Abstract
Advances in cell and gene therapy are opening up new avenues for regenerative medicine. Because of their acquired pluripotency, human induced pluripotent stem cells (hiPSCs) are a promising source of autologous cells for regenerative medicine. They show unlimited self-renewal while retaining the ability, in principle, to differentiate into any cell type of the human body. Since Yamanaka and colleagues first reported the generation of hiPSCs in 2007, significant efforts have been made to understand the reprogramming process and to generate hiPSCs with potential for clinical use. On the other hand, the development of gene-editing platforms to increase homologous recombination efficiency, namely DNA nucleases (zinc finger nucleases, TAL effector nucleases, and meganucleases), is making the application of locus-specific gene therapy in human cells an achievable goal. The generation of patient-specific hiPSC, together with gene correction by homologous recombination, will potentially allow for their clinical application in the near future. In fact, reports have shown targeted gene correction through DNA-Nucleases in patient-specific hiPSCs. Various technologies have been described to reprogram patient cells and to correct these patient hiPSCs. However, no approach has been clearly more efficient and safer than the others. In addition, there are still significant challenges for the clinical application of these technologies, such as inefficient differentiation protocols, genetic instability resulting from the reprogramming process and hiPSC culture itself, the efficacy and specificity of the engineered DNA nucleases, and the overall homologous recombination efficiency. To summarize advances in the generation of gene corrected patient-specific hiPSCs, this review focuses on the available technological platforms, including their strengths and limitations regarding future therapeutic use of gene-corrected hiPSCs.
Introduction: Regenerative Medicine—Cell Plus Gene Therapy
Regenerative medicine aims to replace and/or to regenerate damaged cells, organs, or tissues in order to restore normal function. Cell therapy is an important regenerative medicine approach, in which either differentiated cells or stem cells capable of differentiation are transplanted into an individual with the objective of yielding specific cell types present in the damaged tissue and consequently restoring its function. The most successful example of cell therapy is bone marrow (BM) transplantation, in which the transplanted hematopoietic stem cells (HSCs) are able to regenerate the patient's blood. BM transplantation started in the 1950s and now is a widely established procedure for many hematopoietic diseases (Thomas et al., 1977). Cell therapies for other tissues then followed in the footsteps of the hematopoietic experience. Nowadays, there are numerous ongoing clinical trials using various types of stem cells and some of them are U.S. Food and Drug Administration (FDA)-approved cell-based products (www.fda.gov/BiologicsBloodVaccines/CellularGeneTherapyProducts/ApprovedProducts/default.htm).
Cell replacement can be done with autologous or allogeneic stem cells. When performing allogeneic cell therapy, the risk of immune rejection usually requires the use of immunosuppressive drugs, which can induce toxicity and increase the risk of infections and cancer, which could be life-threatening. This, together with the low availability of suitable donors, makes autologous cell therapy frequently the preferred option for regenerative medicine. However, in the case of monogenic diseases, in which all the cells from the body initially carry the disease-causing mutation in their genomic DNA, a gene correction approach should be considered to generate disease-free autologous cells. Thus, a combination of cell and gene therapy is used. Since the first gene therapy clinical trial in 1990 (Anderson et al., 1990), much effort has been made to develop safer and more efficient approaches.
The first gammaretroviral vectors used in clinical trials were associated with enhancer-mediated cis- and trans-activation, which induced insertional mutagenesis and ended up in leukemia associated with the gene therapy procedure (Hacein-Bey-Abina et al., 2003, 2008; Fischer et al., 2010). In the ongoing clinical trials, lentiviral and retroviral vectors have deletions in their long terminal repeats (LTRs) to minimize trans-activation of the genes surrounding the integration. However, insertional mutagenesis still remains an issue. Other strategies, such as the use of nonintegrating viral vectors, are being studied, as in the case of integration-defective lentiviral vectors (Yanez-Munoz et al., 2006; Matrai et al., 2011), but procedures to maintain nonintegrated DNA in proliferating cells have not yet been developed. The introduction of genetic material in specific, known, and characterized loci of the genome via homologous recombination (HR) would be an ideal option. HR will allow, in principle, specific correction of the mutation without any additional modification in the genome, or introduction of the genetic material in a known and safe genome locus. HR is based on the natural DNA repair process, in which a double-strand break (DSB) is corrected with a homologous DNA sequence. The therapeutic application of HR involves exchanging the mutation for the correct sequence, or even introducing the correct version of the gene in the targeted locus.
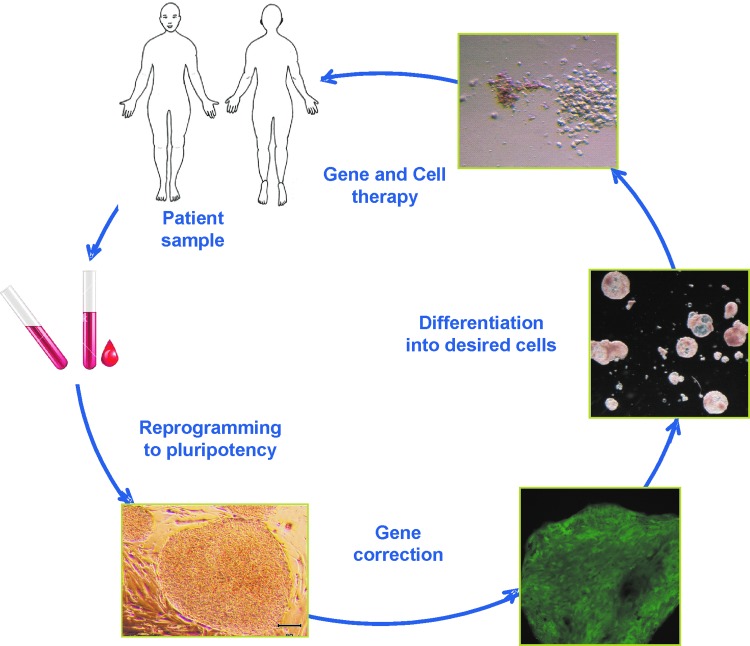
An important consideration is the source of cells for autologous cell therapy. For some purposes, as is the case for hematological diseases, a hematopoietic multipotent stem/progenitor cell present in the adult body can be used. Other examples of these kinds of progenitors in humans include neural stem cells (Galli et al., 2003), mesenchymal stem cells (Deans and Moseley, 2000), and intestinal stem cells (Yui et al., 2012). In the majority of these adult stem cells, an important limitation is that correction of mutations by HR has rarely been described to occur in a manner that retains the multipotentiality of the stem cells. In addition, these kinds of progenitors have been described for only a few tissues in the body. Thus, an autologous stem cell source with wide expansion and differentiation potential is required for future clinical use of HR in the context of regenerative medicine. This issue has been solved with the generation of human induced pluripotent stem cells (hiPSCs) (Takahashi et al., 2007). Human iPSCs offer a powerful novel technology in gene and cell therapies. Their essentially unlimited growth capability allows successfully targeted cell selection, with the possibility that 100% of potentially transplanted cells would be corrected. The fact that they represent a clonal cell population is also advantageous as we can completely interrogate the whole exome or the whole genome for any abnormality that could be accumulated during the entire manipulation procedure, as has been addressed in several reports (Table 1 and Fig. 1). We deal with some of these pioneer works involving hiPSCs and HR technologies in this review.
Table 1.
Human Diseases from Which Human Induced Pluripotent Stem Cells Have Been Generated or Generated and Corrected
| TF delivery method | Efficiency/rounds | Integrative | Cell typea | Disease, phenotype correctiona |
|---|---|---|---|---|
| Lenti/retroviruses | 0.01–1/single | Yes | Fibroblasts (1, 2) B lymphocytes (3) Human adipose stem cells (4) BM-MSCs (5) CD34 progenitors (6) BM-MNCs (7) |
• Not corrected: Leopard syndrome (23), long QT syndrome (24), Timothy syndrome (25), ADA-SCID, Gaucher disease, Duchenne and Becker muscular dystrophy, type 1 diabetes mellitus, trisomy 21, Lesch–Nyhan syndrom (26), CINCA syndrome (27), familial hypercholesterolemia (28), Rett syndrome (29), dyskeratosis congenita (30, 31), familial dilated cardiomyopathy (32), chronic myelogenous leukemia (33), Alzheimer's disease (34), glucose-6-phosphatase deficiency, Crigler–Najjar syndrome, hereditary tyrosinemia type 1 (35) • Corrected by lentiviruses: Fanconi anemia (36) • Corrected by gene editing: Progeria (37), sickle cell anemia (13). X-linked chronie granulamatosis (5). Huntington's disease (38), β-thalassemia (39) |
| Adenoviruses | ∼0.0002/multiple | No | Fibroblasts (8) | |
| Sendai virus | ∼1/single | No | Fibroblasts (9) T cells (10) CD 34 progenitors (11) |
• Corrected by gene editing: α1-Antitrypsin deficiency (40) |
| Excisable lentivirus | 0.01–1/single | Removable | Fibroblasts (12–14) CD34 progenitors (15) |
• Not corrected: Cystic fibrosis (41) • Corrected by lentivirus: β- Thalassemia (12) • Corrected by gene editing: Parkinson's disease (42) |
| Excisable transposons | ∼0.1/single | Removable | Fibroblasts (16) BM-MSCs (5) |
• Corrected by gene editing: Sickle cell anemia (5) |
| Minicircles | ∼0.005/multiple | No | Adipose-derived stromal cells (17) | |
| Episomal vectors | 0.001/multiple | No | Fibroblasts (18) BM- and CB-MNCs (19) |
• Not corrected: Neoplasic bone marrow (19) • Corrected by gene editing: Gyrate atrophy (43) |
| Proteins | 0.001/multiple | No | Fibroblasts (20) | |
| miRNAs | 0.1/single | Yes | Fibroblasts (21) | |
| mRNAs | 1–4/multiple | No | Fibroblasts (22) | • Not corrected: Cystic fibrosis (22) |
ADA-SCID, adenosine deaminase-deficient severe combined immunodeficiency; BM-MNCs, bone marrow-derived mononuclear cells; BM-MSCs, bone marrow-derived mesenchymal stem cells; CINCA syndrome, chronic infantile neurological cutaneous and articular syndrome; miRNAs, micro-RNAs, micro-RNAs; TF, transgene free.
References: (1) Takahasi et al. (2007); (2) Yu et al. (2007); (3) Hanna et al. (2008); (4) Sun et al. (2009); (5) Zou et al. (2011a); (6) Ye et al. (2009); (7) Kunisato et al. (8) Zhou and Freed (2009); (9) Fusaki et al. (2009); (10) Seki et al. (2011); (11) Ban et al. (2011); (12) Papapetrou et al. (2011); (13) Sebastiano et al. (2011); (14) Soldner et al. (2009); (15) Ramos-Mejia et al. (2012); (16) Kaji et al. (2009); (17) Jia et al. (2010); (18) Yu et al. (2009); (19) Hu et al. (2011); (20) D. Kim et al. (2009); (21) Anokye-Danso et al. (2011); (22) Warren et al. (2010); (23) Carvajal-Vergara et al. (2010); (24) Moretti et al. (2010); (25) Yazawa et al. (2011); (26) Park et al. (2008); (27) Tanaka et al. (2012); (28) Cayo et al. (2012); (29) K.Y. Kim et al. (2011); (30) Agarwal et al. (2010); (31) Batista et al. (2011); (32) Sun et al. (2012); (33) Kumano et al. (2012); (34) Israel et al. (2012); (35) Rashid et al. (2010); (36) Raya et al. (2009); (37) Liu et al. (2011); (38) An et al. (2012); (39) Wang et al. (2012); (40) Yusa et al. (2011); (41) Somers et al. (2010); (42) Soldner et al. (2011); (43) Howden et al. (2011).
FIG. 1.
Gene correction approach for a hematopoietic disease, using induced pluripotent stem cells. Color images available online at www.liebertpub.com/hum
Generation of Patient-Specific Pluripotent Stem Cells
Choice of reprogramming platform
Since Yamanaka and colleagues first reported the generation of mouse iPSCs in 2006 (Takahashi and Yamanaka, 2006), and later the groups of Yamanaka (Takahashi et al., 2007) and Thomson (Yu et al., 2007) in human cells in 2007, many laboratories around the world have been able to reprogram a large variety of somatic cells into pluripotent stem cells, from neural stem cells (J.B. Kim et al., 2009) to terminally differentiated B lymphocytes (Hanna et al., 2008). The reproducibility and potentiality (unlimited self-renewal and ability to differentiate into any cell type) of these cells has caused the iPSC field to advance rapidly. hiPSC technology brings together all the potential of human embryonic stem cells (hESCs) in terms of self-renewal and pluripotency without the problems associated with hESC generation (i.e., ethical issues associated with embryo disruption and immunoincompatibility with the recipient of the cells). Therefore, hiPSC technology arises as one of the most promising fields for future cell therapies for many human diseases.
For the generation of hiPSCs, the first reports used gammaretroviruses to express the four defined factors required for reprogramming, OCT4, SOX2, KLF4, and c-MYC (Takahashi et al., 2007), or OCT4, LIN28, NANOG, and SOX2 (Yu et al., 2007), separately in different viral vectors. Because of the nature of the vectors, expression of the factors was silenced after the endogenous pluripotent genes were activated at an adequate level. Safer and more efficient reprogramming approaches have since been developed and many patient-specific hiPSCs have been generated both to model human diseases and correct the diseased hiPSCs through gene therapy approaches. Depending on the cell type being reprogrammed, the number of factors used could be reduced and, more importantly, oncogenes or tumor-related proteins used for reprogramming, such as c-MYC or KLF4, could be removed from the original reprogramming cocktail. This, for example, was the case for reprogramming hematopoietic progenitors (Liu et al., 2012; Meng et al., 2012) or neural stem cells (J.B. Kim et al., 2009), in which these two factors could be removed. Even reprogramming with OCT4 alone was achieved (Thier et al., 2010). However the reprogramming efficiency decreases after removing any reprogramming factor. Although avoiding tumor-related genes increases the safety of the reprogramming process, the safest reprogramming protocol will ultimately involve removable reprogramming transgenes or, even better, nonintegrative systems that will avoid potential adverse effects associated with the integration of the vector sequences in the cell genome (Sommer et al., 2010). Several groups have developed Cre-mediated excisable polycistronic lentiviral vectors (Somers et al., 2010; Papapetrou and Sadelain, 2011) or transposon-based reprogramming systems (Woltjen et al., 2011), which could be removed after obtaining the hiPSC clones. The first truly nonintegrative reprogramming approach described in human cells was reported by the Thomson group using episomal plasmids for expression of the four Yamanaka transcription factors plus NANOG, LIN28, and SV40 large T antigen (SVLT) (Yu et al., 2009). In the same year, reprogramming by recombinant proteins (D. Kim et al., 2009), synthetic mRNAs (Warren et al., 2010), and nonintegrating RNA Sendai viral vectors (Fusaki et al., 2009) was also reported (Table 1). The majority of disease-specific hiPSCs reported until now have been generated with integrative systems, but an increasing number of disease-specific hiPSCs have been generated using these novel and potentially safer approaches (Table 1).
Choice of cell source for reprogramming
As reported by Hanna and colleagues, an adequate level of expression of the reprogramming factors in any cell type would likely allows the creation of an iPSC line (Hanna et al., 2009). The preferred cell source for reprogramming will most likely be the most easily accessible and the one in which the reprogramming factors can be successfully delivered. That is why fibroblasts have been widely used by many groups (Takahashi et al., 2007; Park et al., 2008; Fusaki et al., 2009; Carvajal-Vergara et al., 2010; Liu et al., 2010; Howden et al., 2011; Papapetrou et al., 2011; Sebastiano et al., 2011; Tanaka et al., 2012). Fibroblasts can be easily grown from a small human biopsy and can be efficiently transduced with viral vectors. Another cell source that can be easily obtained and presents several advantages are peripheral blood mononuclear cells (PB-MNCs) (Kunisato et al., 2010). These cells can be obtained from routine blood tests or in patient follow-up, can be frozen and stored, and are easily cultured; in addition, stimulation of the preferred cell type within the PB-MNCs by cytokines is possible. Jaenisch's group (Staerk et al., 2010) showed that by stimulation of PB-MNCs with granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin (IL)-3, and IL-6 before and during the first days of reprogramming, the induction of pluripotency could be prompted in progenitors and cells of myeloid origin, avoiding reprogramming of B or T cells.
However, there are important questions that remain unsolved. For example, is the cell source origin going to influence the characteristics of its corresponding hiPSCs? Several groups have compared hESCs and hiPSCs at the gene expression level and for their genome methylation status. Some authors have found significant differences between hiPSCs and hESCs, including an epigenetic memory of the original cell source in the hiPSCs (Chin et al., 2009; Deng et al., 2009; Doi et al., 2009; Marchetto et al., 2009; Ghosh et al., 2010). Other groups attributed these differences to the intrinsic differences between various hiPSC clones of the same reprogramming experiment or to the various technological platforms that have been used for reprogramming (Guenther et al., 2010; Newman and Cooper, 2010; Bock et al., 2011). This issue has been deeply analyzed, and it has been pointed out that the small number of clones analyzed, in the studies in which a large difference is observed, could have negatively influenced the conclusions (Yamanaka, 2012). In the event that epigenetic memory is proven to be true, would the original cell source have an influence on the differentiation capacity of a specific hiPSC line? According to K. Kim and colleagues, hiPSCs derived from cord blood cells showed a hematopoietic differentiation advantage when compared with hiPSCs derived from keratinocytes (K. Kim et al., 2011). On the other hand, other authors did not find epigenetic memory in hiPSC lines derived from hepatocytes (Ohi et al., 2011). This issue, clearly important for therapeutic applications, will require further study in order to determine to what extent the ultimate transplantable cell type should influence the source of patient-specific cells for reprogramming. As this issue remains unclear, we believe that the cell source should be, first, the most accessible and least invasive, and then, depending on the future use of the hiPSCs, an epigenetically related cell source should be considered if available. This is the case of hiPSCs for cell therapy of blood diseases, in which either fibroblasts or PB-MNCs could be used, based on their accessibility, but PB-MNCs may perhaps prove to be a better option provided they exhibit a differentiation advantage into the hematopoietic lineage.
Correction of Patient-Derived Pluripotent Stem Cells
At present, various strategies have been tested and proven for the correction of patient-specific hiPSCs. Although this review is focused on genetic correction directly in hiPSCs, this is not always achievable, because some genetic diseases imply a reprogramming barrier, as has been the case for Fanconi anemia (FA). In this case, genetic correction was carried out before the generation of FA-hiPSCs (Raya et al., 2009). All the approaches described in this review could also be done before generating patient-specific hiPSCs if the cells of origin allow the culture needed for the genetic correction and selection of corrected cells.
Random integration
The first reports of correction of patient-derived hiPSCs used lentiviral vectors to correct the disease through transgene addition (Raya et al., 2009). However, these vectors, because of their nearly random integration pattern in the genome, are susceptible to transcriptional silencing, depending on whether the integration site resides in a silent or active transcriptional region of chromatin. Furthermore, as mentioned previously, integrated vectors may show enhancer-mediated cis- and trans-activation and might consequently induce insertional mutagenesis. The identification of integrations in safe harbor genomic sites (e.g., far away from genes or coding information) could represent an alternative, safer mode of therapy. The self-renewal and almost indefinite growth properties of hiPSCs enable analysis of the integration sites of these vectors at a clonal level and the selection of those that could be potentially safer (Papapetrou et al., 2011; Bedel et al., 2012). However, the definition of a safe harbor site in the genome is challenging, and it will probably change as we get to know the genome in more depth. Future therapeutic applications of hiPSCs for cell therapy would benefit from a site-specific gene correction approach. The cooperation between hiPSC technology and homologous recombination (HR) has been extensively explored. HR is presented as an exciting and novel alternative to avoid insertional mutagenesis associated with integrative vector-mediated correction.
Site-specific gene editing
Gene editing is a process in which a DNA sequence is replaced or introduced into a specific locus at single-base pair resolution. This precise site-specific introduction requires an accurate recognition mechanism of the target site on the genome. Under normal conditions, maintenance of the integrity of the genome requires repair of the continuous cellular DNA damage with high fidelity. HR is a truly accurate DNA repair mechanism that is basically a “copy and paste” mechanism and is also used to resolve double-strand breaks (DSBs) in the DNA. This process uses an undamaged homologous segment of DNA as a template (conventionally, the sister chromatid) to copy the information across the DSB. Because it copies a normal copy of the undamaged DNA, HR is the most secure process by which to repair DSBs. The fidelity of HR gives the specificity and accuracy that gene editing requires.
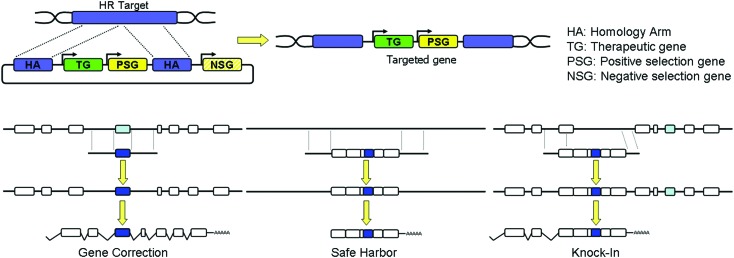
The natural HR process has been exploited by researchers to achieve the desirable site-specific gene editing within a targeted locus by introducing exogenous genomic sequences, homologous to the target locus, flanking the desired DNA material to be inserted. These techniques have been widely used for the generation of knock-out and knock-in transgenic animals (Robbins, 1993). Routinely, homology arms are homologous DNA sequences that cover the target where HR will take place. Between these two arms, a therapeutic or correct sequence of the gene should be found. In addition, drug resistance genes can be introduced between both homology arms for positive selection or suicide genes outside of the homology arms for negative selection. The final structure and complexity of this construction, also called the repair matrix, will vary according to the needs of the researcher (Fig. 2). With the development of disease-specific hiPSCs, this methodology has already been used to correct mutations (Howden et al., 2011; Liu et al., 2011; Ohi et al., 2011).
FIG. 2.
Scheme of the repair matrix, pointing out the various required elements, and of the various disease correction strategies for homologous recombination (HR). Color images available online at www.liebertpub.com/hum
Gene editing via HR in human cells is inefficient and dependent on the generation of a DSB at the specific target site (Carroll, 2011). In the absence of a repair matrix, nonhomologous end joining (NHEJ) is the dominant pathway to solve these DNA lesions in human cells, and its resolution is highly error-prone (Grabarz et al., 2012). In addition, HR varies in different cell types and requires transit through the S–G2 phase of the cell cycle to take place (Delacote and Lopez, 2008). These limitations have typically made gene editing in human cells difficult to achieve. Various approaches have been used to improve gene editing by HR, such as the increase in the length of the DNA sequences homologous to the target site (homology arms) (Song et al., 2010), the use of adeno-associated vectors to more efficiently introduce the repair matrix in the cells (Khan et al., 2010), and the improvement of selection methods for the identification of correctly edited cells, or the stimulation of HR by inducing DSBs using specific DNA nucleases. The use of engineered DNA nucleases that recognize specific sites of the genome is an active area of investigation and is the most commonly reported method for correction of patient-specific hiPSCs.
Engineered DNA nucleases are enzymes that have been developed to induce DSBs specifically at a unique and defined sequence in the cell genome. The rationale for inducing the double-stranded DNA break in the immediate vicinity of the mutant sequences is that these DSBs have been shown to increase the efficiency of homology-directed repair (HDR) by 103- to 104-fold (Porteus and Carroll, 2005). Engineered DNA nucleases are formed by a nuclease domain and a DNA-binding domain, the sequence specificity of which can be artificially modified. The most widely used DNA nucleases are zinc finger nucleases (ZFNs), homing meganucleases (MNs), and transcription activator-like (TAL) effector nucleases (TALENs). They potentially identify a unique sequence within the genome and generate DSBs to induce the recruitment of the cell repair machinery to repair the DSBs, ideally by HR. The DNA-binding domain of ZFNs is derived from DNA-binding zinc finger proteins and is composed of a tandem repeat of Cys2His2 zinc fingers, each of which recognizes three nucleotides. The DNA-binding domain is linked to the nuclease domain of the restriction enzyme FokI. ZFNs work as pairs of two monomers of ZFN in reverse orientation. This ZFN dimer can be designed to bind to a genomic sequence 18–36 nucleotides in length (Porteus and Carroll, 2005; Carroll, 2011). TALENs have a similar structure to ZFNs, but the DNA-binding domain comes from TAL effector proteins. The DNA-binding domain in TALENs is a tandem array of amino acid repeats. Each of these units is able to bind to one of the four possible nucleotides. Thus, the DNA-binding domain can be designed to recognize any desired genomic sequence. TALENs also cleave as dimers (Li et al., 2011). In contrast to these synthetic DNA nucleases, natural MNs are a subset of homing endonucleases. MNs are monomeric proteins that have four DNA-binding domains that recognize a DNA sequence from 14 to 40 nucleotides in length. Directed mutagenesis can be applied to modify the DNA sequence specificity. A unique and specific MN recognition sequence can be found approximately every 300 bp (Paques and Duchateau, 2007).
ZFNs were first developed in 1996 by Y.G. Kim and colleagues and applied for disrupting gene expression by introducing mutations in the selected gene (Kim et al., 1996). More recently, they have been widely used for gene editing in hESCs and hiPSCs. For example, Lombardo and colleagues showed the insertion of the gene encoding green fluorescent protein (GFP) into the CCR5 safe harbor locus in hESCs after inducing HR by ZFN expression; targeted hESCs were able to differentiate into neurons keeping GFP expression (Lombardo et al., 2007). TALENs have been also tested in hESCs and hiPSCs (Hockemeyer et al., 2011). One of the most important potential disadvantages of engineered nucleases is the possibility of their cutting other, related sequences of the genome, the so-called off-target sites. After targeting several loci and comparing HR efficiencies and the presence of off-targets with both types of nucleases, these authors concluded that both ZFNs and TALENs show similar efficiencies and accuracy (Hockemeyer et al., 2011). Nevertheless, an in vitro gene disruption comparison between ZFNs and TALENs showed that TALENs were more efficient and less cytotoxic in this assay (Mussolino et al., 2011). Other authors have reported that targeted efficiency by various nucleases seems to be affected by the epigenetic status of the locus to be targeted (Daboussi et al., 2012). The presence of methylated CpGs (mCpGs) in the TALEN recognition site can dramatically decrease its efficiency (Valton et al., 2012b) and in the case of MNs this also happens if the mCpG is in the central tetrabase of the recognition site (Valton et al., 2012a)
The proof of principle for the clinical application of nuclease-mediated gene editing was tested in hiPSCs from patients affected by various genetic diseases some time later. To correct or insert/express a transgene by HR, three different strategies can be considered (Fig. 2 and Table 2). In the following sections we discuss the various attempts applied for nuclease-based correction.
Table 2.
Summary of the Various Gene Correction Strategies in Terms of Safety and Applicability
| Safety | Recommended scenario | Contraindicated scenario | References | ||
|---|---|---|---|---|---|
| Random integration | Random integration | Risk of insertional mutagenesis | Simple methodology Avoids DSB Promoter mutations |
Negative effect of mutated gene product | Raya et al. (2009) |
| Random integration with safe harbor (SH) clone selection | Definition of safe harbors not clear | Avoids DSB Promoter mutations |
Negative effect of mutated gene product | Bedel et al. (2012); Papapetrou et al. (2011) | |
| Gene editing | Targeted safe harbor integration | Safe for AAVS1 Risks for newly defined safe harbors |
Promoter mutations Many described mutations | Negative effect of mutated gene product Transgene needs to be tightly regulated |
Chang and Bouhassira (2012); Zou et al. (2011b) |
| Targeted knock-in | Safe | Many described mutations within an area of the gene Transgene needs to be tightly regulated |
In some cases incompatible with correction of many isoforms | — | |
| Targeted gene correction | Safe | Unique common mutation in many patients Negative effect of mutated gene product Transgene needs to be tightly regulated Many gene isoforms |
Many described mutations | An et al. (2012); Howden et al. (2011); Liu et al. (2011); Sebastiano et al. (2011); Soldner et al. (2011); Wang et al. (2012); Yusa et al. (2011); Zou et al. (2011a) |
AAVS1, adeno-associated virus integration site 1; DSB, double-strand break.
Targeted safe harbor integration
For safe harbor integration, a complete expression cassette (the therapeutic transgene, promoter, and possibly additional regulatory signals [e.g., enhancer]) is inserted into a specific genome locus that is not susceptible to transgene silencing via epigenetic mechanisms. Ideally, the targeted integration will either not affect expression of the neighboring genes or at least allow modified cells to function normally if the targeting results in disruption of the safe harbor locus. This seems to be the case for AAVS1, CCR5, and ROSA26 loci (Irion et al., 2007; Torres et al., 2011; Yao et al., 2011). Special attention should be taken to avoid targeting loci previously considered nonfunctional but now known to fulfill important regulatory functions as per ENCODE (the Encyclopedia of DNA Elements). One potential advantage of the safe harbor strategy is that there should be significantly less cell-to-cell variation in transgene expression than that resulting from random integration. Although the addition of a promoter to express the therapeutic gene is needed, the main advantage of the safe harbor strategy is the wide variety of diseases that could be treated with a similar repair matrix, only exchanging the therapeutic gene for each disease. Examples are as follows:
X-Linked Chronic Granulomatous Disease: Seminal work published by Malech's group in March 2011 showed, for the first time, ZFN-mediated phenotype correction in neutrophils generated from X-linked chronic granulomatous disease (X-CGD) hiPSCs by inserting a wild-type copy of the CYBB gene (encoding the gp91phox protein) driven by the CAG (cytomegalovirus early enhancer/chicken β-actin) chimeric promoter in the previously described AAVS1 safe harbor locus (Zou et al., 2011b). Puromycin selection was also included in the inserted DNA to select recombined clones. In addition, in some of the AAVS1 alleles that were not targeted, there were mutations associated with NHEJ correction, evidence for cleavage by ZFNs at this site. Having a high number of targeted clones makes it possible to select and grow just those that show no off-target integrations or new mutations. Importantly, after differentiation of the corrected X-CGD hiPSCs, the resulting neutrophils showed equal levels of therapeutic reactive oxygen species (ROS) to neutrophils derived from wild-type hiPSCs.
β-Thalassemia: To achieve a more physiological expression level of the transgene, Chang and Bouhassira (2012) used the specific β-globin promoter for directing expression of the transgene when targeted into the AAVS1 locus. After puromycin selection, all the clones analyzed were targeted at the AAVS1 locus and 50% represented homozygous targeting (i.e., targeting into both AAVS1 loci) as assessed by PCR and Southern blot. Erythroid differentiation of corrected clones showed restoration of hemoglobin quantity and quality without disturbing any AAVS1 locus-neighboring genes.
Targeted correction
Targeted correction typically uses site-specific nucleases designed to recognize a site in the immediate vicinity of the mutation targeted for correction together with a repair matrix precisely matching that of the targeted endogenous sequences, with the exception of the base or bases intended for alteration. The mutant target bases are substituted for by the wild-type bases present in the introduced repair matrix, thus correcting or repairing the gene. In repairing the defective sequence within the endogenous gene locus, the corrected genetic material is maintained within its normal chromatin environment. This ensures the appropriate genetic regulation and expression in the cell. In situations in which the mutant gene product exercises a dominant negative influence over the normal gene product, gene correction may be the only suitable strategy. Gene correction is especially useful for diseases in which the majority of patients have the same well-defined, limited alterations in the DNA sequence, such as sickle cell anemia or cystic fibrosis. When different mutations for the same gene have been reported, gene correction would turn into a patient-specific therapy and therefore the repair matrix, and also perhaps the site-specific nucleases, should be tailor-made for each patient or set of patients. Examples of this approach are as follows:
Parkinson's Disease: hiPSCs from a patient with the A53T mutation in the α-synuclein gene were corrected by ZFN-assisted HR in the mutated locus. In this case, the targeting sequence in the donor vector was approximately 1 kb of the wild-type sequence of the α-synuclein gene with the targeted mutant base in the middle, close to the ZFN cleavage site. As there was no selection cassette in the donor, a selection-free approach was mandatory and the number of clones that had to be analyzed to obtain a correctly targeted clone was higher than with selection-based approaches. This procedure could also be seen as an advantage, as there was just one clonal step instead of two or three, therefore reducing the manipulation steps and the probability of additional genetic alterations.
Sickle Cell Anemia: Two studies have been published for the genetic correction of hiPSCs from patients with sickle cell anemia (Sebastiano et al., 2011; Zou et al., 2011a). The first report showed specific ZFN-mediated gene correction of the βs (A>T) mutation in the HBB locus (Zou et al., 2011a). The authors used a donor vector with a loxP-flanked (“floxed”) hygromycin resistance selection cassette that, after nucleofection of the ZFNs and the repair matrix, allowed the detection of hygromycin-resistant clones. PCR analysis and Southern blotting verified the presence of hiPSC clones correctly targeted within the HBB locus and in no additional loci. After erythrocyte differentiation of the corrected hiPSCs, the authors suspected that the presence of the selection cassette affected expression of the corrected transcript. To avoid possible interference, the selection cassette was excised in 4 of 24 clones by Cre recombinase. Surprisingly, in those clones in which the selection cassette was excised, expression of the corrected gene was still only 25–40% of the expression of the uncorrected allele. The authors speculate that the reduced expression level could be due to two main reasons: either the presence of the remaining loxP sequences after excision of the selection cassette, or the presence of a nucleotide variant (A>G) affecting a GATA-containing 3′ enhancer that may have been generated during HR. This study points out that a selectable cassette could have clear benefits in reducing the number of clones to be analyzed, but it could potentially adversely affect the intended correction by repressing the expression of the transgene unless excised. This work also highlights the importance of investigating the possible acquisition of genetic modifications during reprogramming and/or HR because such mutations could influence the behavior of the corrected hiPSCs. The other gene correction approach for the sickle cell mutation βs was published by Sebastiano and colleagues, following a similar selection-based approach (Sebastiano et al., 2011). They achieved efficient targeting and showed no additional modifications in the nontargeted allele due to NHEJ and no off-target modifications.
β-Thalassemia: The correction of mutations in the β-globin gene was also addressed by Wang and colleagues, who performed genetic correction by ZFN-assisted HR (Wang et al., 2012) and also applied a drug selection procedure to increase targeting efficiency. They were able to differentiate the corrected hiPSCs, as well as uncorrected hiPSCs, to hematopoietic progenitors. Moreover, human β-globin was detected in the peripheral blood of immunodeficient mice transplanted with the corrected hiPSC-derived hematopoietic progenitors, confirming the genetic correction of β-thalassemia.
α1-Antitrypsin Deficiency: ZFN-mediated gene correction was also performed at the α1-antitrypsin (A1AT) locus to correct A1AT deficiency (A1ATD) in hiPSCs derived from a patient with the Glu342Lys point mutation. This approach used a puromycin resistance cassette flanked by piggyBac inverted repeats. Subsequently, the selection cassette was removed from the homozygously targeted clones by piggyBac transposase, obtaining corrected hiPSC clones without any residual sequence footprint. Corrected, excised hiPSC clones were subsequently differentiated into hepatocyte-like cells, confirming the successful correction of A1ATD (Yusa et al., 2011).
Targeted knock-in
In the targeted knock-in strategy, a full or partial cDNA of the therapeutic transgene gene is directly introduced into the endogenous mutant gene locus, generally near the start of the gene in order to precede all or the majority of the mutant exons. Typically, splicing signals are incorporated into the transgene sequences such that expression of the introduced cDNA is regulated by the endogenous regulatory elements of the locus where it is inserted. In principle, this strategy maintains the genetic regulation of the gene and it is applicable to diseases in which a large number of distinct gene mutations occur (in contrast to a single mutant genotype responsible for a significant majority of patients). The knock-in strategy is a highly versatile HR strategy capable, in principle, of treating a large number of patients using a single set of gene-modifying tools (i.e., site-specific nucleases and repair matrices), while preserving the endogenous regulation of the therapeutic gene. Although this strategy has been used to express marker genes led by endogenous promoters (Hockemeyer et al., 2009, 2011; Wang et al., 2011), there is not yet any reported example of this strategy for the correction of patient-specific disease hiPSCs.
We have successfully generated, using Sendai vectorized reprogramming factors, hiPSCs from patients with pyruvate kinase deficiency (PKD), who suffer from nonspherocytic hemolytic anemia. We are pursuing a correction strategy that is capable, in principle, of treating all PKD patients with mutations from exon 3 to the end of the PKLR gene by developing an appropriate repair matrix. Moreover, expression of the corrected R-type pyruvate kinase (RPK) transcript will be regulated under the control of the endogenous PKLR promoter after the knock-in of the partial RPK cDNA into intron 2. If successful, only the corrected RPK protein should be expressed in red blood cells.
Selection of one of these previously described strategies (see the sections Targeted Safe Harbor Integration, Targeted Correction, and Targeted Knock-In, above) requires consideration of both the disease and the number of patients in whom this strategy could be used. For each of these strategies, the type of therapeutic matrix to be used will be different (see Fig. 2).
Risks of Genome Alteration
One of the most important issues in using the aforementioned novel methodologies (i.e., epigenetic reprogramming and site-specific gene correction) will concern ensuring the integrity of the chromosomal DNA. Even though these methodologies have been employed only in the limited number of studies cited previously, it is already clear that genetic abnormalities may be introduced into the hiPSCs either through the reprogramming process, the tissue culture expansion, and/or the gene correction process itself (Blasco et al., 2011; Gore et al., 2011; Pera, 2011).
In the study of gene correction of A1ATD hiPSCs, a complete genome integrity study was performed. Comparative genomic hybridization confirmed that reprogramming and prolonged culture generated amplifications or deletions ranging from 20 kb to 1.3 Mb. But, importantly, there was one corrected line of three lines examined that retained a normal genome. These authors also detected genetic alterations in 2 of 6 lines after HR correction and in 4 of 16 lines after the excision process. They concluded that more genetic alterations were generated during the reprogramming process and the extensive culture of the hiPSCs than during the HR correction (Yusa et al., 2011).
In the study of α-synuclein gene correction (Soldner et al., 2011), the authors examined the hiPSC lines for copy number variation (CNV), because CNVs were previously reported to commonly result from reprogramming as well as from prolonged pluripotent stem cell culture. The authors saw on average 77 CNVs per cell line with an average size of 158 kb. These genetic alterations were most likely generated during reprogramming and culture as there were no substantial differences after gene editing and excision. They also performed whole genome expression array analysis before and after correction and did not detect any expression pattern differences related to gene targeting, indicating that reprogramming itself had a greater impact on genome integrity than the gene-targeting procedures.
In addition to the risks of genomic alteration, it should also be taken into account that a considerable number of hiPSC differentiation protocols include the forced expression of tissue-specific transcription factors (Hanna et al., 2007; Karumbayaram et al., 2009; Belay et al., 2010; Takayama et al., 2012). These procedures constitute an additional step of genome manipulation. Similar procedures of transient expression or genome excision by means of the Cre–loxP system, as done for the expression of hiPSC reprogramming factors, should be used to avoid additional side effects.
Although not related to genome alterations, another potential risk of hiPSC use is their potential immunogenic properties. Some authors have argued the possibility that despite being autologous, hiPSCs could trigger an immune reaction after transplantation. The latest reports regarding this issue have shown that differentiated hiPSCs are not immunogenic at these stages (Araki et al., 2013).
Concluding Remarks
The number of disease-specific hiPSC lines is increasing rapidly. Until now, only a few of them have been genetically corrected by gene-editing approaches. The unlimited proliferating capacity of hiPSCs, while maintaining pluripotent properties, allows for the application of HR techniques and the subsequent selection of properly corrected clones. Selection of the best gene-editing strategy depends on the disease to be corrected (Fig. 2 and Table 2). Targeted correction is the cleanest option. The patient mutation is corrected while leaving no exogenous elements, with the sequence of the corrected locus being indistinguishable from that of a wild-type locus. However, this approach is suitable only for a specific patient or group of patients carrying the same mutation, which limits its use. On the other hand, safe harbor integration is applicable to treat all the genetic diseases already addressed by genetic therapies with retro/lentiviral vectors. However, because the therapeutic gene loses its endogenous regulation, a specific promoter may be required to regulate its expression; in addition, the definition of a safe harbor locus might not be accurate or complete until we have a more in-depth knowledge of regulatory elements within the human genome. The knock-in strategy is an intermediate possibility in which a large number of patients with a defined disease might benefit from this strategy, reducing its development costs. Moreover, the endogenous elements of the locus will regulate expression of the therapeutic gene. However, one concern regarding the knock-in strategy, shared with the safe harbor approach, is that the use of a specific cDNA transgene may exclude the coexpression of various splicing variants. The election among them will depend of knowledge of the targeted locus.
Gene-editing procedures need improvements in terms of efficiency and safety before being applied in humans. The synergy between reprogramming and gene editing is prompting progress in this field of research, in which a wide spectrum of genetic diseases could be treated. Moreover, patient-specific hiPSCs are an ideal platform to improve gene-editing techniques in order to achieve the high efficiency and specificity that gene therapy needs for its future clinical use. There are still bottlenecks for their clinical application. Gene-corrected hiPSCs currently lack robust differentiation procedures to generate a variety of transplantable cells. For example, in the hematology field, the generation of hematopoietic stem cells capable of long-term reconstitution of the whole hematopoietic system has been reported (Amabile et al., 2013). Unfortunately, the need for teratoma formation to obtain functional HSCs in this report avoids its potential clinical application. Another possibility is the transplantation of more mature progenitor cells or terminally differentiated cells capable of long-term survival after infusion such as T cells, erythrocytes, or platelets. Similar strategies could be followed for other tissues. In addition, the development of homologous recombination technology in hiPSCs has broken new ground for its application to other stem cells already used in clinics, such as HSCs (Lombardo et al., 2007). We fully expect that future gene therapy protocols using the aforementioned methodologies will emerge.
Acknowledgments
The authors thank Dr. J.A. Bueren and V.K. Quiceno for careful reading and correction of the manuscript. This work has been supported by grants from the Ministerio de Economía y Competitividad (SAF2011-30526-C02-01), the Fondo de Investigaciones Sanitarias (RD06/0010/0015 and RD12/0019/0023), the Comunidad Autónoma de Madrid (CellCAM S2010/BMD-2420), the PERSIST European Project, the Cystic Fibrosis Foundation (B.R.D.), and the National Heart, Lung, and Blood Institute (B.R.D.). The authors also thank the Fundación Botín for promoting translational research at the Hematopoiesis and Gene Therapy Division-CIEMAT/CIBERER. Z.G. is supported by a fellowship of the Gobierno Vasco (Spain).
Author Disclosure Statement
No competing financial interests exist.
References
- Agarwal S. Loh Y.H. McLoughlin E.M., et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292–296. doi: 10.1038/nature08792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile G. Welner R.S. Nombela-Arrieta C., et al. In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood. 2013;121:1255–1264. doi: 10.1182/blood-2012-06-434407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An M.C. Zhang N. Scott G., et al. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W.F. Blaese R.M. Culver K. The ADA human gene therapy clinical protocol: Points to consider: Response with clinical protocol, July 6, 1990. Hum. Gene Ther. 1990;1:331–362. doi: 10.1089/hum.1990.1.3-331. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F. Trivedi C.M. Juhr D., et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R. Uda M. Hoki Y., et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- Ban H. Nishishita N. Fusaki N., et al. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14234–14239. doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista L.F. Pech M.F. Zhong F.L., et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399–402. doi: 10.1038/nature10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedel A. Taillepierre M. Guyonnet-Duperat V., et al. Metabolic correction of congenital erythropoietic porphyria with iPSCs free of reprogramming factors. Am. J. Hum. Genet. 2012;91:109–121. doi: 10.1016/j.ajhg.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay E. Matrai J. Acosta-Sanchez A., et al. Novel hyperactive transposons for genetic modification of induced pluripotent and adult stem cells: A nonviral paradigm for coaxed differentiation. Stem Cells. 2010;28:1760–1771. doi: 10.1002/stem.501. [DOI] [PubMed] [Google Scholar]
- Blasco M.A. Serrano M. Fernandez-Capetillo O. Genomic instability in iPS: Time for a break. EMBO J. 2011;30:991–993. doi: 10.1038/emboj.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C. Kiskinis E. Verstappen G., et al. Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Zinc-finger nucleases: A panoramic view. Curr. Gene Ther. 2011;11:2–10. doi: 10.2174/156652311794520076. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vergara X. Sevilla A. D'Souza S.L., et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayo M.A. Cai J. Delaforest A., et al. “JD” iPS cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology. 2012;56:2163–2171. doi: 10.1002/hep.25871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.J. Bouhassira E.E. Zinc-finger nuclease mediated correction of α-thalassemia in iPS cells. Blood. 2012;120:3906–3914. doi: 10.1182/blood-2012-03-420703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin M.H. Mason M.J. Xie W., et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi F. Zaslavskiy M. Poirot L., et al. Chromosomal context and epigenetic mechanisms control the efficacy of genome editing by rare-cutting designer endonucleases. Nucleic Acids Res. 2012;40:6367–6379. doi: 10.1093/nar/gks268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans R.J. Moseley A.B. Mesenchymal stem cells: Biology and potential clinical uses. Exp. Hematol. 2000;28:875–884. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- Delacote F. Lopez B.S. Importance of the cell cycle phase for the choice of the appropriate DSB repair pathway, for genome stability maintenance: The trans-S double-strand break repair model. Cell Cycle. 2008;7:33–38. doi: 10.4161/cc.7.1.5149. [DOI] [PubMed] [Google Scholar]
- Deng J. Shoemaker R. Xie B., et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A. Park I.H. Wen B., et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A. Hacein-Bey-Abina S. Cavazzana-Calvo M. 20 years of gene therapy for SCID. Nat. Immunol. 2010;11:457–460. doi: 10.1038/ni0610-457. [DOI] [PubMed] [Google Scholar]
- Fusaki N. Ban H. Nishiyama A., et al. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R. Gritti A. Bonfanti L. Vescovi A.L. Neural stem cells: An overview. Circ. Res. 2003;92:598–608. doi: 10.1161/01.RES.0000065580.02404.F4. [DOI] [PubMed] [Google Scholar]
- Ghosh Z. Wilson K.D. Wu Y., et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore A. Li Z. Fung H.L., et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarz A. Barascu A. Guirouilh-Barbat J. Lopez B.S. Initiation of DNA double strand break repair: Signaling and single-stranded resection dictate the choice between homologous recombination, non-homologous end-joining and alternative end-joining. Am. J. Cancer Res. 2012;2:249–268. [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G. Frampton G.M. Soldner F., et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Garrigue A. Wang G.P., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J. Wernig M. Markoulaki S., et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hanna J. Markoulaki S. Schorderet P., et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J. Saha K. Pando B., et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D. Soldner F. Beard C., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D. Wang H. Kiani S., et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S.E. Gore A. Li Z., et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc. Natl. Acad. Sci. U.S.A. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K. Yu J. Suknuntha K., et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irion S. Luche H. Gadue P., et al. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- Israel M.A. Yuan S.H. Bardy C., et al. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F. Wilson K.D. Sun N., et al. A nonviral minicircle vector for deriving human iPS cells. Nat. Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K. Norrby K. Paca A., et al. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karumbayaram S. Novitch B.G. Patterson M., et al. Directed differentiation of human-induced pluripotent stem cells generates active motor neurons. Stem Cells. 2009;27:806–811. doi: 10.1002/stem.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I.F. Hirata R.K. Wang P.R., et al. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 2010;18:1192–1199. doi: 10.1038/mt.2010.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Kim C.H. Moon J.I., et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.B. Zaehres H. Arauzo-Bravo M.J. Scholer H.R. Generation of induced pluripotent stem cells from neural stem cells. Nat. Protoc. 2009;4:1464–1470. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- Kim K. Zhao R. Doi A., et al. Donor cell type can influence the epigenome and differentiation potential of human induced pluripotent stem cells. Nat. Biotechnol. 2011;29:1117–1119. doi: 10.1038/nbt.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.Y. Hysolli E. Park I.H. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc. Natl. Acad. Sci. U.S.A. 2011;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G. Cha J. Chandrasegaran S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K. Arai S. Hosoi M., et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- Kunisato A. Wakatsuki M. Shinba H., et al. Direct generation of induced pluripotent stem cells from human nonmobilized blood. Stem Cells Dev. 2010;20:159–168. doi: 10.1089/scd.2010.0063. [DOI] [PubMed] [Google Scholar]
- Li T. Huang S. Zhao X., et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G.H. Suzuki K. Qu J., et al. Targeted gene correction of laminopathy-associated LMNA mutations in patient-specific iPSCs. Cell Stem Cell. 2011;8:688–694. doi: 10.1016/j.stem.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Sumer H. Leung J., et al. Late passage human fibroblasts induced to pluripotency are capable of directed neuronal differentiation. Cell Transplant. 2010;20:193–203. doi: 10.3727/096368910X514305. [DOI] [PubMed] [Google Scholar]
- Liu T. Zou G. Gao Y., et al. High efficiency of reprogramming CD34+ cells derived from human amniotic fluid into induced pluripotent stem cells with Oct4. Stem Cells Dev. 2012;21:2322–2332. doi: 10.1089/scd.2011.0715. [DOI] [PubMed] [Google Scholar]
- Lombardo A. Genovese P. Beausejour C.M., et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Marchetto M.C. Yeo G.W. Kainohana O., et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrai J. Cantore A. Bartholomae C.C., et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X. Neises A. Su R.J., et al. Efficient reprogramming of human cord blood CD34+ cells into induced pluripotent stem cells with OCT4 and SOX2 alone. Mol. Ther. 2012;20:408–416. doi: 10.1038/mt.2011.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A. Bellin M. Welling A., et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl J. Med. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- Mussolino C. Morbitzer R. Lutge F., et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M. Cooper J.B. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Ohi Y. Qin H. Hong C., et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou E.P. Sadelain M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat. Protoc. 2011;6:1251–1273. doi: 10.1038/nprot.2011.374. [DOI] [PubMed] [Google Scholar]
- Papapetrou E.P. Lee G. Malani N., et al. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011;29:73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F. Duchateau P. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr. Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- Park I.H. Arora N. Huo H., et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera M.F. Stem cells: The dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- Porteus M.H. Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Ramos-Mejia V. Montes R. Bueno C., et al. Residual expression of the reprogramming factors prevents differentiation of iPSC generated from human fibroblasts and cord blood CD34+ progenitors. PLoS One. 2012;7:e35824. doi: 10.1371/journal.pone.0035824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid S.T. Corbineau S. Hannan N., et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 2010;120:3127–3136. doi: 10.1172/JCI43122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A. Rodriguez-Piza I. Guenechea G., et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J. Gene targeting: The precise manipulation of the mammalian genome. Circ. Res. 1993;73:3–9. doi: 10.1161/01.res.73.1.3. [DOI] [PubMed] [Google Scholar]
- Sebastiano V. Maeder M.L. Angstman J.F., et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T. Yuasa S. Fukuda K. Derivation of induced pluripotent stem cells from human peripheral circulating T cells. Curr. Protoc. Stem Cell Biol. 2011;Chapter 4(Unit4A):3. doi: 10.1002/9780470151808.sc04a03s18. [DOI] [PubMed] [Google Scholar]
- Soldner F. Hockemeyer D. Beard C., et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F. Laganiere J. Cheng A.W., et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers A. Jean J.C. Sommer C.A., et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A. Sommer A.G. Longmire T.A., et al. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H. Chung S.K. Xu Y. Modeling disease in human ESCs using an efficient BAC-based homologous recombination system. Cell Stem Cell. 2010;6:80–89. doi: 10.1016/j.stem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Staerk J. Dawlaty M.M. Gao Q., et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N. Panetta N.J. Gupta D.M., et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N. Yazawa M. Liu J., et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takayama K. Inamura M. Kawabata K., et al. Efficient generation of functional hepatocytes from human embryonic stem cells and induced pluripotent stem cells by HNF4α transduction. Mol. Ther. 2012;20:127–137. doi: 10.1038/mt.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T. Takahashi K. Yamane M., et al. Induced pluripotent stem cells from CINCA syndrome patients as a model for dissecting somatic mosaicism and drug discovery. Blood. 2012 doi: 10.1182/blood-2012-03-417881. [DOI] [PubMed] [Google Scholar]
- Thier M. Munst B. Edenhofer F. Exploring refined conditions for reprogramming cells by recombinant Oct4 protein. Int. J. Dev. Biol. 2010;54:1713–1721. doi: 10.1387/ijdb.103193mt. [DOI] [PubMed] [Google Scholar]
- Thomas E.D. Fefer A. Buckner C.D. Storb R. Current status of bone marrow transplantation for aplastic anemia and acute leukemia. Blood. 1977;49:671–681. [PubMed] [Google Scholar]
- Torres R. Garcia A. Paya M. Ramirez J.C. Non-integrative lentivirus drives high-frequency Cre-mediated cassette exchange in human cells. PLoS One. 2011;6:e19794. doi: 10.1371/journal.pone.0019794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton J. Daboussi F. Leduc S., et al. 5′-Cytosine-phosphoguanine (CpG) methylation impacts the activity of natural and engineered meganucleases. J. Biol. Chem. 2012a;287:30139–30150. doi: 10.1074/jbc.M112.379966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valton J. Dupuy A. Daboussi F., et al. Overcoming transcription activator-like effector (TALE) DNA binding domain sensitivity to cytosine methylation. J. Biol. Chem. 2012b;287:38427–38432. doi: 10.1074/jbc.C112.408864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P. Rodriguez R.T. Wang J., et al. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Zheng C.G. Jiang Y., et al. Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012;22:637–648. doi: 10.1038/cr.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L. Manos P.D. Ahfeldt T., et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltjen K. Hamalainen R. Kibschull M., et al. Transgene-free production of pluripotent stem cells using piggyBac transposons. Methods Mol. Biol. 2011;767:87–103. doi: 10.1007/978-1-61779-201-4_7. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Yanez-Munoz R.J. Balaggan K.S. MacNeil A., et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Yao Y. Nashun B. Zhou T., et al. Generation of CD34+ cells from CCR5-disrupted human embryonic and induced pluripotent stem cells. Hum. Gene Ther. 2011;23:238–242. doi: 10.1089/hum.2011.126. [DOI] [PubMed] [Google Scholar]
- Yazawa M. Hsueh B. Jia X., et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. Zhan H. Mali P., et al. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood. 2009;114:5473–5480. doi: 10.1182/blood-2009-04-217406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. Vodyanik M.A. Smuga-Otto K., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu J. Hu K. Smuga-Otto K., et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S. Nakamura T. Sato T., et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012;18:618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Yusa K. Rashid S.T. Strick-Marchand H., et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Nature. 2011;478:391–4. doi: 10.1038/nature10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W. Freed C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- Zou J. Mali P. Huang X., et al. Site-specific gene correction of a point mutation in human iPS cells derived from an adult patient with sickle cell disease. Blood. 2011a;118:4599–4608. doi: 10.1182/blood-2011-02-335554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J. Sweeney C.L. Chou B.K., et al. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: Functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011b;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]