL mediated gene expression in P. aeruginosa and V. fischeri make it difficult to assess whether observations made are universal across AHL signalling systems or if there are fundamental differences driving their evolution. In this study a combination of in vitro approaches were used to investigate the stability of AHL mediated gene expression in V. fischeri, including competition and long-term subculturing experiments in the absence of selection for the regulated phenotype. Evidence is presented supporting the hypothesis that AHL synthesis is anchored in bacterial genomes through a fitness benefit to individual cells independent of group behaviour.
Methods
Batch culture growth of V. fischeri MJ1 wild type and MJ211 luxI– mutant in isolation and in co-culture
To compare growth of V. fischeri MJ1 [15] and the MJ1 derived luxI knockout mutant V. fischeri MJ211 [16] in isolation, batch cultures were established in triplicate from overnight precultures grown in 100 ml of LB20 medium (1% w/v tryptone, 0.5% w/v yeast extract and 2% NaCl w/v) in a 250 ml Erlenmeyer flask at 30°C and 150 rpm from an initial optical density of 0.01 at 600 nm. Experiments were conducted in the presence and absence of 5 µM N-3-oxohexanoyl-L-homoserine lactone in ethanol purchased from the laboratory of Paul Williams (Nottingham, UK). The unpaired t-test was performed on triplicate readings from all time points to identify statistically significant differences (p<0.05). Triplicate sets of optical density readings from wild type and mutant cultures for each time point were regarded as independent and assumed to be identically normally distributed.
Competition between the wild type and mutant strain was assessed in triplicate 5 ml aliquots in 20 ml plastic screw-capped vials in sequential batch co-culture with starting wild type to mutant ratios of 1∶1 or 1∶10 based on optical density with a starting density of 0.01. Every 24 h over a ten-day period 100 µl of stationary phase culture was transferred to 5 ml of fresh LB20 and the cell concentration of each competitor quantified through plating and counting of bioluminescent (MJ1) and non-bioluminescent (MJ211) colonies. The experimental design is illustrated in Figure 1.
Figure 1. Experimental design for competition experiments.
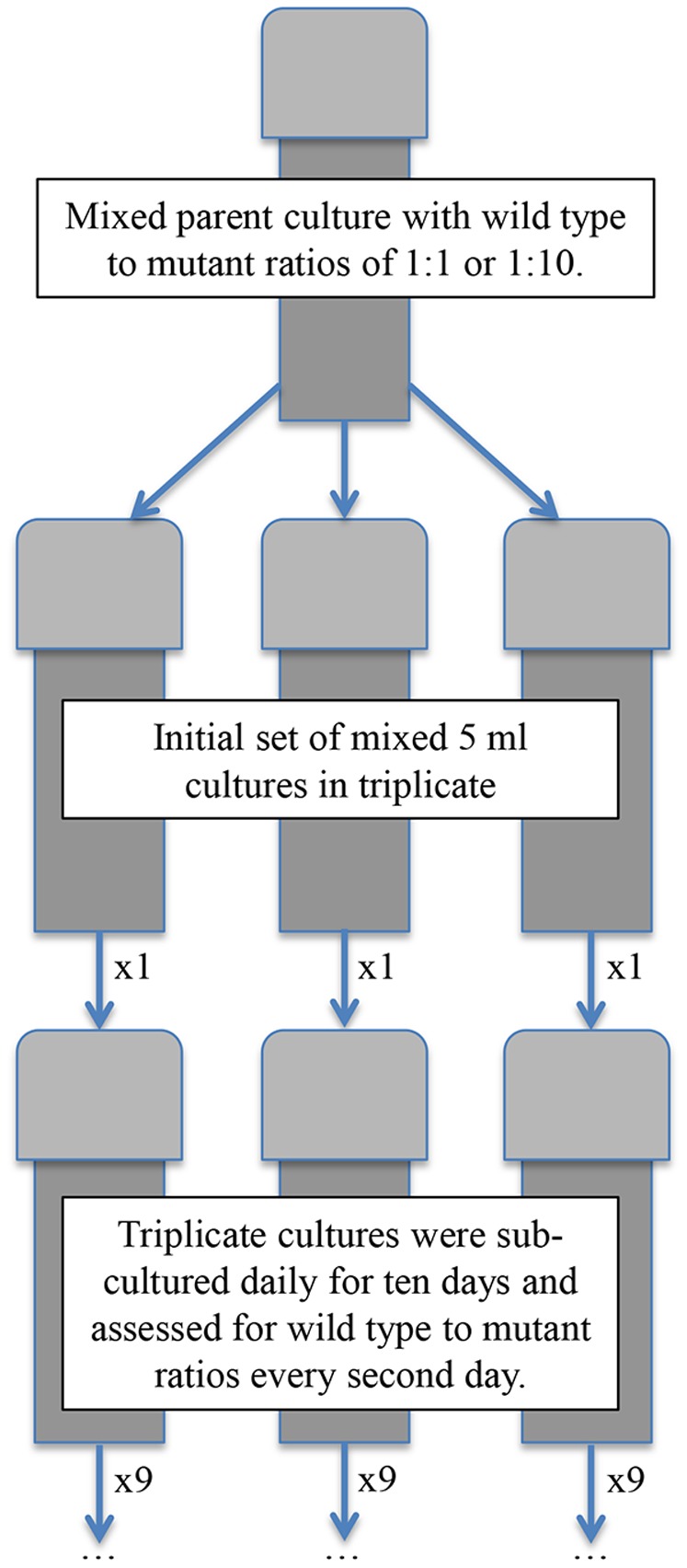
Mixtures of Vibrio fischeri wild type (MJ1) and luxI mutant (MJ211) strains at optical density based starting ratios of 1∶1 or 1∶10 were inoculated (100 µl) into 5 ml of fresh media in triplicate. Each replicate culture was subsequently subcultured daily for ten days and wild type to mutant ratios determined every second day.
In vitro evolution of bioluminescence in V. fischeri MJ1
From a fresh LB20 agar plate inoculated from a glycerol freezer stock maintained in the Centre for Marine BioInnovation (University of NSW, Sydney, Australia) ten colonies of wild type V. fischeri strain MJ1 were used to inoculate 5 ml of AB(VH) medium in 20 ml plastic screw-capped vials and cultured for 24 h shaking at 30°C and 150 rpm. An aliquot (100 µl) of each of the ten stationary phase cultures was then transferred to 5 ml of fresh AB(VH) medium and incubated as above for another 24 h. This sub-culturing routine was carried out every 24 h for 325 days. AB(VH) medium consisted of per liter: 0.3 M NaCl, 0.05 M MgSO4.7H2O, 2 g/L casamino acids (0.2%), 10 mM KH2PO4, 1 mM L-arginine, 2% v/v glycerol, 10 ng/ml riboflavin and 1 µg/ml thiamine [17]. Every 10 – 20 days ten 200 µl aliquots were taken from each of the ten cultures at the end of the logarithmic phase of growth (approximately 15 h into the batch culturing cycle, OD600 = 0.18) and bioluminescence was quantified using a microtiter plate luminometer (Wallac Victor2).
Sequencing dark cultures
Dark V. fischeri cultures were plated out and at least five colonies were randomly picked for sequencing of luxR, luxI, ainR, ainS genes and the luxR-luxI intergenic region using primer sets listed in Table 1. Primers for genes were designed using the V. fischeri ES114 genome to cover the entire coding region. The PCR conditions used for the amplification of genes were an initial denaturation at 95°C for 10 min, followed by 25 cycles of 95°C for 15 s, 56°C for 30 s, and 68°C for 2 min, and a final extension cycle of 5 min at 68°C. PCR products were cleaned using the QIAquick® PCR Purification Kit (Qiagen Pty. Ltd., Australia) and quantified using a Nanodrop spectrophotometer.
Table 1. Primers used for sequencing V. fischeri bioluminescence regulatory genes.
| Target | Primer | Sequence (5′- 3′) |
| luxR | Lux8Fa | TTCCTGGTTCAGAGCCTCAT |
| Lux1445Ra | GCACTCTGTTGACCAAGCAA | |
| luxI | Lux1220Fa | GCAATTCCATCGGAGGAGTA |
| Lux2019Ra | CACTTTTCCATCGTTGACCA | |
| ainR | Ain1064Fb | TGGCTCTTCTTTGACGGTTT |
| AinSRb | AGCTTAAAGAAATTAATGCTCGTCAG | |
| ainS | Ain20Fb | CACGACGAGAACCAAGACCT |
| Ain1243Rb | AACGAATTGCTTCGCATACC | |
| luxR-luxI | EVS109c | CCGCCCTAGGTTATTCAGATAAGCATTGATTAATATC |
| intergenic region | EVS110c | CCGCCCTAGGGCATGCTTAACCTCTATACTCCTCCGATGGAA |
Primers and base numbering based on luxR and luxI (Genbank AF170104.1), bPrimers and numbering based on ainR and ainS (Genbank L37404), c [38].
A sequencing reaction was set up for one of the primers which contained at least 100 ng of PCR products, 1 µl BigDye Terminator 3.1 (Applied Biosystems, United Kingdom), 10 pmol of primer, 3.5 µl 5x buffer (Applied Biosystems, United Kingdom) and molecular grade H2O (Eppendorf, Australia) to a total of 20 µl. The conditions used for the sequencing reaction were an initial step at 96°C for 1 min, followed by 25 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. The sequencing reactions were cleaned up by ethanol precipitation and sequenced with an ABI Prism BigDye kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and an
ABI model 310 genetic analyser (Perkin-Elmer Applied Biosystems) at the Ramaciotti Centre at the University of NSW, Sydney, Australia.
In vitro evolution of bioluminescence in V. fischeri MJ1 with luxR harboured on a plasmid
To assess the evolution of the V. fischeri MJ1 bioluminescence phenotype in the presence of multiple copies of the luxR gene a multi-copy plasmid was constructed harbouring the functional luxR gene from V. fischeri MJ1 and transferred into a dark V. fischeri MJ1 lineage (Lineage 10). The wild-type luxR gene was amplified using pJBA132 (pME6031-luxR-PluxI-RBSII-gfp(ASV)-T0-T1; Tcr; [18]) as the template and primers LuxR-SacI (5′-TTGCCGAGCTC TTTTGCCCAACAGAAAAAGC-3′) and LuxR-KpnI (TTTGCCGGTACC CTCCCTTGCGTTTATTCGAC) incorporating two restriction enzyme sites (underlined) flanking the 1.1 kb PCR product including the luxR promoter region. A 5 kb SacI-KpnI fragment from pLS6 (Cmr, lacZ expression vector derived from pSUP102; [19]) containing a chloramphenicol resistant gene was ligated to the SacI-KpnI digested amplified PCR fragment (1.1 kb) containing the luxR gene. The ligated construct (pLS6luxR) was then chemically transformed into competent Escherichia coli SM10 cells (Thi, thr, leu, tonA, lacy, supE, resA::RP4-2-Tcr::Mu, Kmr; [20]), and used as the donor for conjugal transfer of the construct into recipient V. fischeri MJ1 Lineage 10 (luxR –) cells via filter mating [21]. V. fischeri MJ1 Lineage 10 cells harbouring pLS6LuxR were selected on LB20 plates containing chloramphenicol (2 µg/ml). The presence of the plasmid was confirmed in bioluminescent V. fischeri colonies by plasmid extraction, restriction digestion and gel electrophoresis using standard protocols.
To assess the loss of the bioluminescent phenotype, V. fischeri MJ1 Lineage 10 (pLS6LuxR) colonies were subcultured in 5 ml AB(VH) media every 24 h for 150 days and monitored for changes in bioluminescence as above. As an additional assessment for dark mutants (including luxI mutants) appropriate dilutions of culture were plated out on LB20 (Cm) agar plates every 14 days and screened for the presence of non-bioluminescent colonies.
Results
Growth curve comparisons between Vibrio fischeri strain MJ1 and the AHL synthase mutant strain MJ211
It is generally believed that N-acyl-L-homoserine lactones (AHLs) are secondary metabolites that by definition play no role in cell-structure syntheses and energy transduction in the producer and are therefore not essential for growth and reproductive metabolism [22]. We began our investigation seeking to confirm that deletion of the AHL synthase gene luxI did not compromise the ability of V. fischeri to reproduce by comparing growth of the wild type (strain MJ1) and its AHL synthase mutant (strain MJ211) in batch aerobic cultures in the presence and absence of the cognate AHL N-3-oxohexanoyl-L-homoserine lactone (OHHL) at 5 µM.
Figure 2 illustrates the average increase in optical density of triplicate batch cultures over time. This data reveals that the AHL synthase mutant has very similar growth characteristics to the wild type in batch culture. Small differences were however observed, with the AHL synthase mutant strain generating slightly lower optical densities than the wild type (Fig. 2A). Between 10 and 16 hours in the absence of exogenously added OHHL these differences were statistically significant (Unpaired T test, p <0.05). Statistically significant differences were also observed in the presence of OHHL, however these were observed earlier in the incubation at 4 and 8 hours (Unpaired T test, p <0.05; Fig. 2B). In conclusion, the luxI gene is clearly not essential for the growth of V. fischeri under these conditions but subtle differences in optical density were noted suggesting the luxI gene could confer a detectable fitness advantage.
Figure 2. Impact of luxI deletion on growth of Vibrio fischeri in isolation.
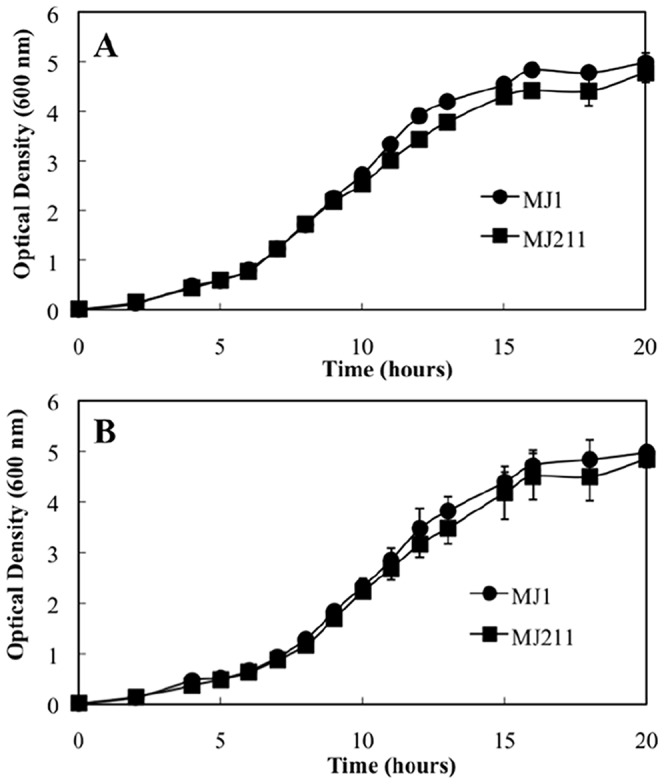
Batch culture growth of V. fischeri wild type strain MJ1 (circles) and luxI deficient mutant strain MJ211 (squares) in the absence (A) and presence (B) of 5 µM N-3-oxohexanoyl-L-homoserine lactone. Values presented are averages of triplicate cultures. Error bars represent standard deviation. Small statistically significant differences were late in the logarithmic phase of growth.
Growth competition between Vibrio fischeri strain MJ1 and the AHL synthase mutant strain MJ211
To further investigate the subtle differences observed in the growth characteristics of wild type V. fischeri and the AHL synthase mutant strain, both were subject to direct competition for resources by co-inoculation into triplicate aerobic batch cultures that were subcultured daily for 10 days. Each strain was quantified every second day by plate counting with the mutant strain distinguished from the wild type based on the luminescence of colonies. Note that the mutant V. fischeri strain MJ211 in co-culture will be exposed to the same extracellular concentration of AHLs as the AHL producing wild type strain MJ1. Hence it is assumed that in the same vessel both strains transcribe the luxCDABEG operon encoding luminescence to the same degree with comparable associated impacts on fitness.
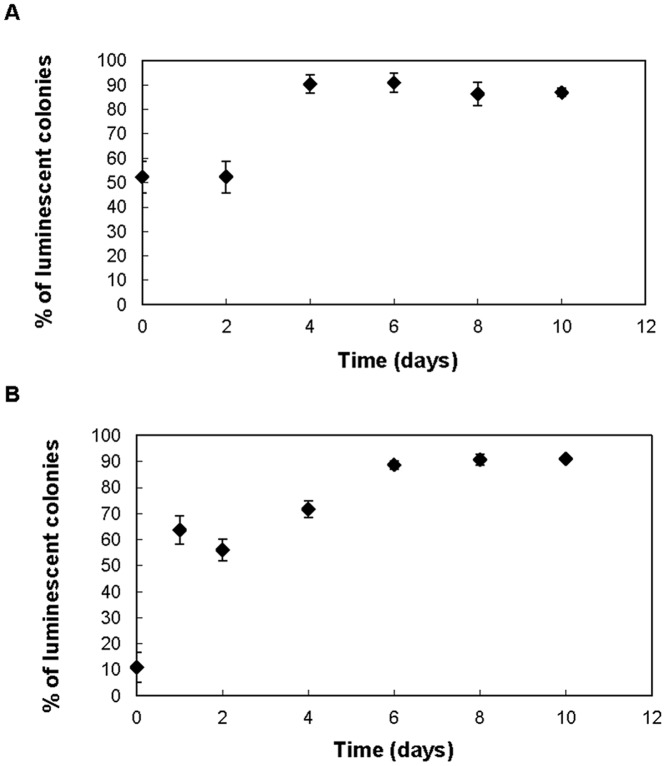
Figure 3 reveals that the luminescent wild type strain grew to dominate all cultures during the course of the experiment irrespective of the starting population ratios. From a starting ratio of 1∶1 it took four days for the ratio to reach 9∶1 (wild type:mutant). From a starting ratio of 1∶10 it took six days for the ratio to reach 9:1 (wild type:mutant). It is recognised that scoring dark colonies as a means of quantifying the luxI mutant MJ211 may include spontaneous mutants of the wild type in genes encoding bioluminescence (eg. luxR, luxICDABEG), although the likelihood is low given the short sub-culturing period. In effect this would overestimate the MJ211 counts and suggest the MJ211 population decline is even more rapid.
Figure 3. Impact of luxI deletion on competition in Vibrio fischeri.
Percentage of luminescence colonies plated from mixed V. fischeri MJ1 (wild type) and MJ211 (luxI mutant) cultures subcultured daily for ten days. The starting ratio of MJ1 to MJ211 was either 1∶1 (Panel A) or 1∶9 (Panel B). Regardless of the initial ratio of wild type (MJ1) to mutant (MJ211) cells, the wild type lineage dominated the cultures within days suggesting the luxI gene represents a selective advantage independent of the bioluminescence phenotype. Average values from triplicate cultures are presented. Error bars represent standard deviation.
In conclusion, in culturing conditions under which there is no known selection for the AHL regulated bioluminescence phenotype, the wild type V. fischeri strain has a clear fitness advantage over the mutant. This result is in conflict with the suggestion that AHL production per se represents a net metabolic burden to producing cells. Further, this result suggests the activity of the luxI gene product confers a fitness advantage independent of the AHL regulated phenotype.
Long term evolution of the quorum sensing system in V. fischeri MJ1
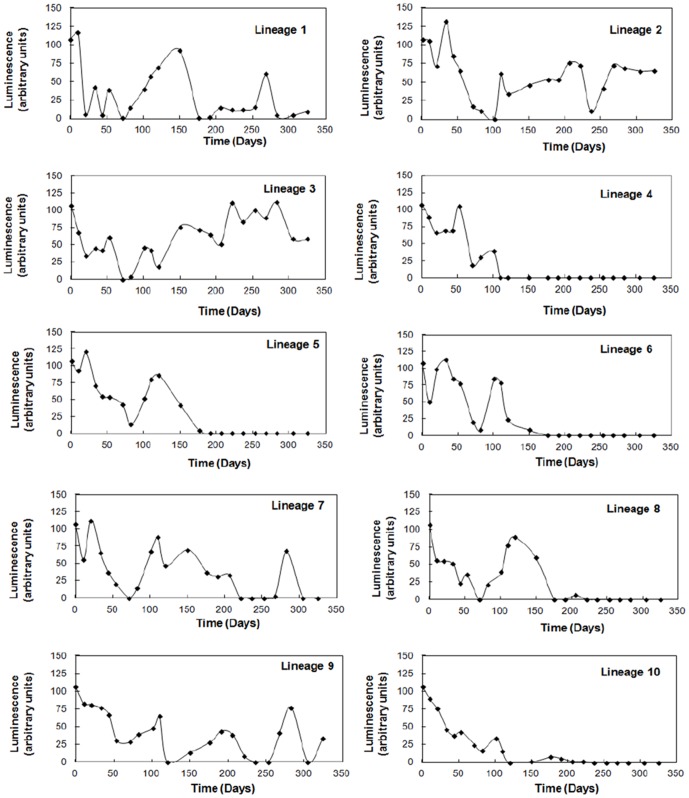
To explore the fate of the quorum sensing system in V. fischeri over thousands of generations, strain MJ1 was inoculated into ten identical aerobic batch cultures that were subsequently subcultured daily for 325 days (approximately 4000–5000 generations). In this experiment the cost of signal synthesis and response could be compared without interference from gene maintenance derived from selection for the regulated phenotype. The bioluminescent output of each culture was monitored every 10–20 days in the late-logarithmic phase of growth throughout the course of the experiment. Figure 4 illustrates the luminescence of each culture in relative light units and reveals fluctuations over several orders of magnitude in light output over time with striking differences observed between lineages. All cultures displayed decreases and increases in luminescence output, whilst five lineages (Lineages 4, 5, 6, 8 and 10) permanently lost the luminescent phenotype during the course of the experiment.
Figure 4. Fate of the bioluminescence phenotype during long term culturing in Vibrio fischeri.
Relative bioluminescence output from ten V. fischeri MJ1 cultures, subcultured daily over 325 days. Large fluctuations are observed in bioluminescence in all cultures. Half of the cultures (4, 5, 6, 8 and 10) irreversibly lost the bioluminescence phenotype during the course of the experiment. Quorum sensing regulatory genes (ainS, ainR, luxI and luxR) and the luxI-luxR intergenic region were sequenced to investigate the cause of the loss of phenotype.
Observations on individual colonies derived from the cultures throughout the experiment revealed diversity in luminescence output (bright, dim and dark) in cultures showing variable luminescence over time. In contrast, colonies derived from lineages 4, 5, 6, 8 and 10 after they had permanently lost the luminescence phenotype were all dark suggesting dark mutants had taken over due to major fitness benefits. This expected result confimed that under the prevailing culture conditions the luminescent phenotype confers a net disadvantage to V. fischeri cells.
To assess whether the loss of luminescence was the result of mutations in genes encoding AHL synthesis or response, the luxR, luxI, ainS and ainR genes were sequenced along with the luxI-luxR intergenic region from at least five colonies in the 250–300 day period for each of the ten lineages. It was observed that four out of the five permanently dark lineages had base pair substitutions or deletions in the luxR gene. Lineage 5 had no mutations in any of the regions sequenced.
Lineage 4 had a four base pair deletion (ATTG) at the end of the sequence (bp 727–730 from the start of the coding sequence or 239–242 in Genbank AF170104.1) removing residue 243 (isoleucine) and creating a frameshift in the remaining 23 bp of the coding region. In a site directed mutagenesis study, Trott and Stevens [23] revealed that residue 243 was crucial for LuxR binding to DNA and activation of the lux operon, thus explaining the origin of the dark phenotype in this lineage.
Lineage 6 had a base pair substitution (GGA > AGA at bp 400 from the start of the coding sequence or bp 569 in Genbank AF170104.1) predicted to result in a change in the protein sequence at residue 134 from glycine (aliphatic) to arginine (basic). This residue forms part of the five-stranded anti-parallel ß5 sheet of the AHL binding cavity [24]. Whilst it is not a conserved residue amongst LuxR homologues, it sits in a cluster of physiologically conserved amino acids [24]. It is therefore likely that this mutation from an aliphatic to basic residue has resulted in loss of AHL binding activity.
Lineage 8 had a base pair substitution (TGC > TAC at bp 584 from the start of the coding sequence or bp 385 in Genbank AF170104.1) predicted to result in a change in the protein sequence at residue 195 from cysteine (sulfur containing) to tyrosine (aromatic). Residue 195 sits in the helix-turn-helix region of LuxR known for DNA binding [24]. This mutation therefore likely impacts on lux gene transcription by interfering with LuxR binding to the lux box.
Lineage 10 had undergone a base pair substitution (ATT > AGT at bp 728 from the start of the coding sequence or bp 241 in Genbank AF170104.1) predicted to result in a change in the protein sequence at residue 243 from isoleucine (aliphatic) to serine (hydroxyl containing). This is the same residue lost in Lineage 4 previously shown to be crucial for LuxR activity [23].
No mutations were observed for the luxI, ainS and ainR genes in all lineages and in contrast to the observations of Bose and colleagues [13] no mutations were observed in the intergenic region encoding the promoter regions for luxI and luxR. This result suggests there was selection against the action of the luxR gene product under the growth conditions tested. The same cannot be said of the luxI gene, adding further weight to the suggestion that expression of the luxI gene and activity of its product represents an unknown selective advantage to the cell.
Evolution of bioluminescence with luxR carried on a multiple copy number plasmid
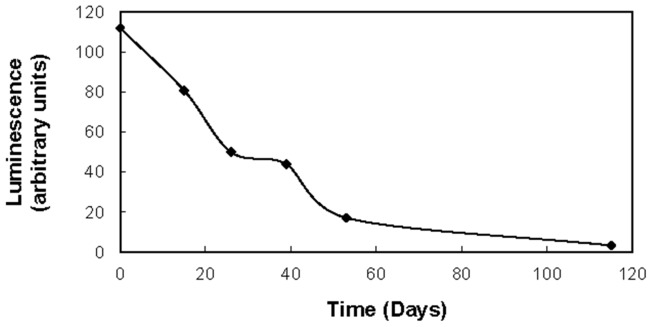
It was reasoned that the absence of luxI mutants in the ten V. fischeri lineages may have resulted from competition from luxR mutants. Cells not responding to the signal would be expected to have a greater selective advantage than cells not producing the signal but still encoding a functional response. To address this, a plasmid (pLS6LuxR) with a functional luxR gene was inserted into a single clone from the permanently dark V. fischeri lineage 10 (luxR mutant) and subcultured daily for 150 days. The culture was monitored for luminescence over time. Figure 5 reveals that the light output decreased gradually over time. Individual cells were assessed for loss of luxI activity resulting in a completely dark phenotype by plating cells and scoring luminescence of the resulting colonies. No dark mutants were observed during the course of the experiment indicating the absence of luxI mutants. The cause of the gradual decline in the luminescence output of the culture is unknown. The fact that all colonies on plates displayed some level of luminescence supports the assertion that the loss of luxR is the primary mechanism by which V. fischeri cells opt out of expression of the cooperative bioluminescence trait.
Figure 5. Impact of luxR complementation on bioluminescene during long term culturing of Vibrio fischeri.
Relative bioluminescence output from the dark V. fischeri MJ1 Lineage 10 with a luxR mutation complimented with a functional luxR gene under control of its own promoter on pLS6luxR. Complementation with luxR initially restored the bioluminescence phenotype to wild type levels, however bioluminescence was observed to decrease during subsequent subculturing for 120 days.
Discussion
Over the past 15 years significant attention has been given to the individual conflicts underlying the cooperative behaviours observed in AHL mediated gene expression in bacteria. This includes both the generation of a communal AHL pool requisite for expression of a trait for which there are ecological benefits only if it is performed en mass and the communal expression of that trait. In this short history, bacterial quorum sensing has provided insight into the evolution of signalling and other cooperative biological phenomena.
Initially, Brookfield [25] and Brown and Johnstone [26] developed bacterial population genetics models of quorum sensing evolution revealing parameter space in which the cooperative traits of signal production and orchestrated phenotype expression were stable despite the presence of signal production or response deficient mutants. It was reasoned that kin selection or indirect fitness benefits were responsible, given the high level of relatedness between clones in a bacterial population. In support of the assumption of existence of signalling deficient mutants in these theoretical works, empirical studies have been published describing the isolation of P. aeruginosa strains with mutations in their quorum sensing machinery, predominantly in lasR [27], [28]. Additionally, studies have been published exploring the fitness costs of signal production and functional gene expression and demonstrating conditions under which the proliferation of quorum sensing cheats is fostered [3], [4], [29]. In this study we have shown that in the absence of selection for the regulated phenotype, wild type V. fischeri cells have a higher reproductive fitness than cells lacking the luxI gene and that luxI mutants do not proliferate over thousands of generations in the presence or absence of functional luxR genes, while luxR mutants in contrast commonly arise. This suggests that the activity of the luxI gene represents a direct fitness benefit selected for independently of the bioluminescent phenotype in this quorum sensing model organism.
Diggle and colleagues [4] rightly pointed out that multiple functions of signalling molecules could alter the relative cost of AHL production, thereby providing an alternative explanation to kin selection for the maintenance of AHL production over evolutionary time. Interestingly, the three examples given to support the point (antibiotic, siderophore and immune modulatory activity) are all cooperative traits conferring indirect fitness benefits, requiring a dense population of cells for the selective advantage of the phenotype to be manifested. In all the literature relating quorum sensing to group selection no mention has been made of the possibility that AHL synthase activity represents a direct fitness benefit, ie. a selective advantage to individual V. fischeri cells independent of the indirect fitness benefits derived from cooperative group behaviour (bioluminescence phenotype).
The experiments conducted in this study do not shed light on what the selective advantage of AHL synthesis to individual V. fischeri cells could be although it is clear that it does not relate to activation of gene transcription through interaction with LuxR. An emerging hypothesis explaining the advantage of AHL synthesis stems from observed links between AHL regulated phenotypes and maintenance of redox homeostasis in the bacterial cytoplasm of V. fischeri and Pseudomonas aeruginosa [30]–[33].
The luxI gene encodes a 193 amino acid protein responsible for conversion of S-adenosylmethionine and hexanoyl-acyl carrier protein to N-3-oxohexanoyl-L-homoserine lactone, 5′-adenosylmethionine and apo-Acyl carrier protein [34]. Because N-3-oxohexanoyl- L-homoserine lactone can diffuse through cell membranes, its synthesis represents a path for excess reducing equivalents to leave the cytoplasm.
Traditional batch culturing conditions using rich media formulations, including those used in this study, are a good example where reduced organic carbon is supplied in excess and oxygen (electron sink) rapidly becomes limiting as cell densities increase. Biofilms are another example in which cells are exposed to excess reducing power (polysaccharides, proteins, glycoproteins, glycolipids and extracellular DNA) in extracellular polymeric matrices [35] with constricted access to electron acceptors based on consumption and diffusion limitations [36], [37]. Future experiments will address the hypothesis that AHL production and release represents a means of removing excess reducing power from the cytoplasm.
In summary, this study has shown that in the absence of selection for the bioluminescence phenotype V. fischeri MJ1 wild type cells outcompete V. fischeri MJ211 luxI deficient cells and that luxI mutations do not arise during subculturing over thousands of generations in the presence or absence of functional LuxR. The appearance of luxR mutants in our experiments is congruent with the hypothesis that this gene is maintained in nature through kin selection. In contrast, these results lend credence to the suggestion that luxI is maintained in the V. fischeri genome, not by kin selection, but by an unknown function of relevance to individual cells.
Funding Statement
This work was partially supported by grants from the Centre for Marine Bio-Innovation, the Environmental Biotechnology Cooperative Research Centre and an Australian Research Council Future Fellowship (FT100100078) awarded to MM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Smith JM, Szathmary E (1995) The Major Transitions in Evolution. Oxford, UK: Oxford University Press.
- 2. Travisano M, Velicer GJ (2004) Strategies of microbial cheater control. Trends Microbiol 12: 72–78. [DOI] [PubMed] [Google Scholar]
- 3. Sandoz KM, Mitzimberg SM, Schuster M (2007) Social cheating in Pseudomonas aeruginosa quorum sensing. Proc Natl Acad Sci U S A 104: 15876–15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Diggle SP, Griffin AS, Campbell GS, West SA (2007) Cooperation and conflict in quorum-sensing bacterial populations. Nature 450: 411–414. [DOI] [PubMed] [Google Scholar]
- 5. Whitehead NA, Barnard AM, Slater H, Simpson NJ, Salmond GP (2001) Quorum-sensing in Gram-negative bacteria. FEMS Microbiol Rev 25: 365–404. [DOI] [PubMed] [Google Scholar]
- 6. Manefield M, Whiteley AS (2007) Acylated homoserine lactones in the environment: chameleons of bioactivity. Philos Trans R Soc Lond B Biol Sci 362: 1235–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darch SE, West SA, Winzer K, Diggle SP (2012) Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc Natl Acad Sci U S A 109: 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fuqua WC, Winans SC, Greenberg EP (1994) Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol 176: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Redfield RJ (2002) Is quorum sensing a side effect of diffusion sensing? Trends Microbiol 10: 365–370. [DOI] [PubMed] [Google Scholar]
- 10.Stabb EV, Schaefer AL, Bose JL, Ruby EG (2008) Quorum signaling and symbiosis in the marine luminous bacterium Vibrio fischeri. In: Winans SC, Bassler BL, editors. Chemical communication among microbes. Washington, DC: ASM Press. pp. 233–250.
- 11. Miyashiro T, Ruby EG (2012) Shedding light on bioluminescence regulation in Vibrio fischeri . Mol Microbiol 84: 795–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bose JL, Rosenberg CS, Stabb EV (2008) Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch Microbiol 190: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bose JL, Wollenberg MS, Colton DM, Mandel MJ, Septer AN, et al. (2011) Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Applied and Environmental Microbiology 77: 2445–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuster B, Perry L, Cooper V, Whistler C (2010) Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis 51: 85–96. [Google Scholar]
- 15. Ruby EG, Nealson KH (1976) Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica; a model of symbiosis based on bacterial studies. Biol Bull 151: 574–586. [DOI] [PubMed] [Google Scholar]
- 16. Kuo A, Blough NV, Dunlap PV (1994) Multiple N-acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri . Journal of Bacteriology 176: 7558–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Greenberg EP, Hastings JW, Ulitzur S (1979) Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Archives of Microbiology 120: 87–91. [Google Scholar]
- 18. Andersen JB, Heydorn A, Hentzer M, Eberl L, Geisenberger O, et al. (2001) gfp-based N-acyl homoserine-lactone sensor systems for detection of bacterial communication. Appl Environ Microbiol 67: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visick KL, Ruby EG (1997) New genetic tools for use in the marine bioluminescent bacterium Vibrio fischeri. In: Hastings JW, Kricka LJ, Stanley PE, editors. Bioluminescence and chemiluminescence. New York, N.Y.: John Wiley and Sons. pp. 119–122.
- 20. Simon R, Priefer U, Puhler A (1983) A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat Biotech 1: 784–791. [Google Scholar]
- 21. de Lorenzo V, Herrero M, Jakubzik U, Timmis KN (1990) Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol 172: 6568–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vining LC (1990) Functions of secondary metabolites. Annu Rev Microbiol 44: 395–427. [DOI] [PubMed] [Google Scholar]
- 23. Trott AE, Stevens AM (2001) Amino acid residues in LuxR critical for its mechanism of transcriptional activation during quorum sensing in Vibrio fischeri . J Bacteriol 183: 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nasser W, Reverchon S (2007) New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Analytical and Bioanalytical Chemistry 387: 381–390. [DOI] [PubMed] [Google Scholar]
- 25. Brookfield JFY (1998) Quorum sensing and group selection. Evolution 52: 1263–1269. [DOI] [PubMed] [Google Scholar]
- 26. Brown SP, Johnstone RA (2001) Cooperation in the dark: signalling and collective action in quorum-sensing bacteria. Proceedings of the Royal Society of London Series B: Biological Sciences 268: 961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heurlier K, Denervaud V, Haas D (2006) Impact of quorum sensing on fitness of Pseudomonas aeruginosa . Int J Med Microbiol 296: 93–102. [DOI] [PubMed] [Google Scholar]
- 28. Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, et al. (2006) Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103: 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rumbaugh KP, Trivedi U, Watters C, Burton-Chellew MN, Diggle SP, et al. (2012) Kin selection, quorum sensing and virulence in pathogenic bacteria. Proc Biol Sci 279: 3584–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, et al. (2007) Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Molecular Microbiology 65: 538–553. [DOI] [PubMed] [Google Scholar]
- 31. Whiteley M, Lee KM, Greenberg EP (1999) Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa . Proc Natl Acad Sci U S A 96: 13904–13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Price-Whelan A, Dietrich LEP, Newman DK (2007) Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. Journal of Bacteriology 189: 6372–6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Kern SE, Newman DK (2010) Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J Bacteriol 192: 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schaefer AL, Val DL, Hanzelka BL, Cronan JE Jr, Greenberg EP (1996) Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci U S A 93: 9505–9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flemming HC, Neu TR, Wozniak DJ (2007) The EPS matrix: the "House of biofilm cells". J Bacteriol 189: 7945–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM (1995) Microbial biofilms. Annu Rev Microbiol 49: 711–745. [DOI] [PubMed] [Google Scholar]
- 37. Stewart PS (2003) Diffusion in biofilms. J Bacteriol 185: 1485–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV (2006) New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72: 802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]