Abstract
Colonization and persistence in the human nasopharynx are prerequisites for Streptococcus pneumoniae disease and carriage acquisition, which normally occurs during early childhood. Animal models and in vitro studies (i.e. cell adhesion and cell cytotoxicity assays) have revealed a number of colonization and virulence factors, as well as regulators, implicated in nasopharyngeal colonization and pathogenesis. Expression of genes encoding these factors has never been studied in the human nasopharynx. Therefore, this study analyzed expression of S. pneumoniae virulence-related genes in human nasopharyngeal samples. Our experiments first demonstrate that a density of ≥104 CFU/ml of S. pneumoniae cells in the nasopharynx provides enough DNA and RNA to amplify the lytA gene by conventional PCR and to detect the lytA message, respectively. A panel of 21 primers that amplified S. pneumoniae sequences was designed, and their specificity for S. pneumoniae sequences was analyzed in silico and validated against 20 related strains inhabitants of the human upper respiratory tract. These primers were utilized in molecular reactions to find out that all samples contained the genes ply, pavA, lytC, lytA, comD, codY, and mgrA, whereas nanA, nanB, pspA, and rrgB were present in ∼91–98% of the samples. Gene expression studies of these 11 targets revealed that lytC, lytA, pavA and comD were the most highly expressed pneumococcal genes in the nasopharynx whereas the rest showed a moderate to low level of expression. This is the first study to evaluate expression of virulence- and, colonization-related genes in the nasopharynx of healthy children and establishes the foundation for future gene expression studies during human pneumococcal disease.
Introduction
Streptococcus pneumoniae (the pneumococcus) is a Gram positive bacterium that causes severe invasive infection such as pneumonia, septicemia, and meningitis especially in children, the elderly and immunocompromised patients [1]–[3]. It has been estimated that the pneumococcus is responsible for 14.5 million cases of disease worldwide and more than 800,000 deaths in children under five each year [4]. Colonization of the nasopharynx is a necessary step along the path to pneumococcal disease (PD) [5], [6]. Upon entering the nasopharynx, and during its residence there, the pneumococcus shares this anatomical and physiological niche with an array of other bacterial inhabitants [7], [8]. Once carriage is established in the nasopharynx, the pneumococcus can remain asymptomatic or migrate through the Eustachian tubes to cause otitis media, descend down the respiratory tract to cause pneumonia, or invade the bloodstream through the respiratory epithelium to cause bacteremia or meningitis [6], [9]. The mechanism(s) behind this migration, preceding disease, is not fully understood.
The prevalence of pneumococcal carriage increases in the first few years of life, peaking at approximately 50–70% in hosts 2–3 years of age and decreasing thereafter until stabilizing at 5–10% in hosts over 10 years of age in industrialized countries [5], [10]–[14]. Studies have reported carriage rates as high as 60% in adults from some developing countries [12], [15]. Carriage studies have classically utilized bacteriologic cultures, and more recently molecular detection using highly sensitive quantitative PCR (qPCR) reactions. These reactions target selected genes found in most screened S. pneumoniae isolates and genome-sequenced strains, (e.g. lytA, ply or cps4A) [16], [17].
To date, at least 93 capsular serotypes have been identified among S. pneumoniae strains [6], [18], [19]. Prevention of PD in children has been achieved by vaccination with pneumococcal conjugate vaccine (PCV), the basis for which is induction of a protective antibody response against the bacterial polysaccharide capsule [6], [20]. Although vaccination has been documented as effective for reducing PD mortality and burden, it seems clear that vaccines with greater coverage, based on proteins (non-capsular antigens) common to all serotypes, will be needed in the future [20].
The ideal protein antigen is one that is present on the cell surface, expressed during nasopharyngeal (NP) carriage and in all stages of the disease (e.g. in lungs during pneumonia) and highly conserved within all serotypes. Animal models of PD and in vitro cultures of human respiratory cells have allowed the identification of a number of factors implicated in colonization of the nasopharynx and in pathogenesis [3], [21]. These factors include the capsular polysaccharide, pneumococcal pneumolysin (Ply), adhesins, several proteins implicated in fratricide and regulators. Some of the best characterized candidates as proteinaceous components of new vaccine formulations are Ply [22], [23], pneumococcal surface protein A (PspA) [24], [25], pneumococcal surface protein C (PspC) [26] and pneumococcal surface antigen A (PsaA). There is a lack of information, however, to evaluate expression of these vaccine candidates or other pneumococcal proteins in the human nasopharynx during carriage, or in any other anatomic site during disease.
A few studies have evaluated the presence of some virulence determinants in pneumococcal strains isolated from disease cases or strains isolated form the nasopharynx of healthy children. For example, the gene pspA, which encodes PspA (a vaccine candidate), was detected in 99% of carriage strains and invasive strains isolated in Spain [27]. The rrgC gene (encoding a protein implicated in biogenesis of the pneumococcal pilus) was amplified by PCR in 21.4% of strains isolated from a Native American population [28] whereas another pilus-associated gene, rlrA, was detected in ∼27% of S. pneumoniae invasive strains isolated in Portugal [29]. Genes encoding colonization factors such as the neuraminidase genes, nanA and nanB, were detected in all carriage strains and 96% of invasive isolates, respectively [30]. Our recent studies also demonstrated that all S. pneumoniae whether isolated from healthy children or from invasive diseases, encode the gene luxS coding for the quorum sensing enzyme LuxS, a regulator of biofilms and nasopharyngeal carriage [31]–[33].
While studies of expression of pneumococcal genes have generally utilized different animal models of carriage and disease [33]–[35], there is virtually no information available to directly measure gene expression in the pneumococcus natural niche, the human nasopharynx. This is in part due to a lack of protocols to isolate high quality pneumococcal RNA from human samples. The potentially low yield of RNA also impairs the use of high-throughput technology for gene expression studies. In this work we developed protocols to analyze pneumococcal RNA that was purified from human NP samples and conducted a study that: 1) identified the bacterial load required to obtain enough pneumococcal nucleic acids for downstream molecular studies, 2) evaluated the prevalence of virulence determinants including genes encoding adhesins, toxins, and regulators implicated in virulence (directly in DNA purified from NP samples collected from healthy children [14]) and 3) investigated the expression of these genes encoding virulence- and colonization-related factors and regulators shown to be important for persistence and virulence.
Materials and Methods
Strains
Streptococci (normal flora strains) and other strains utilized in this study were previously characterized [36] and kindly provided by Dr. Lesley McGee from the Centers for Disease Control and Prevention (CDC). Strains included: S. infantis, S. oralis, S. anguinosus, S. intermedius, S. sobrinus, S. pseudopneumoniae, S. mitis, S. parasanguinis, S. australis, S. mutans, S. peroris, S. oligofermentans, S. intestinalis, S. vestibularis, S. cristatus, S. salivarius, S. gordonii, S. sanguinis, S. sinensis and Dolosigranulum pigrum. Reference, genome-sequenced, S. pneumoniae strain D39 [37] (GenBank accession # NC_008533) and TIGR4 [38] (GenBank accession # NZ_AAGY00000000) were utilized as controls throughout the study.
Nasopharyngeal samples
The NP samples utilized in this work were part of a study of S. pneumoniae colonization conducted in Peru [14]. Children enrolled in the mentioned study were aged 0–3 years of age; more details on the study population can be found in our recent publication [14]. Briefly, samples were collected using rayon swabs and immediately placed in 1 ml of transport medium [skim-milk, tryptone, glucose, and glycerol (STGG) [39] at 4°C and transported to a central laboratory usually within 4 h and then stored at −80°C. The density of S. pneumoniae (CFU/ml) in these NP samples had been previously investigated utilizing a molecular approach [14].
DNA extraction
Strains were grown overnight on blood agar plates, this culture was utilized to prepare a cell suspension in 200 µl of sterile DNA grade water. The suspension was added to 100 µl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 0.04 g/ml lysozyme and 75 U/ml of mutanolysin and then incubated for 1 h at 37°C in a water bath. To extract DNA from NP samples, 200 µl of STGG were added with the above mentioned buffer and incubated under the same conditions. DNA was extracted using the QIAamp DNA Mini protocol, following recommendations of the manufacturer. DNA was finally eluted in 100 µl, quantified using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and kept at −80°C until used.
Conventional PCR and qPCR reactions
Conventional PCR reactions were performed with genomic DNA (∼100 pg) or DNA extracted from NP samples (3 µl) as template, 1 µM of the indicated pair of primers (Table 1), 1× Taq master mix (New England Biolabs) and DNA grade water. All PCR reactions were run in a MyCycler™ Thermal Cycler System (Bio-Rad) under the following conditions: initial denaturing at 95°C for 5 min, followed by 35 cycles of 95°C for 20 s, 55°C for 30 s and 68°C for 1 min, and a final extension at 68°C for 10 min. PCR products were run in 2% agarose gels, stained with ethidium bromide and photographed using a ChemiDoc XRS gel documentation System (Bio-Rad). qPCR reactions were performed with IQ™ SYBR green super mix (BioRad), 300 nM of the indicated primers, and 3 µl of DNA template. Reactions were run in duplicate using a CFX96 Real-Time PCR Detection System (Bio-Rad) and the following conditions; 1 cycle at 55°C for 3 min, 1 cycle at 95°C for 2 min and 40 cycles of 95°C for 15 s, 55°C for 1 min and 72°C for 1 min. Melting curves were generated by a cycle of 95°C for 1 min, 65°C for 1 min and 80 cycles starting at 65°C with 0.5°C increments.
Table 1. Primers utilized in this study.
| Name | Target | Sequence (5′ to 3′) |
| JVS1L | lytA | AGTTTAAGCATGATATTGAGAAC |
| JVS2R | TTCGTTGAAATAGTACCACTTAT | |
| JVS5L | luxS | ACATCATCTCCAATTATGATATTC |
| JVS6R | GACATCTTCCCAAGTAGTAGTTTC | |
| JVS27L | cps4A | CGTCTAAGAGTCAGTCTTTCAATA |
| JVS28R | ATTGATATCCACTCCATAGAGATT | |
| JVS29L | nana | CAGTGATAGAAAAAGAAGATGTTG |
| JVS30R | ATTATTGTAAACTGCCATAGTGAA | |
| JVS31L | mgrA | ATCTGTATCGTCAAGAGTTGTTTA |
| JVS32R | TAAAACCTTTAGTTTAGGCTGATT | |
| JVS35L | 16S rRNA | AACCAAGTAACTTTGAAAGAAGAC |
| JVS36R | AAATTTAGAATCGTGGAATTTTT | |
| JVS53L | comD | AACAGTATGAGAGGGATAGAGGAC |
| JVS54R | GATAAAGGTAGTCCTCGTCAAAAT | |
| JVS55L | comC | ATGAAAAACACAGTTAAATTGGAA |
| JVS56R | TTGTAAAATAAAATCACGGAAGAA | |
| JVS57L | pspA | CATAGACTAGAACAAGAGCTCAAA |
| JVS58R | CTACATTATTGTTTTCTTCAGCAG | |
| JVS59L | Ply | TGAGACTAAGGTTACAGCTTACAG |
| JVS60R | CTAATTTTGACAGAGAGATTACGA | |
| JVS61L | nanB | AACTGTCCATATCTCCTATTTTTC |
| JVS62R | TATTTCTACACCTATCTCACCAGA | |
| JVS63L | lytC | GTCTAGGTTATAGCGGTAAAGAAG |
| JVS64R | GCTCTTATTTACATATTCCCAGTT | |
| JVS65L | pavA | CGATAAAAGCAGTCATAAAATCCT |
| JVS66R | AGGATTGAGAGATTCTGTACTTGG | |
| JVS67L | Iga | CGAATGGCACAAAGATTAAACA |
| JVS68R | TTCTTCCCTTGAAACTGCTCTC | |
| JVS69L | rrgB | CAAAACCACTTGATCCAACAGA |
| JVS70R | ATTACAAATTCTGCCCCAGCTA | |
| JVS73L | cbpA | GCTAATGTAGCGACTTCAGATCAA |
| JVS74R | AGCTTGGAAGAGTTTCTTCACCTA | |
| JVS75L | psaA | CCTGCTGAAAAGAAACTCATTGTA |
| JVS76R | AGGTCTTGATTTGTTCAGGAGTTC | |
| JVS77L | psrP | GCTGCTAGAACTCCAAGTAACACA |
| JVS78R | TCACAAGTTGGAAATACTTCTGGA | |
| JVS79L | codY | TATAACGCATAAAATAGCCAAGCA |
| JVS80R | ATTACATCAATTTTGAAACGCTCA | |
| JVS81L | cbpD | TCCTGTTGATTTAGAACCATTTGA |
| JVS82R | GAGGGAGTGACTTCTTCACAAAAT | |
| JVS83L | Eno | GACGGTACTCCTAACAAAGGTAAA |
| JVS84R | ATAGCTGTAAAGTGGGATTTCAAG | |
| 1406F* | rrn, intergenic spacer region | TGYACACACCGCCCGT |
| 23Sr* | GGGTTBCCCCATTCRG |
From ref (44).
RNA extraction, analysis of RNA preparation, and cDNA synthesis
Once thawed, NP samples (200 µl) had 1 volume of RNAprotect Bacteria® (Qiagen) added and were immediately centrifuged for 15 min at 15000×g in a refrigerated centrifuge (Eppendorf). Total RNA was then extracted from the pellet using the RNeasy Mini Kit (Qiagen) as outlined by the manufacturer and additionally treated with 2 U of DNaseI (Promega) essentially as previously described [31], [40]. Integrity of our RNA preparations [RNA integrity number (RIN)], and RNA concentration of samples, were obtained by using the RNA 6000 Nano kit or RNA 6000 Pico kit electrophoresis system and the 2100 Bioanalyzer (Agilent technologies). Total RNA (500 pg) was cDNA transcribed using the iScript cDNA synthesis kit (Bio-Rad) and following the manufacturer's instructions.
Quantitative RT-PCR (qRT-PCR)
Reactions were performed as described above except that in qRT-PCR reactions we utilized 2 µl of cDNA as template. For the relative quantification of mRNA molecules, purified genomic DNA from S. pneumoniae reference strain TIGR4 or D39 was serially diluted to prepare standards representing 2.14×101, 4.29×101, 4.29×102, 4.29×103, 4.29×104, or 4.29×105 genome copies. A standard curve was constructed and final copies of each gene target, and therefore mRNA copies, were calculated using the Bio-Rad CFX manager software.
RT-PCR reactions
Reactions were performed with 5 ng of DNaseI-treated RNA as template. Those reactions contained 500 nM of lytA primers, 1× PCR master mix (New England Biolabs), molecular grade water and 10 units of AMV retrotranscriptase (Promega). Control RT-PCR reactions were similarly performed, except for the omission of reverse transcriptase. Reactions conditions were the following: initial incubation at 42°C for 30 min and denaturation at 95°C for 5 min, followed by 35 cycles of 95°C for 15 s, 55°C for 30 s and 68°C for 1 min and a final extension at 68°C for 10 min. PCR products were run in 2% agarose gels and stained with SYBR® Safe DNA Gel Stain (Invitrogen).
Results
Detection of the lytA gene and its transcript in NP samples
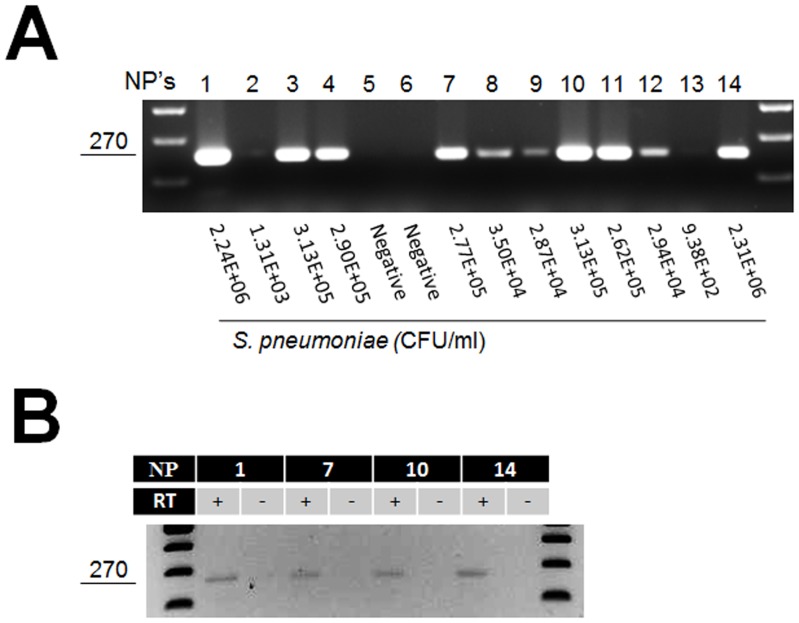
Since expression of S. pneumoniae genes had not been studied in human samples, we first sought to assess whether genes, and expression, could be amplified with conventional PCR and detected by RT-PCR, respectively. To this end, nuclei acids extracted from samples containing different S. pneumoniae loads (e.g. 102, 103, 104, 105, or 106 CFU/ml) previously quantified in our laboratory [14], were utilized. As shown in Fig. 1, only those NPs containing ≥104 CFU/ml allowed the PCR amplification of the lytA gene. In contrast, PCR products were absent when the template was DNA from either negative NP samples or from those containing <104 CFU/ml. Ten NP swabs containing each specific S. pneumoniae density, or negative samples, were further tested and results were similar to those presented in Fig. 1A. Then, RNA was extracted from NP samples and utilized as a template in RT-PCR reactions that allowed for the detection of a lytA message (Fig. 1B). These results indicated that S. pneumoniae DNA or RNA, contained in NP samples with ≥104 CFU/ml, was sufficient to detect by conventional PCR or RT-PCR pneumococcal genes and their transcripts, respectively.
Figure 1. PCR amplification of the lytA gene and RT-PCR detection of its transcript.
(A) DNA was extracted from NP samples and utilized as template in PCR reactions amplifying the lytA gene. The loads of pneumococcus cells in each NP sample is indicated below each lane. The larger the bacterial loads, greater the amount of DNA that should be obtained from each sample. This PCR reaction thus detected DNA from NP samples containing at least ∼2.8×104 CFU/ml (samples 8, 9 and 12). (B) RNA was extracted from the indicated NP sample and was utilized as template in RT-PCR reactions targeting the lytA gene. Reactions were added (+) or not (−) with retrotranscriptase (RT). In both panels, the size of the lytA product is shown at left in base pairs.
Development and validation of a panel of primers to amplify S. pneumoniae genes
The challenge to detect S. pneumoniae genes by PCR, or their transcripts by RT-PCR, in NP samples relies on the potential presence of homologous sequences in species sharing the human upper respiratory tract (including the nasopharynx) with the pneumococcus [41]. Therefore, we first designed and validated, a panel of 21 pair of primers (Table 1) to amplify genes encoding known virulence and colonization factors in the pneumococcus (i.e. ply, nanA, iga, etc.), adhesins (i.e. pavA, pspA, rrgB, etc.) or genes linked to regulatory functions (i.e. luxS, comC, mgrA and codY) [1], [9]. These primers were designed based on sequences available from reference S. pneumoniae strains D39 and TIGR4 [37], [42].
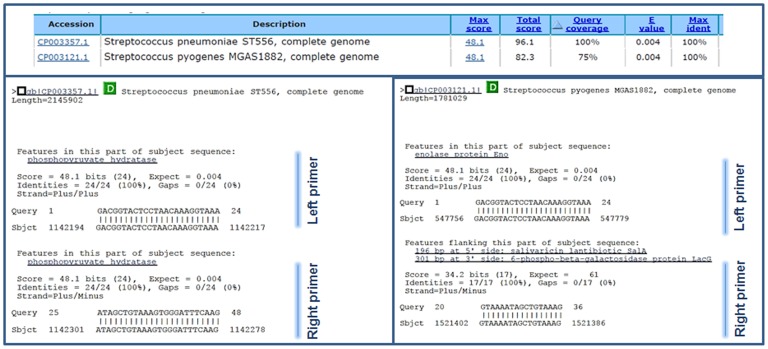
Bioinformatic analysis of both forward and reverse primer sequences revealed that, with some exceptions listed below, most of these Table 1 pair of primers would only hybridize S. pneumoniae sequences. Besides analyzing each primers individually, both forward (i.e. 22 bp) and reverse sequences (i.e. 22 bp) were also placed together (i.e. 44 bp) and then analyzed again using BLAST. The criteria to define in silico hybridization of these concatenated sequences included query coverage >70% and E-score <0.1. Genes/genomes meeting these criteria were individually analyzed to confirm that both primers (i.e. forward and reverse) hybridized within the same gene. For example, Fig. 2 shows that the gene encoding enolase (eno) had a perfect match with S. pneumoniae strain ST556 (query coverage of 100% and an E-score of 0.004). Both forward and reverse sequences were identified within the same (eno) gene (Fig. 2, left bottom panel). Conversely, query coverage of 78% with an E-score of 0.004, which would comply with our criteria for in silico hybridization, was detected in S. pyogenes strain MGAS1882. However, only 24 bp hybridized in a putative eno gene whereas another 17 bp fragment hybridized elsewhere in the genome (Fig. 2, right bottom panel). This in silico hybridization was, therefore, not compatible with a potential PCR product.
Figure 2. In silico analysis of the especificity of primers to amplify the eno gene.
Sequences of primers designed to ampify the S. pneumoniae eno gene were entered into the BLAST website. Among others, total query coverage of 100% and 75% was observed for S. pneumoniae strain ST556 or S. pyogenes MGAS1882, respectively. Left bottom panel shows that both the left and right primers in silico hybridyzed on the S. pneumoniae eno gene. Right bottom panel shows hybridization of only the left primer on the S. pyogenes eno gene. Part of the righ primer in silico hybridized somewhere else in the genome.
Overall, our bioinformatics studies revealed that primers amplifying the gene encoding the enolase (eno) will also hybridize in silico with S. sanguinis, S. parasanguinis, S. mitis, S. oralis and S. gordonii sequences, whereas primers amplifying iga (encoding the IgA protease) will also hybridize with the iga gene from S. oralis. Genes that were also present in other streptococci included the psaA gene found in S. mitis, S. sanguinis and S. oralis, the codY gene that in silico hybridized with S. pseudopneumoniae sequences and cbpD that hybridize with S. mitis and S. pseudopneumoniae sequences. These species are found in the healthy human nasopharynx (Table 2) [7], [41].
Table 2. PCR amplification of S. pneumoniae genes in related species.
| Strain (normal flora of) | Gene target | ||||||||
| lytA | 16S rRNA | eno | iga | psaA | luxS | cbpD | nanB | ||
| 1 | S. infantis (mouth and pharynx) | − | + | + | − | − | − | − | − |
| 2 | S. oralis (mouth) | − | + | + | + | − | − | − | − |
| 3 | S. anguinosus (mouth, pharynx and vagina) | − | + | + | − | − | − | − | − |
| 4 | S. intermedius (mouth, pharynx and intestines) | − | + | − | − | − | − | − | − |
| 5 | S. sobrinus (mouth) | − | + | + | − | − | − | − | − |
| 6 | S. pseudopneumoniae (mouth and pharynx) | − | + | + | − | + | + | + | − |
| 7 | S. mitis (mouth) | − | + | + | − | + | + | − | − |
| 8 | S. parasanguinis (throat) | − | + | + | − | − | − | − | − |
| 9 | Dolosigranulum pigrum (upper respiratory tract) | − | + | − | − | − | − | − | − |
| 10 | S. australis (mouth) | − | + | + | − | + | + | + | − |
| 11 | S. mutans (mouth, teeth) | − | + | + | − | − | − | − | − |
| 12 | S. peroris (teeth and pharynx) | − | + | + | − | + | − | − | − |
| 13 | S. oligofermentans (mouth) | − | + | + | − | + | − | − | − |
| 14 | S. intestinalis (intestines) | − | + | + | − | − | − | − | − |
| 15 | S. vestibularis (mouth) | − | + | − | − | − | − | − | − |
| 16 | S. cristatus (mouth) | − | + | + | − | − | − | − | − |
| 17 | S. salivarius (mouth) | − | + | + | + | − | − | − | − |
| 18 | S. gordonii (teeth, mouth) | − | + | + | − | − | − | − | − |
| 19 | S. sanguinis (mouth, dental plaque) | − | + | + | − | − | − | − | − |
| 20 | S. sinensis (mouth) | − | + | + | − | − | − | − | − |
| 21 | S. pneumoniae (mouth, pharynx, nose) | + | + | + | + | + | + | + | + |
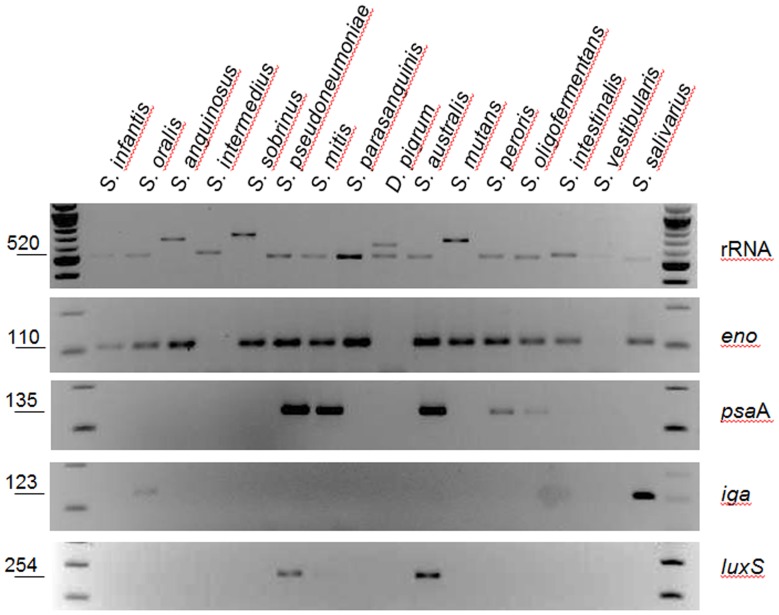
To further assess whether these primers would generate a PCR product we performed PCR reactions using DNA templates from a panel of 20 related species that inhabit the human upper respiratory tract [43]. As expected, the 16S rRNA gene was amplified in all species (Table 2). In contrast to bioinformatics studies that only detected hybridization in 5 species, our PCR studies amplified the eno gene in most of tested strains whereas the iga, psaA, cbpD and luxS genes were detected in only a few species (Fig. 3 and Table 2). The genes lytA, lytC, nanA, nanB, comC, comD, ply, pspA, psrP, codY, rrgB, pavA, cbpA (also known as pspC), cps4A and mgrA were only amplified in control reactions using DNA from S. pneumoniae strain D39 or TIGR4 (i.e. some genes are only encoded by either D39 or TIGR4).
Figure 3. Primers designed to amplify S. pneumoniae genes amplify genes in related species.
Purified DNA (100 pg) from the indicated species was utilized as template in PCR reactions. Specific genes amplified in those reactions are shown at right of each panel. The size of the expected product, in base pairs, is shown to the left.
Together, these results confirmed that 15 pair of primers were specific for S. pneumoniae encoded genes, and therefore could be utilized in reactions containing DNA or RNA purified from NP samples or other anatomic sites.
Detection of S. pneumoniae virulence determinants and regulators in nasopharyngeal samples
Evaluating the presence of genes in a strain isolated from the NP may not represent the whole population of pneumococci found in this anatomic site. This and our unpublished observations that NP samples with high pneumococcal density may contain more than one serotype prompted us to investigate the prevalence of these genes directly in NP samples (N = 50) containing ≥106 CFU/ml of S. pneumoniae load and negative samples (N = 22) [14]. DNA extracted from these samples was used as a template in PCR reactions. As expected, almost all reactions that utilized as template DNA from NPs negative to S. pneumoniae strains were also negative by PCR for S. pneumoniae encoded genes (Table 3). As an internal control that verified the presence of bacterial DNA, and the absence of inhibitors, reactions amplified the bacterial intergenic spacer region from the rrn operon [44] utilizing universal primers (not shown).
Table 3. Molecular detection of S. pneumoniae virulence-related factors and regulators in nasopharyngeal samples.
| Target gene | PCR (% positive) | qPCR (% positive) | |
| Virulence and colonization factors | Sp * negative NP | Sp positive NP | Sp positive NP |
| csp4A | 0 | 90.90 | 97.43 |
| ply | 0 | 90.90 | 100 |
| rrgB | 0 | 13.63 | 92.59 |
| nanA | ND** | ND | 97.43 |
| lytC | ND | ND | 100 |
| nanB | 0 | 81 | 91.89 |
| pspA | 0 | 13.63 | 95.45 |
| psrP | 0 | 42.72 | 45.71 |
| cbpA | 0 | 54.54 | 57.36 |
| pavA | 0 | 90.90 | 100 |
| lytA | 0 | 100 | 100 |
| Regulators | |||
| comD | ND | ND | 100 |
| comC | 0 | 63.63 | 71.05 |
| mgrA | ND | ND | 100 |
| codY | 0 | 86.36 | 100 |
Sp: S. pneumoniae.
ND: not done.
The most prevalent genes in positive NP samples by PCR were lytA, pavA, ply and cps4A (Table 3). An interesting observation was that the pspA gene, which has been described as being encoded by most S. pneumoniae strains [27], was only detected by PCR in ∼13% of the lytA positive NP samples. We hypothesized that the lesser efficiency of some PCR reactions lowered the limit of detection by conventional PCR. To verify this hypothesis, a more sensitive assay (qPCR) was utilized to reveal that ∼92% of samples were pspA positive. Similar findings were obtained when the rrgB gene was screened (Table 3). In summary, the lytA, lytC, pavA, ply, comD, mgrA and codY genes were detected in all samples, whereas the prevalence of other gene targets ranged from >90%<99% in csp4A, rrgB, pspA, nanA, and nanB, to >45%<72% in psrP, cbpA, and comC.
Gene expression of virulence determinants and regulators in the human nasopharynx
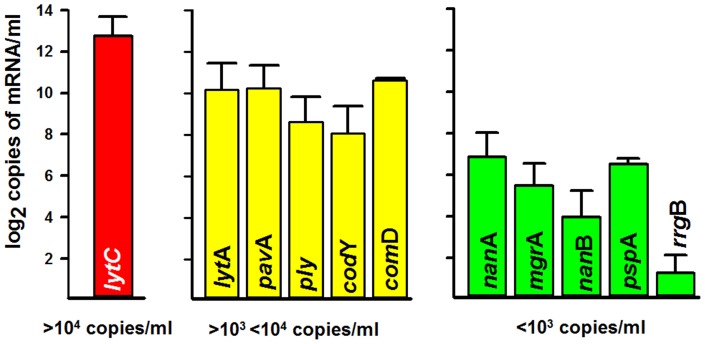
A set of 11 pair of primers, that amplify the most prevalent genes, were chosen to analyze mRNA expression levels in the human nasopharynx. Since not all NP samples contained the target genes, RNA was extracted from 30 samples and its quality and concentration were evaluated using the Agilent RNA 6000 Nano and Pico kits. After synthesizing cDNA, levels of messenger RNA (mRNA) of all screened genes were quantified using a qRT-PCR-based approach. Our studies identified differential gene expression whereby a scale including those showing, in terms of copies/ml of transcript, a low (<103 copies/ml), moderate (>103<104 copies/ml), or high (>104 copies/ml) level of expression could be established. For example, the gene coding for NanA [(nanA) a sialidase implicated in colonization [45], [46]] and the adhesin gene pspA were detected at low levels of expression (Fig. 4).
Figure 4. Gene expression studies.
RNA was obtained from a set of NP samples; cDNA was generated and utilized in quantitative PCR reactions targeting pneumococcal genes. The graphic was adjusted to show a log2 scale of copies of mRNA/ml of the indicated gene. A red bar represent more than 104 copies/ml, yellow bars correspond to those expressing >103<104 copies/ml whereas green bars represent genes whose transcripts were detected at <103 copies/ml. Error bars represent the standard error of the mean calculated using data from all tested NP samples.
The lytA gene, the gene encoding adhesin pavA, the pneumolysin gene (ply) and the comD gene (implicated in competence), showed moderate levels of expression, in comparison with those mentioned above (Fig. 4). In contrast, the lysozyme-encoding gene lytC showed the highest level of expression; most NP samples contained >104 copies/ml of the transcript.
Discussion
We have analyzed in this study, for the first time, expression of important pneumococcal genes when S. pneumoniae is present in the nasopharynx of healthy children. Our results clearly point towards active production of the autolysins LytC and LytA that can be implicated in either fratricide (killing of non-competent pneumococci or other species) or in biofilm formation. We have also demonstrated that transcripts for a vaccine candidate, Ply [22], [47] and an adhesin implicated in carriage and virulence, PavA [48], are detected in pneumoccocal cells residing in the human nasopharynx. We were unable, however, to evaluate mRNA levels of genes encoding other vaccine candidates such as PsaA due to the presence of similar sequences in other species found in the nasopharynx.
Even when we selected NP samples with the highest possible density of pneumococci, the concentration of RNA obtained from our preparations was usually <500 pg/µl. For research purposes, NP swabs collected to detect the pneumococcus are stored in 1 ml of transport medium (STGG) whereby less than 500 ng of bacterial RNA can be obtained with protocols described in this manuscript. As demonstrated, our RNA preparations had enough quality to generate cDNA and analyze expression of a set of selected genes by quantitative RT-PCR. The yield of RNA is, however, an important limitation to the use of bacterial RNA extracted from NP samples for gene expression studies utilizing high-throughput technology (i.e. microarrays, RNA sequencing).
This work provides a panel of primers that were validated against several related strains, most of which are present in the human respiratory tract. Although in silico studies were helpful in determining the specificity of primers designed to amplify pneumococcal genes, studies with DNA purified from those 20 related species revealed more nonspecific products which indicates the need for in vitro studies to validate in silico analyzes. These pneumococcus-specific primers can be utilized to evaluate gene expression in RNA purified from sterile sites (i.e. blood or cerebrospinal fluid) or non-sterile sites (middle ear fluid or bronchoalveolar lavage). For example, a recent study changed our view of the lung microbiome by demonstrating that, although detected with a low density, lungs contain a similar bacterial flora to those of the nasopharynx [49]. Expression of genes in patients experiencing pneumococcal otitis media may also be obtained and compared to those being expressed in the nasopharynx. This information may reveal significant changes in gene expression allowing the pneumococcus to colonize other anatomic sites and to cause disease. These studies are underway in our laboratory.
Studies utilizing conventional PCR and DNA from NP samples to detect pneumococcal genes, correlated with findings utilizing DNA extracted from the strains. For example, the lytA gene could be amplified by PCR in all samples whereas ∼90% of NP samples were positive for the ply gene. qPCR studies found, however, more positive samples for most of the target genes. The more striking results where those obtained with the pspA gene where our PCR studies detected ∼13% positive samples while quantitative PCR detected pspA in >90% of NP samples. This may suggest that a sub-population of pneumococci found in a lower density than that required to be detected by PCR (>104 CFU/ml), but detected by qPCR, could be present in some NP samples. This does not exclude the possibility that in samples, in which results from PCR and qPCR were positive, two populations with both low density and high density encoding the same gene could be present. This hypothesis is also supported by the fact that our NP samples contained a high density of pneumococci (>106 CFU/ml), thereby increasing the chance that those samples would contain more than one serotype or even the same serotype of a genetically different strain.
The gene lytC, which encodes a lysozyme named LytC [50], was detected in 100% of NP samples and was also the gene with the highest level of expression (>104 copies/ml) of the human nasopharynx. Our findings are consistent with a recent observation that a significant increase in anti-LytC antibodies occurs in healthy adult carriers of the pneumococcus in comparison to carriage negative adults [51]. Pneumococcal challenge also induced increased levels of anti-LytC IgG in serum from carriage negative adults [51].
In vitro studies have demonstrated that the mature form of LytC is anchored to the cell envelope [50], and it has an optimal enzymatic activity at ∼30°C, which mimics the temperature in the upper respiratory tract [52]. Recent discoveries have revealed that LytC is one of the most important proteins of the pneumococcal biofilm matrix [53]. LytC has also been implicated in fratricide (i.e. lysis of non-competent cells by competent ones) which has been proposed as a mechanism for predation that contributes to virulence by regulating the release of several virulence factors [54], [55]. Overall, our studies and those mentioned above suggest that LytC is important for the pneumococcus to persist in its human host; however whether LytC is also implicated in pneumococcal disease remains to be investigated.
Our studies also demonstrate a low level of expression of the pspA gene. Carriage studies in human volunteers inoculated with S. pneumoniae strains detected antibodies anti-PspA, indicating that PspA is produced during colonization and/or carriage [51], [56], [57]. This also may indicate that PspA is highly immunogenic since low expression in the human nasopharynx might be enough to stimulate a strong immune response. Another virulence factor that has been associated with antibody production during carriage in children and adults and is an important vaccine candidate is the pneumolysin Ply [51], [58], [59]. Although when purified, Ply may recapitulate lung damage induced by the pneumococcus [60], its role in NP carriage or during biofilm-related otitis media has not yet been fully characterized. Its level of expression in the nasopharynx correlates with a role of Ply in pneumococcal biofilm formation (Shak et al. 2013, unpublished) and production of anti-Ply antibodies in healthy carriers and those experiencing otitis media [58], [59], [61].
In summary, these studies have demonstrated levels of expression of important pneumococcal genes, including vaccine candidates, in the human nasopharynx and have established the basis for future gene expression studies during human pneumococcal disease.
Acknowledgments
We are grateful to Dr. Lesley McGee from CDC for providing all normal flora strains. The authors also thank Dr. Daiichi Morii, Magderie Klugman, Paulina Hawkins and Dr. Sopio Chochua for their invaluable assistance in some molecular reactions. We appreciate the help of Gideon Matzkin for critical reading of the manuscript.
Investigators Group: Vanderbilt University: Dr. Carlos G. Grijalva, Dr. Kathryn M. Edwards, Dr. John V. Williams, and Dr. Marie R. Griffin; and the Instituto de Investigación Nutricional, Lima, Peru: Hector Verastegui, Ana I. Gil, and Dr. Claudio F. Lanata.
Funding Statement
This work was supported by PHS Grant UL1 RR025008 from the Clinical and Translational Science Award program, NIH, National Center for Research Resources (JEV). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jedrzejas MJ (2001) Pneumococcal virulence factors: structure and function. Microbiol Mol Biol Rev 65: 187–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klugman KP, Madhi SA, Albrich WC (2008) Novel approaches to the identification of Streptococcus pneumoniae as the cause of community-acquired pneumonia. Clin Infect Dis 47 Suppl 3S202–206. [DOI] [PubMed] [Google Scholar]
- 3. van der Poll T, Opal SM (2009) Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374: 1543–1556. [DOI] [PubMed] [Google Scholar]
- 4. O'Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, et al. (2009) Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374: 893–902. [DOI] [PubMed] [Google Scholar]
- 5. Bogaert D, De Groot R, Hermans PW (2004) Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4: 144–154. [DOI] [PubMed] [Google Scholar]
- 6. Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, et al. (2012) The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 11: 841–855. [DOI] [PubMed] [Google Scholar]
- 7. Shak JR, Vidal JE, Klugman KP (2013) Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 21: 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dunne EM, Smith-Vaughan HC, Robins-Browne RM, Mulholland EK, Satzke C (2013) Nasopharyngeal microbial interactions in the era of pneumococcal conjugate vaccination. Vaccine 31: 2333–2342. [DOI] [PubMed] [Google Scholar]
- 9. Weiser JN (2010) The pneumococcus: why a commensal misbehaves. J Mol Med (Berl) 88: 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watson K, Carville K, Bowman J, Jacoby P, Riley TV, et al. (2006) Upper respiratory tract bacterial carriage in Aboriginal and non-Aboriginal children in a semi-arid area of Western Australia. Pediatr Infect Dis J 25: 782–790. [DOI] [PubMed] [Google Scholar]
- 11. Jounio U, Juvonen R, Bloigu A, Silvennoinen-Kassinen S, Kaijalainen T, et al. (2010) Pneumococcal carriage is more common in asthmatic than in non-asthmatic young men. Clin Respir J 4: 222–229. [DOI] [PubMed] [Google Scholar]
- 12. Adetifa IM, Antonio M, Okoromah CA, Ebruke C, Inem V, et al. (2012) Pre-vaccination nasopharyngeal pneumococcal carriage in a Nigerian population: epidemiology and population biology. PLoS One 7: e30548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hussain M, Melegaro A, Pebody RG, George R, Edmunds WJ, et al. (2005) A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol Infect 133: 891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, et al. (2013) Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children. Pediatr Infect Dis J 32: 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roca A, Hill PC, Townend J, Egere U, Antonio M, et al. (2011) Effects of community-wide vaccination with PCV-7 on pneumococcal nasopharyngeal carriage in the Gambia: a cluster-randomized trial. PLoS Med 8: e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park HK, Lee HJ, Kim W (2010) Real-time PCR assays for the detection and quantification of Streptococcus pneumoniae . FEMS Microbiol Lett 310: 48–53. [DOI] [PubMed] [Google Scholar]
- 17. Carvalho Mda G, Tondella ML, McCaustland K, Weidlich L, McGee L, et al. (2007) Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45: 2460–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Song JH, Dagan R, Klugman KP, Fritzell B (2012) The relationship between pneumococcal serotypes and antibiotic resistance. Vaccine 30: 2728–2737. [DOI] [PubMed] [Google Scholar]
- 19. Yother J (2011) Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65: 563–581. [DOI] [PubMed] [Google Scholar]
- 20. Rodgers GL, Klugman KP (2011) The future of pneumococcal disease prevention. Vaccine 29 Suppl 3C43–48. [DOI] [PubMed] [Google Scholar]
- 21. Kadioglu A, Weiser JN, Paton JC, Andrew PW (2008) The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6: 288–301. [DOI] [PubMed] [Google Scholar]
- 22. Salha D, Szeto J, Myers L, Claus C, Sheung A, et al. (2012) Neutralizing Antibodies Elicited by a Novel Detoxified Pneumolysin Derivative, PlyD1, Provide Protection against Both Pneumococcal Infection and Lung Injury. Infect Immun 80: 2212–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Denoel P, Philipp MT, Doyle L, Martin D, Carletti G, et al. (2011) A protein-based pneumococcal vaccine protects rhesus macaques from pneumonia after experimental infection with Streptococcus pneumoniae. Vaccine 29: 5495–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seo SU, Kim JJ, Yang H, Kwon HJ, Yang JY, et al. (2012) Effective protection against secondary pneumococcal pneumonia by oral vaccination with attenuated Salmonella delivering PspA antigen in mice. Vaccine 30: 6816–6823. [DOI] [PubMed] [Google Scholar]
- 25. Vadesilho CF, Ferreira DM, Moreno AT, Chavez-Olortegui C, Machado de Avila RA, et al. (2012) Characterization of the antibody response elicited by immunization with pneumococcal surface protein A (PspA) as recombinant protein or DNA vaccine and analysis of protection against an intranasal lethal challenge with Streptococcus pneumoniae . Microb Pathog 53: 243–249. [DOI] [PubMed] [Google Scholar]
- 26. Moreno AT, Oliveira ML, Ho PL, Vadesilho CF, Palma GM, et al. (2012) Cross-reactivity of antipneumococcal surface protein C (PspC) antibodies with different strains and evaluation of inhibition of human complement factor H and secretory IgA binding via PspC. Clin Vaccine Immunol 19: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rolo D, Ardanuy C, Fleites A, Martin R, Linares J (2009) Diversity of pneumococcal surface protein A (PspA) among prevalent clones in Spain. BMC Microbiol 9: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Basset A, Trzcinski K, Hermos C, O'Brien KL, Reid R, et al. (2007) Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J Clin Microbiol 45: 1684–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aguiar SI, Serrano I, Pinto FR, Melo-Cristino J, Ramirez M (2008) The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettigrew MM, Fennie KP, York MP, Daniels J, Ghaffar F (2006) Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect Immun 74: 3360–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP (2011) The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79: 4050–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vidal JE, Howery KE, Ludewick HP, Nava P, Klugman KP (2013) Quorum-sensing systems LuxS/autoinducer 2 and Com regulate Streptococcus pneumoniae biofilms in a bioreactor with living cultures of human respiratory cells. Infect Immun 81: 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Joyce EA, Kawale A, Censini S, Kim CC, Covacci A, et al. (2004) LuxS is required for persistent pneumococcal carriage and expression of virulence and biosynthesis genes. Infect Immun 72: 2964–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, et al. (2004) Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun 72: 5582–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, et al. (2006) Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol 61: 1196–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carvalho Mda G, Bigogo GM, Junghae M, Pimenta FC, Moura I, et al. (2012) Potential nonpneumococcal confounding of PCR-based determination of serotype in carriage. J Clin Microbiol 50: 3146–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, et al. (2007) Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 39. O'Brien KL, Bronsdon MA, Dagan R, Yagupsky P, Janco J, et al. (2001) Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 39: 1021–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vidal JE, Ohtani K, Shimizu T, McClane BA (2009) Contact with enterocyte-like Caco-2 cells induces rapid upregulation of toxin production by Clostridium perfringens type C isolates. Cell Microbiol 11: 1306–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, et al. (2011) Microbial communities of the upper respiratory tract and otitis media in children. MBio 2: e00245–00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, et al. (2001) Complete genome sequence of a virulent isolate of Streptococcus pneumoniae . Science 293: 498–506. [DOI] [PubMed] [Google Scholar]
- 43. Lemon KP, Klepac-Ceraj V, Schiffer HK, Brodie EL, Lynch SV, et al. (2010) Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio 1: e00129–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borneman J, Triplett EW (1997) Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol 63: 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, et al. (2009) Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis 199: 1497–1505. [DOI] [PubMed] [Google Scholar]
- 46. Parker D, Soong G, Planet P, Brower J, Ratner AJ, et al. (2009) The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun 77: 3722–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Douce G, Ross K, Cowan G, Ma J, Mitchell TJ (2010) Novel mucosal vaccines generated by genetic conjugation of heterologous proteins to pneumolysin (PLY) from Streptococcus pneumoniae . Vaccine 28: 3231–3237. [DOI] [PubMed] [Google Scholar]
- 48. Pracht D, Elm C, Gerber J, Bergmann S, Rohde M, et al. (2005) PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation. Infect Immun 73: 2680–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, et al. (2011) Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 184: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garcia P, Paz Gonzalez M, Garcia E, Garcia JL, Lopez R (1999) The molecular characterization of the first autolytic lysozyme of Streptococcus pneumoniae reveals evolutionary mobile domains. Mol Microbiol 33: 128–138. [DOI] [PubMed] [Google Scholar]
- 51. Ferreira DM, Neill DR, Bangert M, Gritzfeld JF, Green N, et al. (2013) Controlled Human Infection and Rechallenge with Streptococcus pneumoniae Reveals the Protective Efficacy of Carriage in Healthy Adults. Am J Respir Crit Care Med 187: 855–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sommer F, Kroger R, Lindemann J (2012) Numerical simulation of humidification and heating during inspiration within an adult nose. Rhinology 50: 157–164. [DOI] [PubMed] [Google Scholar]
- 53. Domenech M, Garcia E, Prieto A, Moscoso M (2013) Insight into the composition of the intercellular matrix of Streptococcus pneumoniae biofilms. Environ Microbiol 15: 502–516. [DOI] [PubMed] [Google Scholar]
- 54. Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Havarstein LS (2009) Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155: 2223–2234. [DOI] [PubMed] [Google Scholar]
- 55. Guiral S, Mitchell TJ, Martin B, Claverys JP (2005) Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A 102: 8710–8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wright AK, Ferreira DM, Gritzfeld JF, Wright AD, Armitage K, et al. (2012) Human nasal challenge with Streptococcus pneumoniae is immunising in the absence of carriage. PLoS Pathog 8: e1002622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McCool TL, Cate TR, Moy G, Weiser JN (2002) The immune response to pneumococcal proteins during experimental human carriage. J Exp Med 195: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simell B, Korkeila M, Pursiainen H, Kilpi TM, Kayhty H (2001) Pneumococcal carriage and otitis media induce salivary antibodies to pneumococcal surface adhesin a, pneumolysin, and pneumococcal surface protein a in children. J Infect Dis 183: 887–896. [DOI] [PubMed] [Google Scholar]
- 59. Rapola S, Jantti V, Haikala R, Syrjanen R, Carlone GM, et al. (2000) Natural development of antibodies to pneumococcal surface protein A, pneumococcal surface adhesin A, and pneumolysin in relation to pneumococcal carriage and acute otitis media. J Infect Dis 182: 1146–1152. [DOI] [PubMed] [Google Scholar]
- 60. Rubins JB, Duane PG, Charboneau D, Janoff EN (1992) Toxicity of pneumolysin to pulmonary endothelial cells in vitro. Infect Immun 60: 1740–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rapola S, Kilpi T, Lahdenkari M, Makela PH, Kayhty H (2001) Antibody response to the pneumococcal proteins pneumococcal surface adhesin A and pneumolysin in children with acute otitis media. Pediatr Infect Dis J 20: 482–487. [DOI] [PubMed] [Google Scholar]