Abstract
Despite the successful identification of several relevant genomic loci, the underlying molecular mechanisms of schizophrenia remain largely unclear. We developed a computational approach (NETBAG+) that allows an integrated analysis of diverse disease-related genetic data using a unified statistical framework. The application of this approach to schizophrenia-associated genetic variations, obtained using unbiased whole-genome methods, allowed us to identify several cohesive gene networks related to axon guidance, neuronal cell mobility, synaptic function and chromosomal remodeling. The genes forming the networks are highly expressed in the brain, with higher brain expression during prenatal development. The identified networks are functionally related to genes previously implicated in schizophrenia, autism and intellectual disability. A comparative analysis of copy number variants associated with autism and schizophrenia suggests that although the molecular networks implicated in these distinct disorders may be related, the mutations associated with each disease are likely to lead, at least on average, to different functional consequences.
A pressing challenge of human genetics is to combine diverse diseaserelated genetic variations to illuminate pathways and networks affected in common disorders. Schizophrenia represents an important example of a common psychiatric disorder in which a statistically significant contribution to disease susceptibility has now been demonstrated for different types of genetic variations. Specifically, several genomic loci associated with common human polymorphisms have been implicated by genome-wide association studies (GWAS)1–4, a contribution from de novo and rare copy number variants (CNVs) has been established5–7, and a significant contribution from de novo single nucleotide variants (SNVs) was demonstrated in a recent study based on exome sequencing in two populations8.
Biological networks provide a natural framework for integration of diverse genetic variations associated with such a complex and multifactorial phenotype as schizophrenia9,10. To identify affected molecular networks, we have developed an algorithm (NETBAG+) that searches for cohesive clusters of genes perturbed by disease-associated genetic variations (Fig. 1a). The approach is based on the previously described phenotype network11, which assigns every pair of human genes a score proportional to the likelihood ratio that these genes are involved in the same genetic phenotype (Online Methods). The phenotype network was used previously to identify a functionally cohesive gene cluster perturbed by de novo CNVs in autism11. The new NETBAG+ approach is able to integrate data from multiple types of genetic variation: SNVs, CNVs and GWAS-implicated loci. The greedy search algorithm identifies highly connected gene clusters that are affected by genetic variations, and the significance of the identified clusters is then established using an appropriate randomization (Online Methods). Although we and others have previously developed several methods to identify and analyze disease-related gene networks11–15, to our knowledge NETBAG+ is the first principled approach for integration of diverse sources of genome-wide genetic variation under a unified framework. The statistical power of this integrative approach stems from the convergence of different types of genetic variations on a set of interrelated molecular processes.
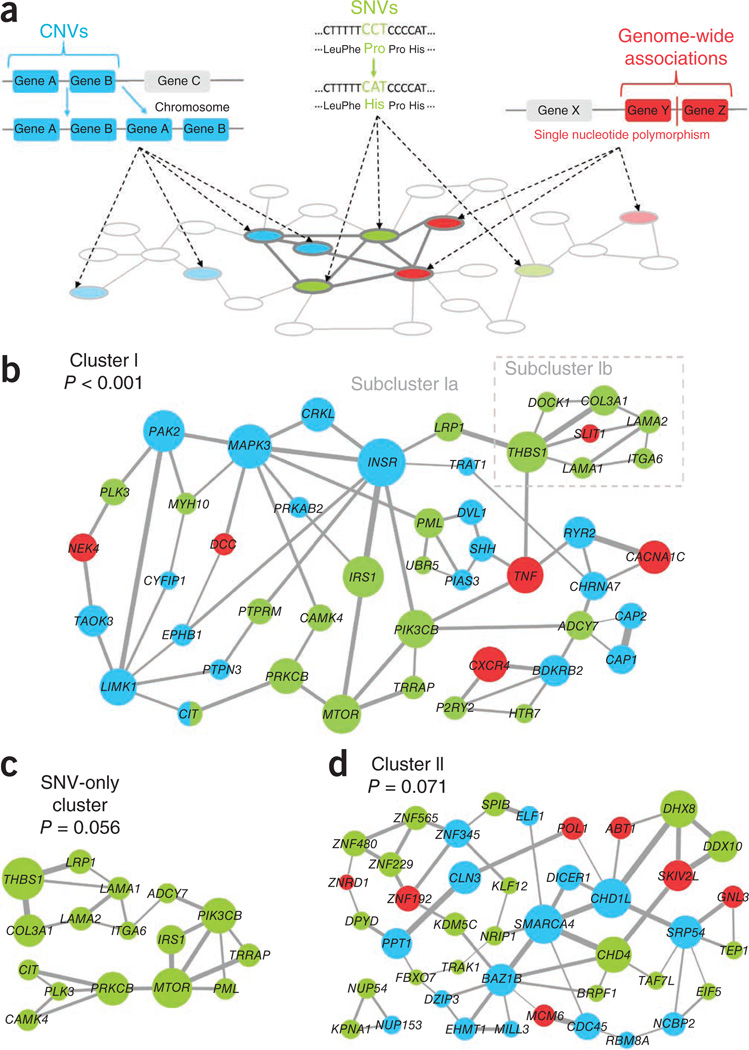
Figure 1.
The NETBAG+ approach and the identified schizophrenia gene clusters. (a) The NETBAG+ algorithm: different types of genetic variations are mapped to a phenotype network (pale gray) in which every pair of genes is assigned a score proportional to the likelihood ratio that those genes share a genetic phenotype. Strongly interconnected clusters (dark gray) are identified among disease-associated genes. Cluster scores are based on the weighted sum of edges between all genes in the cluster; this score is proportional to the likelihood that all cluster genes share the same phenotype. Cluster significance is then established by an appropriate randomization (Online Methods). (b) Cluster results from the combined set of schizophrenia-associated genetic variations: genes from de novo CNVs are in blue, genes from non-synonymous de novo SNVs are in light green and genes from GWAS-implicated regions in dark red. Edge widths are proportional to the strength of the likelihood score between the two genes, and node sizes are proportional to the gene’s contribution to the overall cluster score (Online Methods). For simplicity, only the strongest two edges from each gene are shown. Cluster I was the best cluster from the combined set of all schizophrenia genetic variations (P < 0.001). (c) The best cluster found when using only genes affected by non-synonymous de novo SNVs (P = 0.056). (d) Cluster II, the best cluster from the combined set of all schizophrenia genetic variations when the genes forming cluster I were removed from the input data (P = 0.071).
Here we applied the NETBAG+ algorithm to integrate several unbiased whole-genome data sets associated with schizophrenia. We identified several cohesive gene networks related to the disorder and characterized their biological and cellular functions. We also investigated the expression of the network genes in the brain. Finally, we examined the relationship between the genes forming the identified schizophrenia networks and genes associated with other neurodevelopmental disorders, such as autism and intellectual disability.
RESULTS
Gene clusters affected by schizophrenia-associated variations
We considered non-synonymous de novo SNVs from recent studies8,16, de novo CNVs from published genome-wide scans7,17–23 and genomic regions implicated by GWAS1–4,24–28. In total, this set contained 1,044 genes (159 from non-synonymous de novo SNVs, 712 from de novo CNVs, 173 from GWAS) from 213 genomic locations. In searching for cohesive gene clusters, the algorithm was allowed to pick any gene affected by a de novo SNV, any gene in a de novo CNV (one gene per CNV) or any gene in a GWAS-implicated region (one gene per region).
On the basis of the aforementioned input data, NETBAG+ identified a significant gene cluster (P < 0.001) containing in total 47 genes (22 from SNVs, 20 from CNVs, 6 from GWAS regions) (Fig. 1b). The identified cluster contained two weakly connected subclusters (subcluster Ia and subcluster Ib). In addition to combining all genetic data (SNVs, CNVs and GWAS regions), we also performed NETBAG+ searches using different combinations of genetic variations as the algorithm input (Supplementary Table 1). For example, we obtained a marginally significant (P = 0.056) cluster using only de novo SNVs (Fig. 1c); all genes in this cluster were also members of the cluster obtained using the combined data (cluster I). The highest significance was achieved when all types of genetic variations were considered together (Supplementary Table 1). Thus, different sources of genetic variations appear to reinforce each other, increasing the overall cluster significance. After masking the genes forming cluster I—that is, removing these genes from the input data—the NETBAG+ algorithm was able to identify another marginally significant cluster, cluster II (Fig. 1d, P = 0.071). Notably, cluster I and cluster II included three of the four genes (LAMA2, TRRAP, DPYD) with recurrent non-synonymous SNVs in the cohort analyzed in a recent study8 (Fisher’s exact test, one-tailed, P = 0.05), supporting the NETBAG+ clustering results and also providing more evidence that these genes are involved in schizophrenia pathophysiology.
In contrast to the results for non-synonymous SNVs and CNVs from schizophrenia patients, we detected no significant clusters in various control sets (Supplementary Table 1). For example, there were no significant clusters identified when searching genes affected by de novo non-synonymous SNVs observed in a control population8, synonymous de novo SNVs observed in schizophrenia patients8, or non-synonymous de novo SNVs observed in unaffected siblings of autism patients in two recently published studies29,30. Furthermore, we identified no significant clusters when the aforementioned sets were combined with de novo CNVs seen in unaffected siblings of autism patients in another recent study31 (Online Methods).
Biological processes associated with schizophrenia clusters
To determine functions of genes forming the identified schizophrenia clusters, we used two computational tools (FuncAssociate32 and DAVID33) that identify over-represented Gene Ontology (GO) terms in a given gene set. These analyses showed that the genes in cluster I participate in several important neurodevelopmental processes, such as axon guidance, neuron projection development, and cell migration and locomotion (Table 1 and Supplementary Tables 2 and 3). The GO analysis also implicated several cellular pathways, including signaling through essential second messengers: calcium, cyclic AMP and inositol trisphosphate. Separate analysis of genes forming subclusters Ia and Ib (Supplementary Tables 2 and 3) showed that the former was enriched for gene functions related to signaling and axon guidance, the latter for functions related to neuron mobility and locomotion.
Table 1.
GO terms associated with cluster I
| N | X | Padj | GO identifier | GO term |
|---|---|---|---|---|
| FuncAssociate | ||||
| 16 | 326 | <0.001 | GO:0007411 | Axon guidance |
| 11 | 335 | <0.001 | GO:0040012 | Regulation of locomotion |
| 7 | 108 | <0.001 | GO:0000187 | Activation of MAPK activity |
| 8 | 193 | <0.001 | GO:0001666 | Response to hypoxia |
| 9 | 295 | <0.001 | GO:0030334 | Regulation of cell migration |
| 9 | 333 | <0.001 | GO:0051960 | Regulation of nervous system development |
| 8 | 289 | 0.001 | GO:0019932 | Second-messenger-mediated signaling |
| 6 | 132 | 0.001 | GO:0008286 | Insulin receptor signaling pathway |
| 8 | 307 | 0.001 | GO:0050767 | Regulation of neurogenesis |
| 7 | 227 | 0.001 | GO:0071375 | Cellular response to peptide hormone stimulus |
| 6 | 155 | 0.001 | GO:0010975 | Regulation of neuron projection development |
| 7 | 253 | 0.002 | GO:0045664 | Regulation of neuron differentiation |
| 3 | 16 | 0.015 | GO:0035004 | Phosphatidylinositol 3-kinase activity |
| 4 | 54 | 0.018 | GO:0051896 | Regulation of protein kinase B signaling cascade |
| 5 | 119 | 0.021 | GO:0007204 | Elevation of cytosolic calcium ion concentration |
| 4 | 58 | 0.024 | GO:0007190 | Activation of adenylate cyclase activity |
| 7 | 323 | 0.046 | GO:0032870 | Cellular response to hormone stimulus |
| 6 | 217 | 0.048 | GO:0048011 | Nerve growth factor receptor signaling pathway |
| DAVID | ||||
| 7 | 107 | 8.85E-05 | GO:0007411 | Axon guidance |
| 8 | 169 | 8.94E-05 | GO:0030334 | Regulation of cell migration |
| 9 | 256 | 1.09E-04 | GO:0031175 | Neuron projection development |
| 8 | 184 | 1.33E-04 | GO:0000165 | MAPKKK cascade |
| 8 | 193 | 1.70E-04 | GO:0007409 | Axonogenesis |
| 9 | 339 | 6.14E-04 | GO:0048666 | Neuron development |
| 6 | 96 | 6.47E-04 | GO:0009894 | Regulation of catabolic process |
| 7 | 163 | 9.33E-04 | GO:0030425 | Dendrite |
| 9 | 342 | 0.001 | GO:0043005 | Neuron projection |
| 7 | 183 | 0.001 | GO:0006874 | Cellular calcium ion homeostasis |
GO annotation terms that were over-represented among genes in cluster I (Fig. 1b) on the basis of the analysis with FuncAssociate32 and DAVID33. N is the number of cluster genes annotated with a given GO term and X is the total number of human genes with that GO annotation. Padj values in the table represent P-values adjusted for multiple hypothesis testing by the Benjamini-Hochberg procedure in DAVID and using simulations32 in FuncAssociate. Repetitive and broad GO terms—that is, terms associated with many human genes—are not listed in the table; for a full list of all significant terms, see Supplementary Tables 2 and 3.
The genes forming cluster II (Supplementary Tables 2 and 3) were enriched for functions related to chromosomal organization and chromosomal remodeling. Notably, a similar GO enrichment analysis of all genes affected by non-synonymous de novo SNVs or de novo CNVs did not identify any significantly enriched functional terms. Thus, the developed computational approach reveals cohesive functional networks hidden within the genomic loci affected in schizophrenia.
Temporal expression of genes in schizophrenia clusters
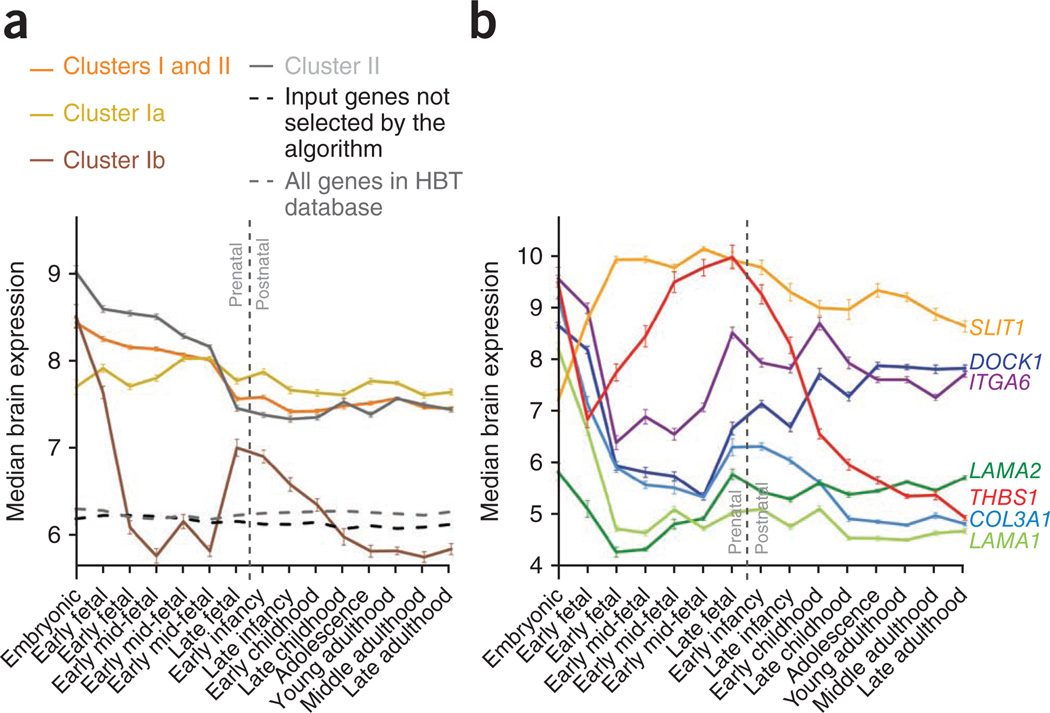
Complementary to curated gene ontology terms, another important descriptor of biological function is temporal gene expression profile. To investigate brain-related gene expression, we took advantage of the Human Brain Transcriptome (HBT) database34 and calculated the median brain expression profiles for the genes forming the identified clusters across 15 developmental stages from embryonic to late adulthood (Fig. 2a; average expression profiles are shown in Supplementary Fig. 1). The level of brain expression for all genes forming the identified clusters was significantly higher than expression of all genes in the HBT database (Wilcoxon rank-sum test, P < 1 × 10−20) and all genes used as the input for NETBAG+ but not selected by the algorithm (P < 1 × 10−20). Moreover, the expression of the cluster genes was higher during prenatal than the postnatal developmental stages (P < 1 × 10−20). This result is in agreement with significant enrichment of nonsynonymous de novo mutations in genes with high prenatal expression observed in a recent study8, and it suggests that prenatal genetic insults are particularly important for the etiology of schizophrenia.
Figure 2.
Temporal gene expression profiles in the brain across developmental stages for genes forming the identified clusters. Gene expression data were obtained from the Human Brain Transcriptome database (http://hbatlas.org/). Median expression levels for each gene were quantile normalized values and log2-transformed across all samples. (a) Temporal profiles of the median gene expression for the schizophrenia clusters shown in Figure 1. Temporal profiles of the average gene expression are shown in Supplementary Figure 1. Error bars represent s.e.m. across all applicable genes. (b) Temporal expression profiles for individual genes forming subcluster Ib. Five genes in this subcluster (DOCK1, ITGA6, LAMA2, THBS1 and COL3A1) independently exhibited U-shaped expression profiles; that is, high expression during embryonic development followed by a decrease in early or mid-fetal development and then an increase during late fetal development or infancy. Error bars represent s.e.m. across samples.
Of note, genes forming subcluster Ia, subcluster Ib and cluster II showed distinct expression profiles. Subcluster Ia contains many genes with broad brain-related functions that are essential across all developmental periods. The median gene expression in this subcluster was very uniform across the developmental stages considered, but with higher levels during prenatal periods (P = 1 × 10−6). Genes forming cluster II are primarily responsible for chromosomal organization and remodeling; their expression is likely to be particularly important during periods of neuronal development and differentiation. Naturally, the median expression profile for the cluster II genes was much higher in prenatal than postnatal developmental stages (P < 1 × 10−20). Although the genes forming subcluster Ib also displayed higher prenatal expression (P = 5 × 10−11), their median expression profile showed a prominent decrease between early fetal and late mid-fetal stages, approximately corresponding to the period between 10 and 20 weeks after conception. Several genes (DOCK1, ITGA6, COL3A1, LAMA2, THBS1) in this subcluster independently showed U-like expression profiles (Fig. 2b). This observation suggests that in the context of this subcluster, specific processes occurring early or late in corticogenesis may be predominantly affected in schizophrenia.
Processes perturbed in schizophrenia-derived neurons
To further validate biological processes implicated by considering diverse genetic variations associated with schizophrenia, we considered expression data from a recent study35. In that study, fibroblasts from schizophrenia patients were reprogrammed into pluripotent stem cells and subsequently differentiated into neurons. The analysis implicated a set of 596 genes with significantly altered expression levels in patient-derived neurons compared to neurons derived from matched controls.
The functional analysis of the differentially expressed genes with DAVID identified multiple significant GO terms (Table 2). Many of the identified terms matched the terms associated with the functional clusters implicated by our analysis of genetic variations (Table 1): neuronal differentiation, cell migration and motility, axonogenesis, neuron projection development and differentiation. This suggests that multiple lines of evidence converge on similar functions and processes.
Table 2.
GO terms associated with expression changes in neurons derived from schizophrenia patients (DAVID)
| N | X | Padj | GO identifier | GO term |
|---|---|---|---|---|
| 18 | 166 | 0.01 | GO:0050767 | Regulation of neurogenesis |
| 22 | 244 | 0.01 | GO:0000904 | Cell morphogenesis involved in differentiation |
| 20 | 192 | 0.011 | GO:0051960 | Regulation of nervous system development |
| 16 | 133 | 0.013 | GO:0045664 | Regulation of neuron differentiation |
| 22 | 256 | 0.018 | GO:0031175 | Neuron projection development |
| 19 | 209 | 0.025 | GO:0048667 | Cell morphogenesis involved in neuron differentiation |
| 18 | 193 | 0.027 | GO:0007409 | Axonogenesis |
| 23 | 307 | 0.03 | GO:0048870 | Cell motility |
| 23 | 307 | 0.03 | GO:0051674 | Localization of cell |
| 24 | 342 | 0.032 | GO:0043005 | Neuron projection |
| 16 | 159 | 0.039 | GO:0030424 | Axon |
| 9 | 59 | 0.039 | GO:0050769 | Positive regulation of neurogenesis |
In a recent study35 fibroblasts from schizophrenia patients and controls were reprogrammed into pluripotent stem cells that were subsequently differentiated into neurons. The table shows GO terms identified by DAVID33 that are enriched among 596 genes with significantly altered expression levels in schizophrenia-derived neurons. N is the number of cluster genes annotated with a given GO term and X is the total number of human genes with that GO annotation. Padj values in the table represent P-values adjusted by Benjamini-Hochberg procedure in DAVID. Repetitive and broad GO terms (that is, terms associated with many human genes) are not listed in the table; for a full list of all significant terms, see Supplementary Tables 2 and 3.
Relation of schizophrenia clusters to related disorders
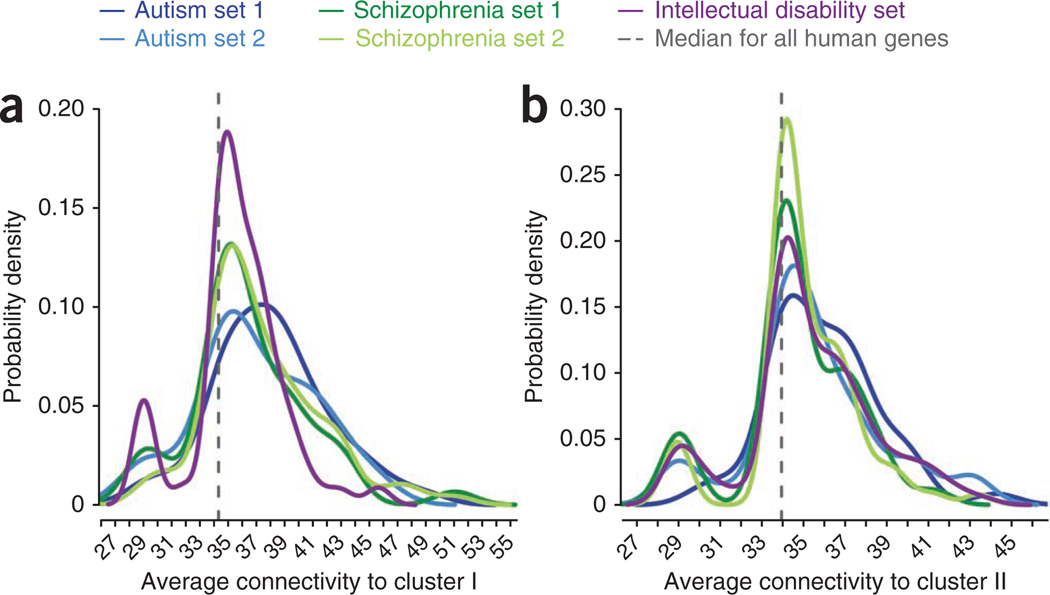
As we and others demonstrated previously, genes implicated in diverse psychiatric and neurological disorders are often closely related in terms of their biological and molecular function12,13,36. We explored the relationships between the cluster genes (Fig. 1) and genes previously implicated in schizophrenia, autism and intellectual disability using the strength of their connections (that is, likelihood ratio scores) in the NETBAG+ phenotype network (Online Methods). For this analysis, we took each gene in each curated set and calculated its connectivity strength to the schizophrenia cluster genes. We then compared the distribution of these connectivities to the connectivities between the schizophrenia cluster genes and all genes sequenced in a recent study8 (Fig. 3 and Table 3). This analysis demonstrated that genes in cluster I were strongly related to two curated sets of schizophrenia-implicated genes37–39 (Wilcoxon rank-sum test, P = 3 × 10−4 and P = 9 × 10−12). We also observed a significant relationship (P = 1 × 10−6) to a curated set of genes associated with intellectual disability40. As expected, we found no significant relationship to either of two control sets8: synonymous schizophrenia de novo SNVs (P = 0.9) or de novo SNVs in unaffected controls (P = 0.3).
Figure 3.
Distributions of connectivity strengths between schizophrenia clusters and genes previously implicated in schizophrenia and other related disorders. (a) Distributions of connectivity strengths between cluster I and disease sets. (b) Distributions of connectivity between cluster II and disease sets. The x axes show corresponding likelihood scores in the NETBAG+ phenotypic network. Disease sets shown in the figure are an autism network from the analysis of de novo CNVs11, a curated set of autism genes40, two lists of schizophrenia genes37–39 and a list of intellectual disability genes40. The distributions were smoothed using a Gaussian kernel. Vertical dashed lines indicate the median connectivity strength between the schizophrenia clusters identified in the present study and all human genes sequenced in a recent study8.
Table 3.
Connectivity strengths between schizophrenia clusters and other disease sets
| Gene sets | Number of genes | P-value to cluster I | P-value to cluster II |
|---|---|---|---|
| Autism set 1, based on CNV cluster from previous analysis11 | 45 | 3 × 10−10 | 0.0006 |
| Autism set 2, based on a literature review40 | 36 | 6 × 10−5 | 0.02 |
| Schizophrenia set 1, based on a meta-analysis37 | 42 | 0.0003 | 0.16 |
| Schizophrenia set 2, based on a meta-analysis38,39 | 75 | 1 × 10−11 | 0.019 |
| Intellectual disability set, based on a literature review40 | 110 | 2 × 10−6 | 0.0003 |
| Synonymous schizophrenia de novo SNVs from a recent study8 | 25 | 0.9 | 0.7 |
| De novo SNVs in unaffected controls from a recent study8 | 18 | 0.3 | 0.2 |
Statistical significance of functional relationship between schizophrenia clusters and genes previously implicated in schizophrenia and related disorders. Each P-value in the table quantifies the difference of two distributions: the distribution of connectivity strengths between a schizophrenia cluster and a given gene set, and the distribution of connectivity strengths between the schizophrenia cluster and all human genes sequenced in a recent study8. The NETBAG+ phenotypic network was used to calculate the connectivity strengths between each pair of genes. P-values were calculated using the Wilcoxon rank-sum test. Corresponding distributions are plotted in Figure 3.
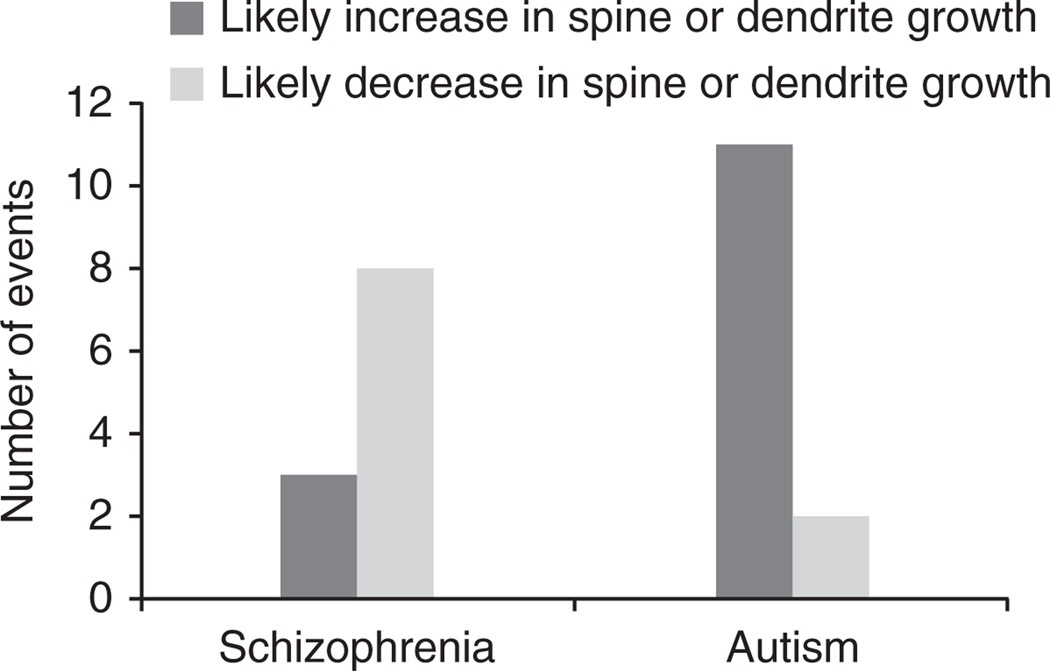
This observation raises a question: how can mutations in related and overlapping genes lead to different clinical phenotypes? Although a detailed understanding of this question will certainly require extensive clinical and biological research, we decided to gain an initial insight by focusing on a distinct phenotype previously considered by us and others: growth of dendrites and dendritic spines. Most excitatory glutamatergic synapses in the human brain are formed on dendritic spines, and their structural aberrations have been implicated in several psychiatric and neurological disorders41,42. Likely impact on the growth of dendrites or dendritic spines by a gene in a CNV can be investigated on the basis of the corresponding dosage change—a deletion or a duplication. Using this approach, we previously noted that CNVs associated with autism should primarily lead to an increase in spine or dendritic growth11. Notably, a similar analysis in schizophrenia based on known mutant phenotypes for CNV-associated cluster genes (Supplementary Table 4) revealed the opposite effect (Fig. 4): a majority of schizophrenia-associated CNVs should lead to a decrease in growth of dendrites or spines. A spine density increase in autism43 and decrease in schizophrenia44 was observed in postmortem brain analyses. We note that many mutations leading to a decrease in spine density were also observed in autism45, and an increase in spine density can actually lead to weaker synaptic connections, for example due to immature spine morphology46. Clearly, changes in spine and dendritic growth are not the only factors contributing to distinct clinical phenotypes. Nevertheless, our analysis does suggest that mutations associated with different neurodevelopmental disorders may lead, at least on average, to different functional consequences.
Figure 4.
Likely impact of genes from de novo CNVs in autism and schizophrenia on growth of dendrites or dendritic spines. Using the dosage changes (deletion or duplication) for CNV-associated genes in the schizophrenia and autism11 clusters, we explored available literature for phenotypes related to growth changes of dendrites or dendritic spines. This analysis showed that whereas de novo CNVs in autism primarily lead to an increase in growth of dendrites or dendritic spines, de novo CNVs in schizophrenia lead, on average, to the opposite effect. The difference in the phenotypic impact for the two disorders was significant (Fisher’s exact test, two-tailed, P = 0.01; Barnard’s exact test, two-tailed, P = 0.007). Genes that were considered in the analysis, their corresponding CNVs and predicted functional impact are provided in Supplementary Table 4.
DISCUSSION
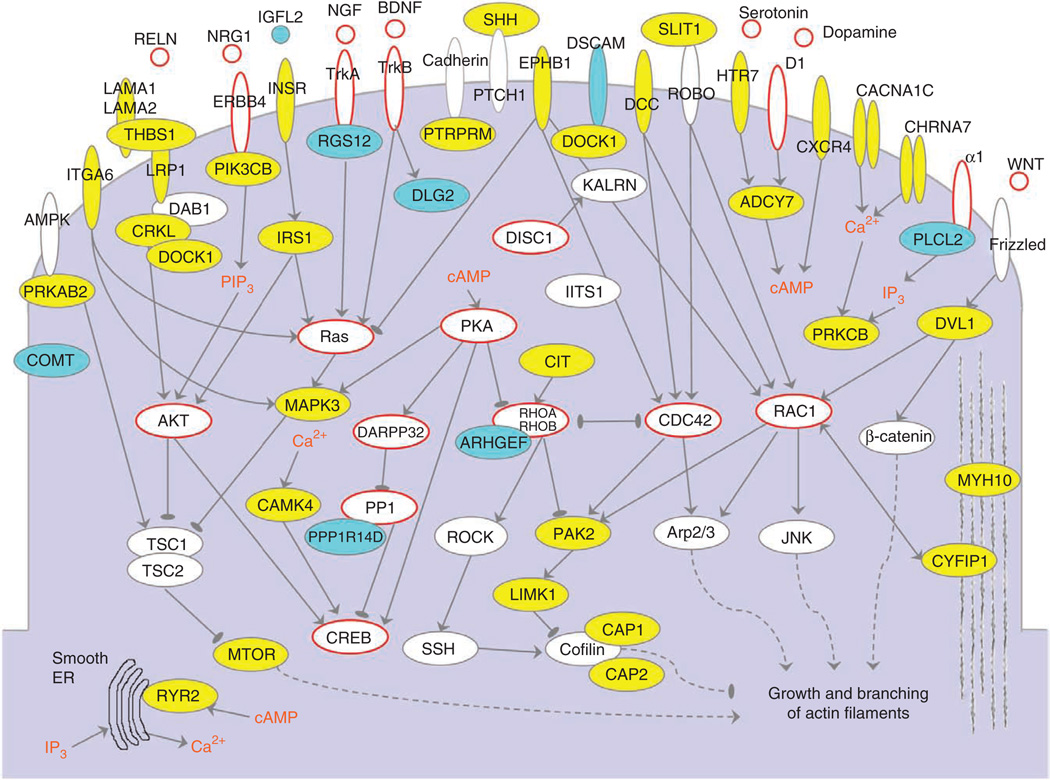
It is worthwhile to consider the genes forming the identified clusters not only as a network of binary interactions (Fig. 1) but also in the context of relevant signaling pathways (Fig. 5). Individual components of the presented network are active in diverse developmental and functional contexts, such as cell motility, axonal guidance and synaptogenesis. Several conceptual signaling levels can be delineated in the network. The first layer is formed primarily by a diverse array of receptors and channels, ranging from receptors involved in axonal guidance (such as ephrins and DCC) to ionotropic and metabotropic neurotransmitter receptors (such as CHRNA7 and HTR7). The second signaling layer is formed by cellular kinases, phosphatases and GTPases that are, either directly or indirectly, regulated by the first signaling layer. The third layer consists of regulatory (such as CREB) or structural (such as Cofilin) proteins involved in neurite outgrowth, synaptogenesis and synaptic plasticity. In addition to the aforementioned horizontal layers, several well-defined top-down pathways that were previously discussed in connection with schizophrenia and other brain-related diseases can be recognized47,48. These include the reelin, WNT and insulin signaling pathways; pathways involving Akt and phosphatidylinositol 3-OH kinase, MAP kinase, and mTOR signaling; and the protein kinase C and protein kinase A pathways. Considering the remarkable diversity of the implicated molecular circuits, it is likely that many hundreds of genes (>800, according to a recent estimate8) may ultimately contribute to the etiology of schizophrenia.
Figure 5.
Genes forming cluster I in the context of cellular signaling pathways. Proteins encoded by cluster genes are shown in yellow, and those corresponding to other relevant genes that were present in the input data but not selected by the NETBAG+ algorithm are shown in cyan. Proteins and signaling molecules that were not part of the input data but were previously implicated in schizophrenia are circled in red. ER, endoplasmic reticulum; IP3, inositol-1,4,5-trisphosphate; PIP3, phosphatidylinositol-1,4,5-trisphosphate.
Although genetic variations considered here differ in their type and origin, in combination they perturb a complex but interrelated set of molecular processes. This functional convergence allows the presented integrative approach to identify the cohesive functional networks. A similar convergence, resulting from common biological mechanisms underlying disease phenotypes, should also occur in many other human disorders. If this is indeed the case, it is likely that genetic data collected using unbiased whole-genome approaches and analyzed by proper computational methods will soon reveal the underlying molecular networks for a significant fraction of common human maladies, thus realizing an important goal of the human genome project.
ONLINE METHODS
Schizophrenia-associated genetic variation
We used three types of genetic variation: 159 non-synonymous de novo SNVs from two recent studies8,16, de novo CNVs from several previous analyses7,17–23 and 14 genomic regions that were implicated by SNPs (P < 5 × 10−8) in recent genome-wide association studies1–4,24–28 (GWAS). We considered all genes affected by non-synonymous de novo SNVs, all genes that overlap the de novo CNVs events according to the human genome NCBI build 36 and—following previous studies—all genes overlapping a region 250 kb in either direction from SNPs implicated by GWAS; similar results were obtained using calculations with distances of 100 kb and 450 kb from GWAS-implicates SNPs (Supplementary Table 1). In total, our set contained 1,044 genes from 213 genomic regions: 159 from SNVs, 712 from CNVs, and 173 from loci implicated by GWAS.
Phenotype network
The NETBAG+ algorithm is based on our previously described phenotype network11 in which all pairs of human genes are connected by weighted edges proportional to the likelihood that the genes share a genetic phenotype. These likelihood scores are based on a naive Bayesian integration of various protein-function descriptors. The functional descriptors used to build the phenotype network are: shared annotations in Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), protein domains from the InterPro database, tissue expression from the TiGER database; direct protein-protein interactions, or shared interaction partners in a number of databases (BIND, BioGRID, DIP, HPRD, InNetDB, IntAct, BiGG, MINT and MIPS); phylogenetic profiles and chromosomal co-clustering across sequenced genome49.
NETBAG+ algorithm
Genes affected by the considered genetic variations were mapped to the phenotype network. Clusters were assigned a score based on a weighted sum of their edges11, representing the likelihood that all cluster genes participate in the same genetic phenotype. Starting from each input gene, a greedy search algorithm was used to find high-scoring clusters of every size. A cluster significance was determined based on a distribution of cluster scores obtained by applying the same greedy search algorithm to randomized data. To generate random data sets, we selected genes with average connection strengths in the phenotype network similar to the corresponding disease-associated input genes. This ensures that overall connectivity of disease genes does not drive cluster significance. The average connection strength was calculated by averaging the 20 strongest edges from a particular gene to all other network genes. For a cluster of a given size, we assigned a size-specific P-value based on randomized clusters of the same size. To correct for multiple hypothesis testing (due to considering clusters at multiple sizes), we considered the best P-value from each random trial regardless of cluster size and used this distribution to assign a corrected (global) P-value to the size-specific P-value. Throughout the paper, we used this corrected P-value to characterize cluster significances. We ignored clusters with five genes or less to ensure that our analysis was not influenced by very small gene clusters with strong connections.
Cluster functional analysis
To establish specific biological functions associated with the schizophrenia clusters, we used two computational tools, FuncAssociate and DAVID, to find over-represented GO terms. For clarity, we only show GO terms associated with fewer than 350 human genes (Supplementary Table 2 for FuncAssociate and Supplementary Table 3 for DAVID). In the tables, we report P-values corrected for multiple hypothesis testing.
Expression changes in schizophrenia-derived neurons
We considered expression data from a recent study35. In that study fibroblasts from schizophrenia patients and controls were reprogrammed into pluripotent stem cells and subsequently differentiated into neurons. This analysis implicated a set of 596 genes with significantly altered expression levels in patient-derived neurons.
Likely impact of CN V events on dendrites and dendritic spines
To assess the impact of cluster genes associated with de novo CNVs on the growth of dendrites and dendritic spines, we performed a literature analysis. CNV polarity (deletion or duplication) allowed us to determine a likely change in the corresponding gene dosage. CNV-associated genes were taken from either the schizophrenia clusters identified in the present study or the autism cluster identified in our previous work11. For the two genes with both duplication and deletion events (CRKL and PIAS3), we used the reported CNV frequency5 in both disorders to determine the predominant polarity associated with each disease. The information about CNV-associated genes, polarities and phenotypes reported in the literature is provided in Supplementary Table 4.
Validation and analysis of the identified clusters
In order to validate the NETBAG+ phenotype network, the identified clusters and the associated biological functions, we performed several additional analyses.
First, we demonstrated that the phenotype network and scoring method can be used to rank genes responsible for a diverse set of genetic phenotypes. For this task, we considered known disease genes from the OMIM database, excluding diseases that were used in training of the phenotype network, diseases with less than three associated genes and diseases with somatic mutations such as cancer. In total, we considered 74 genetic phenotypes with 338 associated genes (Supplementary Table 5). For each gene in the test set, we randomly selected 99 decoy human genes with comparable network connectivity. We then ranked these 100 genes on the basis of the strength of connections in the phenotype network to the remaining OMIM genes responsible for the same phenotype. The results of this prioritization test showed that the phenotype network and the scoring method perform well in ranking disease genes. The correct gene was ranked as the top gene (out of 100 genes) in 39% of the cases, in the top three in 53% of the cases and in the top ten in 66% of the cases (Supplementary Fig. 2). This demonstrates that the network and the scoring method are not specific to schizophrenia or brain disorders and perform well across diverse phenotypes.
Second, we examined direct protein-protein interactions between genes in the identified clusters annotated in BioGRID, HPRD and DIP (Supplementary Fig. 3). We performed a commonly used permutation test to understand whether clusters identified in our analysis were more densely connected than in structurally equivalent random networks. To generate structurally equivalent random networks, the real protein-protein network was permuted by swapping known interaction pairs, while conserving the number of connections (degree) of each gene. Thirteen known interactions exist between the 47 genes in cluster I, and five interactions exist between the 42 genes in cluster II. After permutation, there were fewer interactions on average, 8.74 (P = 0.11, Z-score = 1.36) for cluster I and 2.8 (P = 0.17, Z-score = 1.21) for cluster II. Consequently, there is only a marginal significance for the inter-connectivity of the genes forming the clusters in the real network compared to random networks. This result illustrates that integrative methods (such as NETBAG+) are more powerful in establishing the significance of functional connectivities in disease clusters compared to protein-protein interactions alone.
Third, we applied our algorithm to an independent set of schizophrenia-related CNVs. This set contained rare inherited CNVs, which are more likely to contain a smaller fraction of causative events, and de novo CNVs associated with childhood-onset schizophrenia (COS)6. Overall, the independent set included 48 CNV events (35 inherited and 13 de novo COS events) containing in total 244 genes. Using this set, NETBAG+ detected a small, but marginally significant (P = 0.05), cluster of ten genes (Supplementary Fig. 4). We used DAVID to identify GO terms associated with the alternative cluster (Supplementary Table 3). This analysis showed that the alternative cluster is associated with many biological and cellular functions that are also associated with the clusters identified in our main analysis: insulin receptor signaling, axonogenesis, regulation of cell mobility and locomotion, neuron morphogenesis and differentiation, and neuron projection development. Consequently, the alternative set of CNVs provides an independent confirmation that multiple functions identified in the paper are indeed likely to be affected in schizophrenia.
Finally, we performed a manual literature review of all 159 genes with de novo SNVs from recent studies8,16. Brief functional descriptions (obtained primarily from GenBank and NCBI) for these genes are shown in Supplementary Table 6. Using the literature information, we observed that our clusters are enriched in genes with known brain and neuronal functions. Specifically, the identified clusters contained 26 genes (out of 56 in total) with brain or neural functions (Fisher’s exact test P = 10−4, Barnard’s exact test P = 2 × 10−5).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all participating families and to clinical collaborators J.L. Roos and H. Pretorius, as well as to nursing sisters R. van Wyk, C. Botha and H. van den Berg for subject recruitment and evaluation. We would also like to sincerely thank M. Wigler, D. Geschwind, G. Fischbach and all members of the Vitkup laboratory for discussions. This work was supported in part by a grant from the Simons Foundation (SFARI award number SF51), US National Centers for Biomedical Computing (MAGNet) grant U54CA121852 to Columbia University, US National Institute of Mental Health grants MH061399 (to M.K.) and MH077235 (to J.A.G.) and the Lieber Center for Schizophrenia Research at Columbia University. S.R.G. was supported in part by US National Institute of General Medical Sciences training grant T32 GM082797. B.X. was partially supported by a US National Alliance for Research in Schizophrenia and Depression (NARSAD) Young Investigator Award.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONRIBUTIONS
S.R.G. and J.C. performed computational analysis, interpreted the results and wrote the manuscript. T.S.B. contributed to the computational analysis. B.X., J.A.G. and M.K. designed the study, contributed data, interpreted the results, and contributed to functional analysis and manuscript writing. D.V. designed the study, supervised the project, interpreted the results and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Donovan MC, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 3.Ripke S, et al. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yue WH, et al. Genome-wide association study identifies a susceptibility locus for schizophrenia in Han Chinese at 11p11.2. Nat. Genet. 2011;43:1228–1231. doi: 10.1038/ng.979. [DOI] [PubMed] [Google Scholar]
- 5.Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- 7.Xu B, et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat. Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- 8.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Gen. 2012 Oct 3; doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girard SL, Dion PA, Rouleau GA. Schizophrenia genetics: putting all the pieces together. Curr. Neurol. Neurosci. Rep. 2012;12:261–266. doi: 10.1007/s11910-012-0266-7. [DOI] [PubMed] [Google Scholar]
- 10.Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, “just the facts”: what we know in 2008 part 1: overview. Schizophr. Res. 2008;100:4–19. doi: 10.1016/j.schres.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Gilman SR, et al. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70:898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proc. Natl. Acad. Sci. USA. 2008;105:4323–4328. doi: 10.1073/pnas.0701722105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh KI, et al. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iossifov I, Zheng T, Baron M, Gilliam TC, Rzhetsky A. Genetic-linkage mapping of complex hereditary disorders to a whole-genome molecular-interaction network. Genome Res. 2008;18:1150–1162. doi: 10.1101/gr.075622.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossin EJ, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girard SL, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat. Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- 17.Bassett AS, et al. Clinically detectable copy number variations in a Canadian catchment population of schizophrenia. J. Psychiatr. Res. 2010;44:1005–1009. doi: 10.1016/j.jpsychires.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guilmatre A, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch. Gen. Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirov G, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 20.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malhotra D, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulle JG, et al. Microdeletions of 3q29 confer high risk for schizophrenia. Am. J. Hum. Genet. 2010;87:229–236. doi: 10.1016/j.ajhg.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefansson H, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirov G, et al. A genome-wide association study in 574 schizophrenia trios using DNA pooling. Mol. Psychiatry. 2009;14:796–803. doi: 10.1038/mp.2008.33. [DOI] [PubMed] [Google Scholar]
- 25.Lencz T, et al. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Mol. Psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stefansson H, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sullivan PF, et al. Genomewide association for schizophrenia in the CATIE study: results of stage 1. Mol. Psychiatry. 2008;13:570–584. doi: 10.1038/mp.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70:886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 32.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arguello PA, Gogos JA. Genetic and cognitive windows into circuit mechanisms of psychiatric disease. Trends Neurosci. 2012;35:3–13. doi: 10.1016/j.tins.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 37.Allen NC, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat. Genet. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 38.Sun J, Kuo PH, Riley BP, Kendler KS, Zhao Z. Candidate genes for schizophrenia: a survey of association studies and gene ranking. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2008;147B:1173–1181. doi: 10.1002/ajmg.b.30743. [DOI] [PubMed] [Google Scholar]
- 39.Jia P, Sun J, Guo AY, Zhao Z. SZGR: a comprehensive schizophrenia gene resource. Mol. Psychiatry. 2010;15:453–462. doi: 10.1038/mp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res. Brain Res. Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- 42.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 2011;14:285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 44.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch. Gen. Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 45.Peça J, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irwin SA, et al. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am. J. Med. Genet. 2001;98:161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 47.Kvajo M, McKellar H, Gogos JA. Molecules, signaling, and schizophrenia. Curr. Top. Behav. Neurosci. 2010;4:629–656. doi: 10.1007/7854_2010_41. [DOI] [PubMed] [Google Scholar]
- 48.Pickard B. Progress in defining the biological causes of schizophrenia. Expert Rev. Mol. Med. 2011;13:e25. doi: 10.1017/S1462399411001955. [DOI] [PubMed] [Google Scholar]
- 49.Chen L, Vitkup D. Predicting genes for orphan metabolic activities using phylogenetic profiles. Genome Biol. 2006;7:R17. doi: 10.1186/gb-2006-7-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.