Abstract
Perception of sound is a fundamental role of the auditory system. Traveling with the force of their mechanical energy, sound waves are captured by the ear and activate the sensory pathway of this complex organ. The hair cells, specialized sensory cells within the inner ear, function to transmit the mechanical energy into electrical nerve stimuli that reach the brain. A large number of proteins are responsible for the overarching tasks required to maintain the complex mechanism of sound sensation. Many hearing disorders are due to single gene defects, inherited in a Mendelian fashion, enabling clinical diagnostics. However, at the same time, hearing impairment is genetically heterogeneous, with both common and rare forms occurring due to mutations in over 100 genes. The crosstalk between human and mouse genetics has enabled comprehensive studies on gene identification and protein function, taking advantage of the tools animal models have to offer. The aim of the following review is to provide background and examples of human deafness genes and the discovery of their function in the auditory system.
Keywords: deafness, Usher syndrome, myosin VIIA, cadherin 23, stereocilin, microRNA-96
Hearing loss: a Mendelian disorder
Hearing loss is the most common sensory loss, affecting 1-2 out of 1000 newborns and over half of the population by the age of 80 (US National Institute on Deafness and Other Communication Disorders, http://www.nidcd.nih.gov/health/statistics/hearing.html). Severe hearing loss that occurs during infancy can have dramatic effects on speech acquisition and literacy. Deafness with a later onset might have a deleterious effect on the quality of life of an affected individual. Hearing loss may be sensorineural, caused by damage to the inner ear or auditory nerve, or conductive, due to reduction of sound waves passing through the outer and middle ear (reviewed in 1). Sensorineural hearing loss can be present at birth or acquired at later stages in life. Among the factors causing acquired hearing loss are side effects of different infections and diseases and ototoxic drugs, as well as exposure to excessive noise, either social or industrial. Approximately 60-70% of hearing loss is due to genetic factors and is genetically heterogeneous, with single gene defects in many different genes (Figure 1).2 Furthermore, genetic hearing impairment can occur either as syndromic hearing loss (SHL), associated with other abnormalities, or non-syndromic hearing loss (NSHL), as an isolated phenotype, which accounts for about 70% of the cases. Vestibular dysfunction may be associated with NSHL as well. Hearing loss can be inherited either in a dominant or recessive mode, whether it is syndromic or non-syndromic.
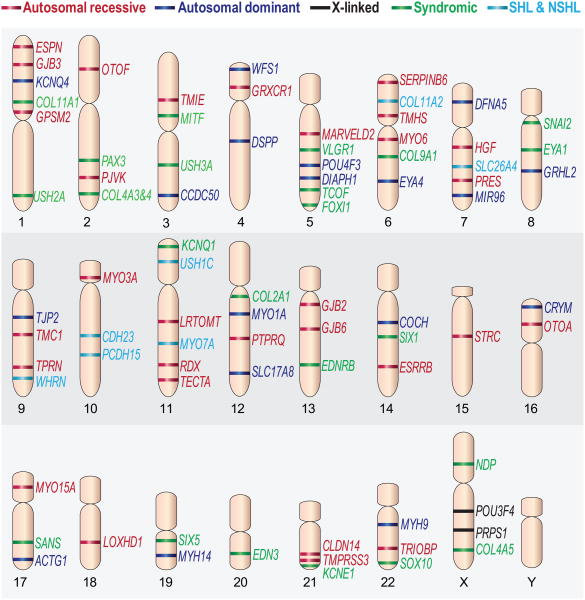
Figure 1.
A scheme representing the chromosomal location of genes with mutations that cause hearing loss. The genes are classified as non-syndromic autosomal recessive (red), non-syndromic autosomal dominant (blue), X-linked genes (black), syndromic (green), and genes associated with both syndromic and non-syndromic hearing loss (light blue). Previously published in 8, 70 with permission by authors.
A comprehensive listing of all genes and the proteins they encode is no longer practical to complete in a review, as it was just a few years ago. The breadth and variability of phenotypes, mutations, mechanisms and pathways is remarkable. As the inner ear is an organ with diverse cell types, many cellular targets are susceptible to damage due to genetic mutations. In this review, we concentrate on one of the most sensitive sites to genetic mutations, the hair cell, which is a sensory cell that functions to detect and amplify the mechanical signal evoked by sound and translate it into the electrical signal transmitted to the brain. We define several characteristics and qualities of hereditary hearing loss. Four genes are described as examples of Mendelian phenotypes that helped decipher major mechanisms or functions of the inner ear and have shed light on auditory pathways. In each case the mouse was instrumental in defining the mechanism. Each represents a different prototype of the variability and heterogeneity of this disorder, due to a single gene defect.
Hearing loss: from past to present
Descriptions of hereditary hearing loss go back as far as the 16th century.3 In the early days of the 17th century Paulus Zacchias (1584-1659), an Italian physician, stated that marriage of deaf people should be avoided, as they would give birth to deaf children like themselves. This statement suggests that Zacchias was aware of the hereditary factor in congenital deafness. In those days, however, it was still believed that hearing is the “gate of the mind” and therefore congenital deafness leads to defective intelligence.4 By 1880 evidence for both autosomal dominant and autosomal recessive inheritances of hearing loss were presented, although these terms were not determined yet. X-linked inherited deafness is presumed to first be described in 1863, although it was not fully characterized until 1930 by Dow and Poynter.5 Prevention of marriage between deaf individuals was also proposed in legislation later on, in 1883, by Alexander Graham Bell. The approach of banning hearing impaired individuals from society was taken even further under Nazi Germany, who embraced eugenics. During Nazi control 1,600 deaf individuals were murdered.3 Today, there are many specialized schools and communities around the world for the deaf, providing cultural centers and institutions of higher education, such as Gallaudet University in Washington DC, USA (http://www.gallaudet.edu/).
Syndromic hearing loss
SHL is known to be associated with defects in many different organs.6 Forms of dominant SHL include Wardenburg syndrome (deafness, structural defects derived from the neural crest and pigmentation anomalies); Branchial-oto-renal syndrome (Branchial fistulas, renal anomalies, and abnormal development of the ear); Stickler syndrome (distinctive facial abnormalities, eye problems, hearing loss and joint problems) and Neurofibromatosis 2 (characterized by tumors, including acoustic neuromas). Recessive SHL includes the deafness-blindness of Usher syndrome, Pendred Syndrome (inner ear and enlargement of the thyroid gland, known as goiter); Jervell and Lange-Nielsen syndrome (characterized by electrocardiographic changes as well as hearing loss); and Cockayne syndrome (described with dwarfism, retinal atrophy and deafness). X-linked syndromic deafness includes Alport syndrome (also known as Hereditary Nephritis) and Norrie Disease (congenital ocular symptoms with progressive hearing loss). It is difficult to state how many forms of SHL there are. Hearing impairment is often overlooked with more difficult disorders present in a patient. Toriello once estimated that “new” syndromes are being described weekly.7 At least 32 genes have been identified for SHL (Hereditary Hearing loss Homepage, http://hereditaryhearingloss.org).8
Non-syndromic hearing loss
Since the early 1990s, 149 NSHL loci have been identified, and are marked as DFNA for autosomal dominant, DFNB for autosomal recessive deafness, and DFNX for X-linked loci (Hereditary Hearing loss Homepage).8 The first dominant locus, DFNA1, was identified in 19929 in a large kindred from Costa Rica. Five years later it was discovered that the gene DIAPH1, which encodes the protein diaphanous that regulates actin polymerization, is responsible for the hearing loss in this family.10 DFNB1, the first recessive deafness locus identified, was revealed in 1994 in two consanguineous families from Tunisia that were affected by this form of deafness.11 The gene, GJB2, responsible for the hearing loss in these families was identified soon after, in 1997, encoding the gap-junction protein connexin 26 (Cx26).12 Today, GJB2 is the most common deafness-causing gene, known to account for up to 50% of the cases of non-syndromic sensorineural hearing loss in some populations.13 In 1995 the first sex-linked hearing loss-causing gene was identified, POU3F4,14 which encodes a transcription factor with a POU domain, mapped to the interval of Xq21 and found to be responsible for DFNX2. DFNX2 was formerly known as DFN3, but when a Y-linked deafness locus was identified15 the nomenclature was changed to provide differentiation between the sex chromosomes.16
Major breakthroughs defining auditory function
Several major discoveries uncovered mechanisms of auditory function, combining gene and protein networks. Myosin VIIA mutations, associated with both Usher syndrome and NSHL, encodes a protein that is part of the Usher network of proteins that interact with one another and help define functions of the stereocilia. Cadherin 23 and protocadherin 15 mutations are associated with both syndromic and non-syndromic forms of hearing loss. Most compelling, these proteins together form the tip link between stereocilia, a major site of mechanotransduction. Mutations in stereocilin are associated with NSHL, and the loss of this protein leads to structural changes in the outer hair cells, implicating this protein in hair cell function. Finally, an entirely new paradigm in hearing loss was demonstrated with the discovery of mutations in a microRNA (miRNA). Together, these examples provide a snapshot of several different scenarios of this hereditary disease, which together form the complex function of the auditory system.
DFNB2/USH1B: myosin VIIA
An understanding of Usher syndrome on a molecular level has been achieved by defining part of the protein network in the inner ear that is responsible for development and mechanisms of the hair bundle. Usher syndrome is the most common cause of combined deafness-blindness known in humans, affecting 1:25,000 children.17, 18 It is defined by congenital, bilateral deafness followed by a later onset of vision loss, caused by retinitis pigmentosa (RP). The retinal degeneration is a consequence of photoreceptor cell death beginning from the periphery of the retina. Patients suffer from night blindness, followed by narrowing of the visual field (“tunnel vision”), leading to absolute blindness.19 Usher syndrome is heterogeneous, both clinically and genetically, with at least 12 chromosomal loci involved in the three types of USH syndrome, Usher type 1 (USH1), USH2 and USH3 (further subdivided, when relevant, to A-H). The different types are distinguishable from each other mainly by the severity and onset of the deafness and variability of vestibular defects, while all have RP. USH1 patients are profoundly deaf from birth, and have vestibular dysfunction. The deafness in USH2 is less severe, and the vestibular function is normal. USH3 is characterized by mild progressive hearing loss with occasional vestibular dysfunction.20
The first USH1 gene that was identified, for USH1B, is myosin VIIa (MYO7A)21. The protein is an unconventional myosin that moves towards the plus end of actin, from the stereocilia base to its tip. It has been described as a dimeric motor and is known to require high concentrations of ATP for its motile activity along actin filaments.22 As part of the myosin superfamily, myosin VIIa has the common structure of myosins, a motor head domain that includes the actin-binding and the ATP-binding sites, a neck domain with five IQ motifs that are expected to bind myosin-light chains, and the variable tail domain. The tail region contains the functional domains that determine myosin VIIa specificity.23 It contains two large tandem repeats that are separated by SH3 domains. These repeats consist of a MyTH4 (myosin tail homology 4) domain and a FERM domain (4.1, ezrin, readixin, moesin), which is thought to be responsible for the attachment of the protein to the plasma membrane.18 The tail portion of myosin VIIa is known to bind the scaffolding proteins harmonin24 and SANS18, as well as Pcdh1525, which are all part of the USH protein network, in addition to binding to the F-actin filaments (Figure 2).22, 26 These interactions form the protein network of Usher syndrome. Myosin VIIa is also known to interact with vezatin,27 which attaches it to the cadherin-catenin complex, increasing cell-cell adhesion. In the inner ear these two proteins have a membrane to membrane interaction in the links that interconnect the bases of adjacent stereocilia. Recently, a direct interaction between Cdh23 and myosin VIIa has been reported.28 This finding enhances the relationship within the Usher protein network, as all of its components have been shown to interact with one another.
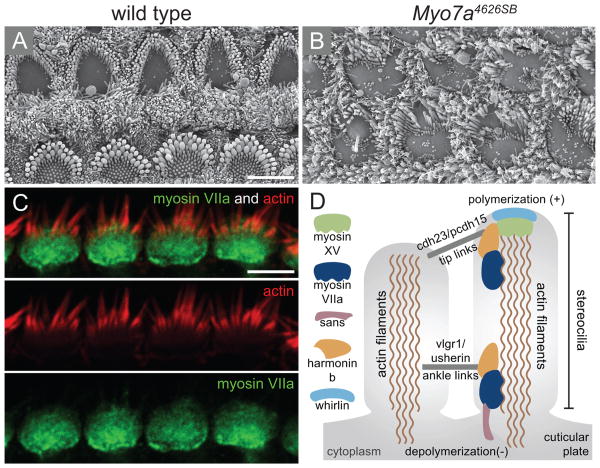
Figure 2.
Phenotypic effect of mutations in MYO7A. (A-B) Scanning electron micrographs of wild type (A) and (B) null allele of myosin VIIa (Myo7a4626SB) that expresses less than 1% of myosin VIIA protein, showing a near complete loss of stereocilia structure and disorganisation of the remaining hair bundle. (C) Immunolocalization of myosin VIIa is shown in the cuticular plate. (D) Schematic representation of the network of proteins that are involved in Usher syndrome within the stereocilia. Myosin VIIA is connected to the actin filaments as well as to harmonin b and to the tip links that are composed of cadherin 23 (cdh23) and protocadherin (pcdh15). Scale bars: 5 μm in (A) applies to B, 5 μm in (C). Figures (A-B) were received from Tim Self and Karen Steel and (D) was previously published in Friedman et al. 200771 with permission by authors.
In addition to its involvement in Usher syndrome, mutations in myosin VIIA causes non-syndromic deafness, either dominant (DFNA1129) or recessive (DFNB230, 31). Myosin VIIa was also found to be mutated in the mouse, shaker1 (sh1), causing progressive degeneration of the cochlea (Figure 2) together with vestibular dysfunction.32 From this mouse model, myosin VIIa was found to be expressed in the cochlea as early as embryonic day 16.5.33 The protein is expressed in the stereocilia and cuticular plate of the hair cells (Figure 2).34 The shaker-1 (sh1) gene has seven different alleles, and in some of the mutated alleles (Myo7a6J and Myo7a816SB in particular) the mutation causes abnormal stereocilia development in both inner and outer hair cells.33 There is a high correlation between the location of the mutation within the protein and its severity. Mutations causing ultrastructural changes in the stereocilia were either deletions or missense mutations in the myosin motor-head core. Mutated forms of myosin VIIa fail to localize correctly to the stereocilia.35 These mutated forms are responsible for the more severe phenotypes.
DFNB12/USH1D: Cadherin 23 and DFNB23/USH1F: Protocadherin 15
Mutations in CDH23 (Cadherin-related 23) are responsible for both DFNB12 and USH1D.36 Mutations were identified in a number of consanguineous families from Pakistanian and Indian origin36 as well as a Cuban family.37 The protein encoded by CDH23 is comprised of 3354 amino acids and predicted to have 27 cadherin repeats that are involved in Ca2+-dependent cell adhesion, an N′ terminal signal peptide and one transmembrane domain.38 Null mutations in this gene are associated with USH1D, whereas missense mutations are associated with DFNB12.36, 39 The waltzer mouse phenotype is due to mutations in Cdh23, and serves as a mouse model for USH1D and DFNB12.40, 41 These mice have hearing loss and vestibular dysfunction, caused by disorganization of their stereocilia (Figure 3).41 Another mouse that suffers from progressive hearing loss and carries a mutation in Cdh23 is the ENU-induced mouse salsa.39 As opposed to the waltzer mouse, the hair bundle in the salsa mouse is intact and only the stereocilia tip-links are lost, presumably causing loss of tension within the stereocilia. Based on these two mouse models, it was suggested that USH1D, caused by null mutations in CDH23, is due to hair bundle developmental defects, while DFNB12, with the missense mutations, is caused by tip link defects. Cadherin 23 was shown to have a role in maintaining the hair bundle during development,41 but in adults has the role of comprising the tip links, transmitting the force to the ion channels. The tip links are extracellular filaments connecting between adjacent stereocilia and control the opening of mechanosensory transduction channels (Figure 3).42 The mechanosensory channels are located near the stereocilia tips. Sound vibrations causes deflection of the hair bundle toward the longest stereocilia. This deflection increases the tension in the tip link and the hair cells will respond by opening or closing the ion transduction channels,43 leading to an influx of cations into the stereocilia and resulting in depolarization of the hair cell.
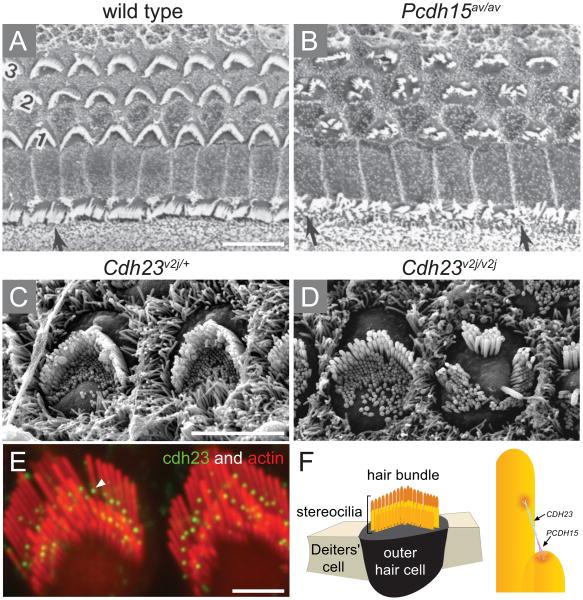
Figure 3.
The tip link of adjacent stereocilia is assembled by cadherin 23 and protocadherin. (A-D) Scanning electron micrographs of cochlear sensory epithelium of Ames waltzer (av) and waltzer (v) mice carrying mutations in Pcdh15 and Cdh23, respectively. (A) Wild type sensory epithelium is organized with one row of inner hair cell and three rows of outer hair cells with a typical ‘V’ shape arrangement of their stereocilia. (B) A spontaneous mutation of Pcdh15 leads to severely impaired organization of the hair bundles of both inner (indicated by arrows) and outer hair cells that lost the ‘V’ shape structure within their apical surface. (C) High power images of outer hair cells highlights the polarity nature of these cells characterized by hair bundles that point towards the same direction to the kinocilium. (D) The outer hair cells of Cdh23v2j/v2j mutant mice lost their planar cell polarity, showing groups of stereocilia of the surface of the cell facing different directions. Moreover, the essential staircase arrangement of the stereocilia is no longer maintained within the bundle. (E) Immunolocalization of cadherin 23 (green) is defined to the tip of the stereocilia (red) in adult guinea pig. (F) Schematic representation of an outer hair cell surrounded by two supporting cells. The hair bundle is composed of rows of stereocilia in gradual heights which create the typical staircase structure of the bundle (yellow). The tip links that connect adjacent stereocilia of lower and higher rows are composed of cadherin 23 and protocadherin 15. Ages: (A-B) P10; (C-D) P4. Scale bars: 10 μm in (A, B); 5 μm (C, D); 2 μm (E). Figures (A-B) are reprinted with permission from Alagramam et al. 2001;47 figures (C-D) from Di Palma et al. 200141 and figures (E-F) from Kazmierczak et al. 2007.48
Another component of the tip links is encoded by PCDH15 (Protocadherin-15),44 which has 11 calcium-binding cadherin repeats and is responsible for USH1F and DFNB23, as was identified in two Pakistani families.45 In the syndromic form, affected members are profoundly deaf and suffer from progressive RP. The mouse model for USH1F is Ames waltzer, which is profoundly deaf with disorganized abnormal stereocilia (Figure 3).46, 47 Protocadherin 15 forms the lower part of the tip link and interacts with cadherin 23, which forms the upper part, together creating the tip-link filament,48 which is about 180nm long. The interaction between these two proteins is made through their extra-cellular cadherin domains, at their N′ terminus, and is Ca2+ dependent. The salsa mutation also affects the interaction between cadherin 23 and protocadherin 15, even though the mutation affects an amino acid that is outside the interaction site.39 The studies of these two proteins revealed the structure of the tip links, one of the most important components of the auditory hair bundle, which is important for both maintaining the highly organised structure of the stereocilia and playing a role in the mechano-electrical (MET) process in the inner ear.
DFNB16: stereocilin
Stereocilin is the gene responsible for DFNB16, recessively-inherited NSHL, found in two families of Pakistani and French origin.49 The affected individuals have postlingual hearing loss from early childhood, which is more severe at higher frequencies. The gene encodes an ∼1800 amino acid long protein that is localized at the tips of the stereocilia of the outer hair cells (Figure 4). The protein was first hypothesized to be responsible for the attachment of the tectorial membrane to the sensory cells in the inner ear.50 A mouse model lacking stereocilin51 provided a clearer picture of the role of stereocilin in the inner ear. The tectorial membrane in these mice is intact, but hair bundle imprinting, which represents the anchoring of outer hair cells stereocilia to the tectorial membrane, was not detectable. Outer hair cells derived from Strc-/- mice lack the horizontal top connectors. The distal ends of the stereocilia are less connected in the mutants than in wild type mice, suggesting that the top connectors have a role in the cohesion of the outer hair cell hair bundle of adult mice. In the cochlea, the outer hair cells acts as an amplifier, assisting in sharpening the sound-induced mechanical stimulation that reaches the cochlea, while the inner hair cells serve to transmit the sensory information into the brain. The outer hair cells, in addition to sound amplification, generate mechanical and electrical waveform distortions, essential for hearing in noisy environments.52 This distortion creates a masking effect that contributes to speech understanding and simplicity. The studies of the Strc-/- mutant mice suggest that the top connectors may either generate distortion by themselves due to non-linear stiffness, or they may alter hair-bundle displacements, leading to distortion of sound waveforms. This work suggests a role for stereocilin in a major mechanism of outer hair cell function, revealed through genetic mutations.
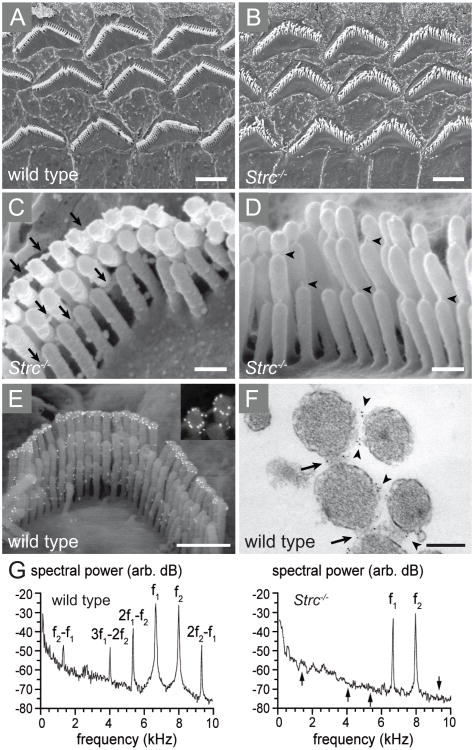
Figure 4.
Stereocilin assembles the horizontal top connectors and is suggested to be essential for the nonlinearity underlying cochlear waveform distortions. (A-D) Scanning electron micrographs (SEM) of cochlear sensory outer hair cells of stereocilin deficient mice, Strc-/-. (A) Low power image of the three rows of outer hair cells in a wild type mouse. (B) The stereocilia of Strc-/- outer hair cells retain the typical ‘V’ shape but are dispersed from one another. (C) Higher magnification image of a single outer hair cell of Strc-/- mice shows that the top connectors between stereocilia (arrows) are absent in the mutant hair bundles. (D) Despite the missing top connectors, the tip links of Strc-/- hair bundles are maintained (arrow heads). (E-F) The localization of stereocilin is detected by immunogold labelling on scanning electron micrographs of a wild type outer hair cell (E), in a ring-shaped labelling around the tips of the tallest stereocilia row. Labelling can also be detected between the stereocilia within the top connectors, in a transmission electron micrograph of a stereocilin-labelled wild type outer hair cell hair bundle (F), marked with arrows. (G) Cochlear microphonic (CM) shown by frequency spectra. The black arrows within the Strc-/- mice spectra indicate the frequency points where prominent intermodulations are apparent. Ages: (A-E) P14 (F) P22. Scale bars: 2.5 μm in (A, B), 0.25 μm (C, D), 1 μm (E) and 0.2 μm in (F). Figures (A-G) are reprinted with permission from Verpy et al. 2008.51
DFNA50: miR-96
MiRNAs are small non-coding RNAs 21-23 nucleotides long, which regulate gene expression through the RNA interference (RNAi) mechanism, known to affect proliferation, differentiation and developmental processes in eukaryotes.53 Mir genes are included within introns of protein-coding genes or in intragenic regions. Intronic miRNAs are transcribed along with the hosting protein-coding gene. A miR is first transcribed to a long RNA molecule, the pri-miRNA, which is further processed by the nucleases Drosha and Dicer1 through the pre-miRNA intermediate to generate the mature miRNA.54, 55 The seed sequence (nucleotides 2-8) of the mature miRNA is a key determinant of which mRNAs will be regulated by each miRNA. MiR genes are often organised in clusters that are co-transcribed as polycistronic genes to a single pri-miRNA from which a couple of mature miRNAs will originate.56
MiR-96 is part of the miR-183/96/182 cluster, which is expressed in the mouse inner ear, retina, and additional tissues.57-59 In the inner ear, it is mainly expressed in developing hair cells. The locus DFNA50 segregated in a Spanish family with postlingual, progressive, NSHL at all frequencies.60 Further analysis revealed a mutation in miR-96 (+13 G>A), causing replacement of the fourth nucleotide within the conserved seed region of the mature miR-96 sequence, which segregated with the hearing impairment in the family.61 Another mutation in the seed region of miR-96 (+14 C>A) was found in another Spanish family with autosomal dominant, progressive, high-frequency hearing loss. The fact that in both families the hearing loss was postlingual suggests that miR-96 may have a regulatory role in maintaining gene expression required for normal function, rather than inner ear development. Notably, this is the first study showing a nucleotide substitution in the seed region of a miRNA that is responsible for a human Mendelian disease.61
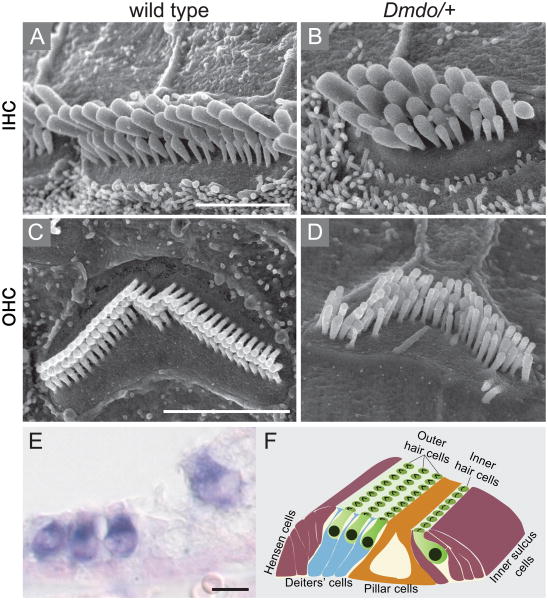
N-ethyl-N-nitrosourea (ENU) is a mutagen used to screen for new dominant or recessively-inherited mutations. Diminuendo (Dmdo) is an ENU-induced mouse model, with an A>T substitution in the miR-96 seed62 (Figure 5). These mice show progressive hearing impairment in heterozygotes and profound deafness in homozygotes, associated with hair cell defects. In homozygotes, most of the hair cells are degenerated by 4 weeks of age. Luciferase assays in transfected non-inner ear cells demonstrated that five mRNAs, Aqp5, Celsr2, Myrip, Odf2 and Ryk, are regulated by miR-96.61, 62 These targets are suggested to be over-expressed in the inner ears of humans or mice with mutations in the miR-96 seed. However, while the level of Aqp5 and Celsr2 mRNAs was notably up-regulated in the cochlear organ of Corti of the mutant Dmdo mice compared to wild type, the differences in protein expression levels were small.62 MiR-96 may function to fine-tune the expression level of these putative targets. It is also possible that other post translational regulation might play a role in regulating AQP5 and CELSR2 protein expression levels. These changes may indirectly affect the expression of many other genes, which are not direct targets of mir-96. A genome-wide approach revealed nearly 100 affected transcripts that were either down- or up-regulated in the organ of Corti of mutant mice compared to wild type mice. In particular, five genes that are well known in the inner ear were down-regulated (prestin, oncomodulin, Pitpnm1, Gfi1 and Ptprq), although none of them has the seed region for mir-96. This suggested a downstream effect of epigenetic down-regulation caused by the miRNA that may help explain the mechanism behind the hearing impairment of these mice.
Figure 5.
miR-96 is required for normal morphology and function of the auditory system. (A-D) Scanning electron microscopy (SEM) images of cochlear sensory hair cells of diminuendo mice depleted of miR-96. Wild type inner (A) and outer (C) hair cells exhibit normal morphology of the hair bundles with actin-rich stereocilia protrusions arranged in a typical staircase manner. (B and D) Created by ENU mutagenesis, a point mutation within the seed of miR-96 in Dmdo mice leads to altered morphology of the auditory hair cells. (B) The inner hair cells of the Dmdo/+ mutant mice show additional rows of stereocilia with a bulged and rounded cell surface. (D) Dmdo/+ outer hair cells lost their staircase arrangement while stereocilia of the first row of the bundle are taller than the middle rows. (E) In situ hybridization (ISH) revealed the spatial expression of miR-96 within the auditory apparatus defined to the cochlear inner and outer hair cells (dark blue). (F) Schematic illustration combining the top and cross view of the cochlear sensory epithelium, also known as the organ of Corti. The hair cells (green) are shown in their typical arrangement of one inner and three outer rows. The hair bundle of each hair cell is apparent on its surface (black). A wide array of supporting cells are essential surrounding the hair cells including Dieters' (blue) Hensens and inner sulcus cells (purple). The pillar cells (brown) create the tunnel of Corti that separate between the inner and outer rows of hair cells. Ages: (A-D) P28 (E) P0. Scale bars: 3 μm in (A-D), 5 μm in (E). Figures (A-E) are reprinted with permission from Lewis et al. 200962 and figure (F) was previously published in Dror and Avraham 2009.72
Conclusion
The advancements in the field of hereditary hearing loss have been dramatic since the mapping of the first chromosomal loci for X-linked deafness in 1988, SHL in 1990 and NSHL in 1992. Since then, a month has rarely gone by without the discovery of a new gene or allele for hearing loss. Along with the fact that connexin 26 accounts for a majority of congenital NSHL, these advances have provided new opportunities for diagnostics. Early identification of specific mutations using molecular diagnostics alleviates the need for other clinical tests.63 It also facilitates predictions regarding the potential progressivity of the hearing loss (if not early onset profound) and other abnormalities, in the case of SHL. For example, children born with the PCDH15 R245X mutation are profoundly deaf, but the RP does not arise until about the age of 10.64 Early diagnosis allows the family and physicians to be aware of impending blindness, which may dictate future treatment, e.g. cochlear implantation.
Efforts are currently underway worldwide to develop therapeutics based on gene therapy, stem cells and/or regenerative medicine. Caution must be taken, however, in raising immediate hopes in these areas for patients, since while there has been progress in basic research towards inner ear therapeutics,65, 66 major technological and other hurdles remain.67
Most important, there are many single-gene defects for inherited hearing loss yet to be discovered. While over 50 genes have been found for NSHL since the discovery of connexin 26, most mutations are rare and occur in one or a few families, making diagnostics today limited to only a handful of genes. Massively parallel sequencing (or deep sequencing) has been applied in a few cases towards gene discovery for hearing loss.68, 69 As targeted genome capture, whole exome sequencing and deep sequencing of entire genomes becomes more affordable, these techniques will be applied more to hereditary hearing loss as well, enabling further gene and mutation discovery. Most compelling, the complex genetic and protein interactions of the auditory system will be elucidated through studies of classical Mendelian phenotypes.
Acknowledgments
Research in the Avraham laboratory is funded by the Israel Science Foundation Grant 1486/07, the National Institutes of Health (NIDCD) R01DC005641, the Israel Ministry of Health, and the European Commission FP6 Integrated Project Eumodic 037188. We would like to thank Kumar Algaramam, Morag Lewis, Konrad Noben-Trauth, Ulrich Mueller, Elizabeth Verpy, Christine Petit, Tim Self, and Karen Steel for sharing figures with us.
References
- 1.Petit C, Richardson GP. Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci. 2009;12:703–710. doi: 10.1038/nn.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petit C, Levilliers J, Hardelin JP. Molecular genetics of hearing loss. Annu Rev Genet. 2001;35:589–646. doi: 10.1146/annurev.genet.35.102401.091224. [DOI] [PubMed] [Google Scholar]
- 3.Gorlin RJ. Genetic Hearing Loss-A Brief History. In: Gorlin RJ, Toriello HV, Cohen MM, editors. Hereditary Hearing Loss and its Syndromes. Oxford University Press; New York: 1995. pp. 3–4. [Google Scholar]
- 4.Cranefield PF, Federn W. Paulus Zacchias on mental deficiency and on deafness. Bull N Y Acad Med. 1970;46:3–21. [PMC free article] [PubMed] [Google Scholar]
- 5.Dow GS, Poynter CI. The Dar family. Eugen News. 1930;15:128–130. [Google Scholar]
- 6.Gorlin RJ, Toriello HV, Cohen MM, editors. Hereditary Hearing Loss and its Syndromes. Oxford University Press; New York: [Google Scholar]
- 7.Toriello HV. New syndromes from old: evaluation of heterogeneity and variability in syndrome definition and delineation. Am J Med Genet Suppl. 1988;4:55–70. doi: 10.1002/ajmg.1320310511. [DOI] [PubMed] [Google Scholar]
- 8.Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. 2010 http://hereditaryhearingloss.org. Update: 1.10.2010.
- 9.Leon PE, Raventos H, Lynch E, Morrow J, King MC. The gene for an inherited form of deafness maps to chromosome 5q31. Proc Natl Acad Sci U S A. 1992;89:5181–5184. doi: 10.1073/pnas.89.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch ED, Lee MK, Morrow JE, Welcsh PL, Leon PE, King MC. Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science. 1997;278:1315–1318. [PubMed] [Google Scholar]
- 11.Guilford P, Ben Arab S, Blanchard S, Levilliers J, Weissenbach J, Belkahia A, Petit C. A non-syndrome form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nat Genet. 1994;6:24–28. doi: 10.1038/ng0194-24. [DOI] [PubMed] [Google Scholar]
- 12.Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 13.Kenneson A, Van Naarden Braun K, Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med. 2002;4:258–274. doi: 10.1097/00125817-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 14.de Kok YJ, van der Maarel SM, Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME, Ropers HH, Cremers FP. Association between X-linked mixed deafness and mutations in the POU domain gene POU3F4. Science. 1995;267:685–688. doi: 10.1126/science.7839145. [DOI] [PubMed] [Google Scholar]
- 15.Wang QJ, Lu CY, Li N, Rao SQ, Shi YB, Han DY, Li X, Cao JY, Yu LM, Li QZ, Guan MX, Yang WY, Shen Y. Y-linked inheritance of non-syndromic hearing impairment in a large Chinese family. J Med Genet. 2004;41:e80. doi: 10.1136/jmg.2003.012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen MB, Wang Q, Willems PJ. Sex-linked deafness. Clin Genet. 2008;73:14–23. doi: 10.1111/j.1399-0004.2007.00913.x. [DOI] [PubMed] [Google Scholar]
- 17.El-Amraoui A, Petit C. Usher I syndrome: unravelling the mechanisms that underlie the cohesion of the growing hair bundle in inner ear sensory cells. J Cell Sci. 2005;118:4593–4603. doi: 10.1242/jcs.02636. [DOI] [PubMed] [Google Scholar]
- 18.Reiners J, Nagel-Wolfrum K, Jurgens K, Marker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 19.van Soest S, Westerveld A, de Jong PT, Bleeker-Wagemakers EM, Bergen AA. Retinitis pigmentosa: defined from a molecular point of view. Surv Ophthalmol. 1999;43:321–334. doi: 10.1016/s0039-6257(98)00046-0. [DOI] [PubMed] [Google Scholar]
- 20.Williams DS. Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 2008;48:433–441. doi: 10.1016/j.visres.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- 22.Inoue A, Ikebe M. Characterization of the motor activity of mammalian myosin VIIA. J Biol Chem. 2003;278:5478–5487. doi: 10.1074/jbc.M210489200. [DOI] [PubMed] [Google Scholar]
- 23.Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 24.Adato A, Michel V, Kikkawa Y, Reiners J, Alagramam KN, Weil D, Yonekawa H, Wolfrum U, El-Amraoui A, Petit C. Interactions in the network of Usher syndrome type 1 proteins. Hum Mol Genet. 2005;14:347–356. doi: 10.1093/hmg/ddi031. [DOI] [PubMed] [Google Scholar]
- 25.Senften M, Schwander M, Kazmierczak P, Lillo C, Shin JB, Hasson T, Geleoc GS, Gillespie PG, Williams D, Holt JR, Muller U. Physical and functional interaction between protocadherin 15 and myosin VIIa in mechanosensory hair cells. J Neurosci. 2006;26:2060–2071. doi: 10.1523/JNEUROSCI.4251-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petit C. Usher syndrome: from genetics to pathogenesis. Annu Rev Genomics Hum Genet. 2001;2:271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- 27.Kussel-Andermann P, El-Amraoui A, Safieddine S, Nouaille S, Perfettini I, Lecuit M, Cossart P, Wolfrum U, Petit C. Vezatin, a novel transmembrane protein, bridges myosin VIIA to the cadherin-catenins complex. EMBO J. 2000;19:6020–6029. doi: 10.1093/emboj/19.22.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahloul A, Michel V, Hardelin JP, Nouaille S, Hoos S, Houdusse A, England P, Petit C. Cadherin-23, myosin VIIa and harmonin, encoded by Usher syndrome type I genes, form a ternary complex and interact with membrane phospholipids. Hum Mol Genet. 2010;19:3557–3565. doi: 10.1093/hmg/ddq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SD. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin VIIA gene. Nat Genet. 1997;17:268–269. doi: 10.1038/ng1197-268. [DOI] [PubMed] [Google Scholar]
- 30.Liu XZ, Walsh J, Mburu P, Kendrick-Jones J, Cope MJ, Steel KP, Brown SD. Mutations in the myosin VIIA gene cause non-syndromic recessive deafness. Nat Genet. 1997;16:188–190. doi: 10.1038/ng0697-188. [DOI] [PubMed] [Google Scholar]
- 31.Weil D, Kussel P, Blanchard S, Levy G, Levi-Acobas F, Drira M, Ayadi H, Petit C. The autosomal recessive isolated deafness, DFNB2, and the Usher 1B syndrome are allelic defects of the myosin-VIIA gene. Nat Genet. 1997;16:191–193. doi: 10.1038/ng0697-191. [DOI] [PubMed] [Google Scholar]
- 32.Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type VII myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- 33.Self T, Mahony M, Fleming J, Walsh J, Brown SD, Steel KP. Shaker-1 mutations reveal roles for myosin VIIA in both development and function of cochlear hair cells. Development. 1998;125:557–566. doi: 10.1242/dev.125.4.557. [DOI] [PubMed] [Google Scholar]
- 34.Hasson T, Walsh J, Cable J, Mooseker MS, Brown SD, Steel KP. Effects of shaker-1 mutations on myosin-VIIa protein and mRNA expression. Cell Motil Cytoskeleton. 1997;37:127–138. doi: 10.1002/(SICI)1097-0169(1997)37:2<127::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS, Zafar AU, Khan SN, Sabar F, Javid FT, Wilcox ER, Tsilou E, Boger ET, Sellers JR, Belyantseva IA, Friedman TB. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat. 2008;29:502–511. doi: 10.1002/humu.20677. [DOI] [PubMed] [Google Scholar]
- 36.Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, Wayne S, Bellman S, Desmukh D, Ahmed Z, Khan SN, Kaloustian VM, Li XC, Lalwani A, Riazuddin S, Bitner-Glindzicz M, Nance WE, Liu XZ, Wistow G, Smith RJ, Griffith AJ, Wilcox ER, Friedman TB, Morell RJ. Usher syndrome 1D and nonsyndromic autosomal recessive deafness DFNB12 are caused by allelic mutations of the novel cadherin-like gene CDH23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del CSCM, Vila MC, Molina OP, Gal A, Kubisch C. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed ZM, Riazuddin S, Riazuddin S, Wilcox ER. The molecular genetics of Usher syndrome. Clin Genet. 2003;63:431–444. doi: 10.1034/j.1399-0004.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- 39.Schwander M, Xiong W, Tokita J, Lelli A, Elledge HM, Kazmierczak P, Sczaniecka A, Kolatkar A, Wiltshire T, Kuhn P, Holt JR, Kachar B, Tarantino L, Muller U. A mouse model for nonsyndromic deafness (DFNB12) links hearing loss to defects in tip links of mechanosensory hair cells. Proc Natl Acad Sci U S A. 2009;106:5252–5257. doi: 10.1073/pnas.0900691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson SM, Householder DB, Coppola V, Tessarollo L, Fritzsch B, Lee EC, Goss D, Carlson GA, Copeland NG, Jenkins NA. Mutations in Cdh23 cause nonsyndromic hearing loss in waltzer mice. Genomics. 2001;74:228–233. doi: 10.1006/geno.2001.6554. [DOI] [PubMed] [Google Scholar]
- 41.Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, Kachar B, Steel KP, Noben-Trauth K. Mutations in Cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for Usher syndrome type 1D. Nat Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- 42.Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, Gillespie PG, Muller U. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428:950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- 43.Gillespie PG, Muller U. Mechanotransduction by hair cells: models, molecules, and mechanisms. Cell. 2009;139:33–44. doi: 10.1016/j.cell.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Riazuddin S, Griffith AJ, Frolenkov GI, Belyantseva IA, Richardson GP, Friedman TB. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci. 2006;26:7022–7034. doi: 10.1523/JNEUROSCI.1163-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Riazuddin S, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raphael Y, Kobayashi KN, Dootz GA, Beyer LA, Dolan DF, Burmeister M. Severe vestibular and auditory impairment in three alleles of Ames waltzer (av) mice. Hear Res. 2001;151:237–249. doi: 10.1016/s0378-5955(00)00233-1. [DOI] [PubMed] [Google Scholar]
- 47.Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27:99–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- 48.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, Kachar B. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449:87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 49.Verpy E, Masmoudi S, Zwaenepoel I, Leibovici M, Hutchin TP, Del Castillo I, Nouaille S, Blanchard S, Laine S, Popot JL, Moreno F, Mueller RF, Petit C. Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet. 2001;29:345–349. doi: 10.1038/ng726. [DOI] [PubMed] [Google Scholar]
- 50.Jovine L, Park J, Wassarman PM. Sequence similarity between stereocilin and otoancorin points to a unified mechanism for mechanotransduction in the mammalian inner ear. BMC Cell Biol. 2002;3:28. doi: 10.1186/1471-2121-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verpy E, Weil D, Leibovici M, Goodyear RJ, Hamard G, Houdon C, Lefevre GM, Hardelin JP, Richardson GP, Avan P, Petit C. Stereocilin-deficient mice reveal the origin of cochlear waveform distortions. Nature. 2008;456:255–258. doi: 10.1038/nature07380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldstein JL. Auditory nonlinearity. J Acoust Soc Am. 1967;41:676–689. doi: 10.1121/1.1910396. [DOI] [PubMed] [Google Scholar]
- 53.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 54.Friedman LM, Avraham KB. MicroRNAs and epigenetic regulation in the mammalian inner ear: implications for deafness. Mamm Genome. 2009;20:581–603. doi: 10.1007/s00335-009-9230-5. [DOI] [PubMed] [Google Scholar]
- 55.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 58.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 59.Sacheli R, Nguyen L, Borgs L, Vandenbosch R, Bodson M, Lefebvre P, Malgrange B. Expression patterns of miR-96, miR-182 and miR-183 in the development inner ear. Gene Expr Patterns. 2009;9:364–370. doi: 10.1016/j.gep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Modamio-Hoybjor S, Moreno-Pelayo MA, Mencia A, del Castillo I, Chardenoux S, Morais D, Lathrop M, Petit C, Moreno F. A novel locus for autosomal dominant nonsyndromic hearing loss, DFNA50, maps to chromosome 7q32 between the DFNB17 and DFNB13 deafness loci. J Med Genet. 2004;41:e14. doi: 10.1136/jmg.2003.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 62.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brownstein Z, Avraham KB. Deafness genes in Israel: implications for diagnostics in the clinic. Pediatr Res. 2009;66:128–134. doi: 10.1203/PDR.0b013e3181aabd7f. [DOI] [PubMed] [Google Scholar]
- 64.Ben-Yosef T, Ness SL, Madeo AC, Bar-Lev A, Wolfman JH, Ahmed ZM, Desnick RJ, Willner JP, Avraham KB, Ostrer H, Oddoux C, Griffith AJ, Friedman TB. A mutation of PCDH15 among Ashkenazi Jews with the type 1 Usher syndrome. N Engl J Med. 2003;348:1664–1670. doi: 10.1056/NEJMoa021502. [DOI] [PubMed] [Google Scholar]
- 65.Gubbels SP, Woessner DW, Mitchell JC, Ricci AJ, Brigande JV. Functional auditory hair cells produced in the mammalian cochlea by in utero gene transfer. Nature. 2008;455:537–541. doi: 10.1038/nature07265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shibata SB, Raphael Y. Future approaches for inner ear protection and repair. J Commun Disord. 2010;43:295–310. doi: 10.1016/j.jcomdis.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, Ahmed ZM, Riazuddin S, Khan SN, Friedman TB. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, Kanaan M. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010;68:293–308. doi: 10.1016/j.neuron.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 71.Friedman LM, Dror AA, Avraham KB. Mouse models to study inner ear development and hereditary hearing loss. Int J Dev Biol. 2007;51:609–631. doi: 10.1387/ijdb.072365lf. [DOI] [PubMed] [Google Scholar]
- 72.Dror AA, Avraham KB. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet. 2009;43:411–437. doi: 10.1146/annurev-genet-102108-134135. [DOI] [PubMed] [Google Scholar]