Abstract
Background
We examined the microbiota of bronchoalveolar lavage (BAL) samples with next-generation sequencing (NGS) technology to determine whether its results correlate with those of standard culture methods or affect patient outcome or both.
Methods
We collected BAL samples in the surgical intensive care unit (SICU) as part of the standard of care for intubated individuals who had a Clinical Pulmonary Infection Score (CPIS) ≥6 points. A portion of the BAL fluid was sequenced for the 16S region of ribosomal deoxyribonucleic acid (rDNA) with the Roche 454 FLX Titanium sequencer. Sequences were analyzed through a data-analysis pipeline to identify the appropriate taxonomic designation (∼species) of each 16s sequence. The bacterial microbiota of each BAL sample was compared with the bacteria identified in the sample through standard culture methods. Correlations between the taxonomic diversity of the microbiota and clinical outcome were examined through linear regression and Pearson correlation.
Results
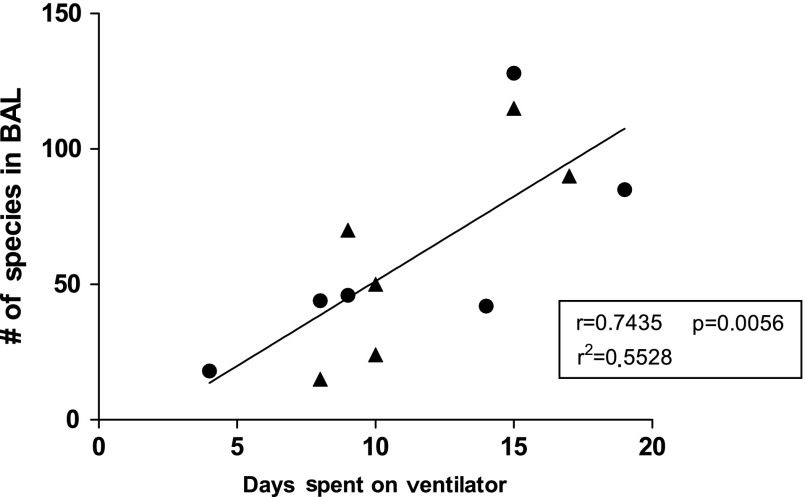
Bronchoalveolar lavage samples from 12 individuals in the SICU who had a CPIS ≥6 points were examined through 454 pyrosequencing. The number of phylotypes (∼species) in the samples ranged from 15 to 129. The number of phyla in the BAL samples ranged from 3 to 14. There was little correlation between the bacteria identified by NGS and those identified with standard culture methods. The same predominant bacterial strain was identified by both culture and sequencing in only a single sample. The correlation between patient days on a ventilator and the number of species in BAL samples was significant (r=0.7435, p=0.0056; r2=0.5528).
Conclusions
Increasing diversity of the bacterial microbiota in BAL samples correlates with the duration of mechanical ventilation. Bacteria identified through standard culture methods were not well correlated with the findings of NGS.
Respiratory infection in mechanically ventilated trauma patients presents substantial barriers to recovery. Trauma patients who develop ventilator-associated or community-acquired pneumonia have an increased hospital length of stay and more days of mechanical ventilation than do similarly injured patients who do not develop pneumonia [1–3]. A crucial aspect of the proper management of pneumonia is early detection and proper identification of the causative pathogen, which allows aggressive, targeted antibiotic therapy. However, traditional culture-based detection methods require the growth of a bacterial pathogen for a period of days before it can be identified, and analysis of bronchoalveolar lavage (BAL) samples reveals that many patients who exhibit the clinical signs of a respiratory infection remain culture-negative or yield results described as “normal flora” [4].
Culture-independent methods exist for the specific characterization of bacterial species in a variety of tissue and fluid samples [5]. However, these methods were until recently too laborious and costly to constitute feasible clinical alternatives to bacterial culture. Recent advances in deoxyribonucleic acid (DNA) sequencing technology, also called next-generation sequencing (i.e., 454 pyrosequencing), generate results for substantially longer base sequences in DNA than do older techniques, and are particularly well suited for the specific identification of bacteria in clinical samples. By locating bar-coded primers in the highly conserved regions flanking the bacterial gene that encodes 16s ribosomal ribonucleic acid (rRNA), investigators were able to identify simultaneously the individual species components of mixed bacterial populations with high sensitivity and at a lower cost than for previous analytic methods [7]. The advent of 454 pyrosequencing has revolutionized thinking about bacterial taxa that inhabit the human body [8–12]. This technology has also disproved certain dogmas, including the belief that the lung had a sterile mucosal surface [13–15].
Studies published previously examined the human bacterial microbiota of secretions and organ surfaces [10,11], including the respiratory tract [13,14]. Studies of the microbiome relative to asthma [14] and chronic obstructive pulmonary disease [14] as compared with those in healthy controls demonstrated that the respiratory tract is in fact not sterile. Although the predominant phyla identified were relative to disease state (i.e., asthma), the respiratory tract was also found to be home to a wide array of bacteria. Bacterial abundance varies extensively among subjects, ranging from a small number of species/phylotypes to colonization by multiple species. In a study of sputum samples from patients with lower respiratory tract infections done with 454 pyrosequencing of 16S rDNA, Zhou et al. found discordant results with regard to the pathogens identified by traditional culture methods. In a portion of their samples, they were unable to identify any pathogens with traditional bacterial culture. When they examined the same samples with pyrosequencing, they were able to determine a dominant pathogen as well as a complex bacterial community [15].
In the present study we used 454 pyrosequencing to analyze the microbiota in BAL samples from mechanically ventilated patients in a surgical intensive care unit (SICU). Our objective was to examine the bacterial diversity of BAL samples as related to clinical outcomes.
Patients and Methods
Study participants
Patients in the SICU of Parkland Memorial Hospital in Dallas, TX, were enrolled in the study under a protocol approved by the institutional review board of the University of Texas–Southwestern Medical Center. As part of the standard of care, patients with a Clinical Pulmonary Infection Score (CPIS) ≥6 points underwent BAL [16,17]. Samples of BAL fluid were collected with an unprotected catheter in accordance with standard operating procedures developed by a large-scale collaborative project named Inflammation and the Host Response to Injury [18].
DNA extraction
A 1-mL aliquot of unmodified BAL fluid was stored at −80°C for molecular analysis. The remaining BAL fluid was submitted to the clinical pathology laboratory of Parkland Memorial Hospital for microbial identification. After thawing, samples were centrifuged at 14,000 rpm for 30 sec and resuspended in 500 mcL of RLT buffer (Qiagen, Valencia, CA) (with β-mercaptoethanol). A sterile 5-mm steel bead (Qiagen) and 500-mcL sterile 0.1-mm glass beads (Scientific Industries, Bohemia, NY) were used for complete bacterial lysis in a Qiagen TissueLyser (Qiagen) that was run at 30Hz for 5 min. The lysed samples were then centrifuged briefly and 100 mcL of 100% ethanol was added to a 100-mcL aliquot of the sample supernatant. This mixture was added to a DNA spin column, and DNA recovery protocols were followed as described in the instructions for use of the QIAamp DNA Mini Kit (Qiagen), beginning at step 5 of the tissue-treatment protocol. Deoxyribonucleic acid was eluted from the column with 30 mcL of water, and the samples were diluted to a final concentration of 20 ng/mcL. The DNA in the samples was quantified with a Nanodrop spectrophotometer (Nyxor Biotech, Paris, France).
Pyrosequencing
Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) was done as described previously [6,19–23], but with the new technique of bacterial tag-encoded FLX titanium amplicon pyrosequencing (bTETAP). This technique is based on bTEFAP but utilizes titanium-containing reagents and titanium-based procedures, with a one-step PCR, a mixture of hot start and hot start high-fidelity Taq polymerases, and amplicons originating from the 28F 5′GAGTTTGATCNTGGCTCAG to 519R 5′GTNTTACNGCGGCKGCTG regions of the rRNA of Escherichia coli, and provides results for the V1–V3 regions of the 16S ribosome.
Analysis of bacterial diversity data
Following pyrosequencing of the 16S rRNA in the patient specimens, all unsuccessful sequence readings, low-quality sequence ends, and tags were removed, and any non-bacterial ribosome sequences and chimeras were removed from the specimens with the use of software described previously [6,19–22]. Sequences shorter than 350 bp according to the bTEFAP method were excluded. To determine the identity of the remaining bacteria in each specimen, 16S rRNA sequences were first examined with a distributed BLASTn .NET (nucleotide BLAST) algorithm [24] against a database of high-quality 16s bacterial sequences derived from the National Center for Biotechnology Information, and characterized as being of high quality according to the criteria of the Ribosomal Data Project ver. 9 [25]. The resulting BLASTn outputs were compiled with a .NET and C# analysis pipeline and validated through taxonomic distance methods, and were analyzed through data reduction as described previously [12,26]. Rarefaction to estimate maximum diversity was done to yield high-quality readings, with 16S rRNA sequences trimmed to 220 bp and depleted of non-ribosomal sequences and chimeras, as described previously [27].
Bacterial identification
On the basis of the BLASTn-derived sequence-identity method described above, which uses the percent of the full-length query sequence that aligns with a given database sequence, and with validation through taxonomic distance methods, bacteria were classified at the appropriate taxonomic levels according to the following criteria: (1) sequences having>97% identity (<3% divergence) with known or well-characterized 16s sequences were considered species-specific; (2) those with 95%–97% identity were considered genus-specific; (3) those with 90%–95% identity were considered as belonging to a bacterial family; and (4) those with 80%–90% identity were resolved as being within an order. After resolution according to these parameters, the percentage of each of the bacteria identified in a BAL sample was analyzed individually to provide information about their relative abundance within and among the BAL samples according to the relative numbers of sequence readings in a given sample. The results of the evaluations at a given taxonomic level other than the species level represent all sequences resolved to the level of identification of their primary genera or closest relative (where indicated). Measurements of alpha diversity were made with Quantitative Insights into Microbial Ecology (QIIME) software (open source software, University of Colorado, Boulder, CO; www.qiime.org) [28] on a rarified set of 1,590 sequences from each BAL sample over 10 iterations with the results averaged for each sample. The species operational taxonomic units (sOTUs) for the bacteria within a sample were calculated with 3% divergence to determine the predicted numbers of OTUs. Shannon a diversity (index to quantify the uncertanity of predicting the next entity in sequence) and chao1 (a richness index) indices were also calculated for each sample subjected to QIIME [29,30]. Bacterial identification and quantification according to the culture procedure used as the standard of care were done by the pathology department of Parkland Memorial Hospital according to standardized protocols [31].
Statistical analysis
Statistical analyses were done with Prism 5 (GraphPad Software San Diego, CA) and the predictive analystics software (PASW) statistical package 17.0 (IBM, Armonk, NY). Regression statistics were calculated by simple linear regression, and correlation values were estimated with the Pearson correlation coefficient, r. The Fisher exact test was used to compare the next-generation sequencing (NGS) method, through 454 pyrosequencing, with culture as a means of bacterial identification.
Results
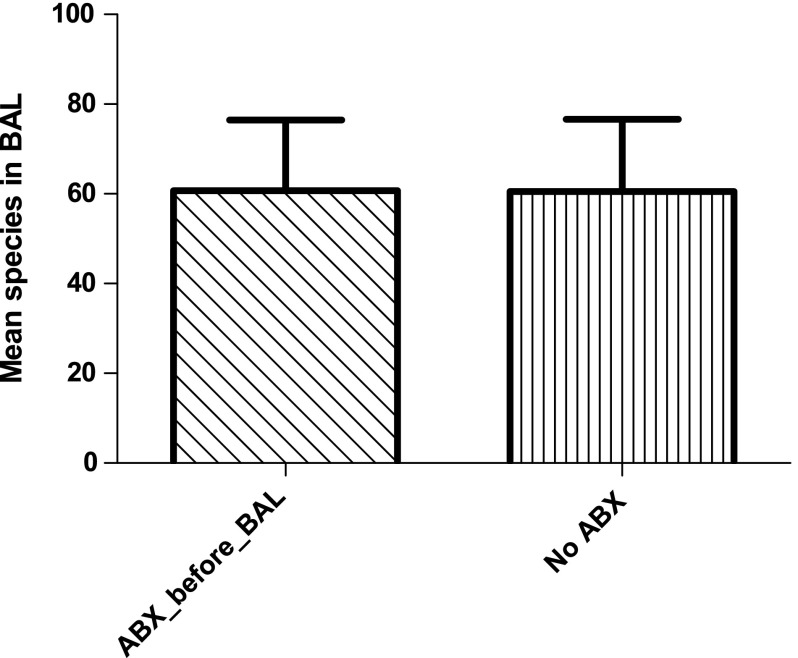
Table 1 presents demographic data for the 12 subjects enrolled in the study. The subjects ranged in age from 18 to 64 years, with a mean (±SD) of 45.6±17.5 years. The study sample was primarily male (83%) and Caucasian (75%). Total intensive care unit length of stay ranged from 4 to 36 d (mean: 20.4±13.5 d) and the range for days of ventilation was 4–19 d (mean: 11.5±4.4 d). Six of the subjects were receiving broad-spectrum antibiotic treatment before BAL was done. The difference in number of bacterial species identified in the antibiotic-treated group of subjects was not significantly different from that in the group not given antibiotic treatment (p=0.364; Fig. 1). The total duration of antibiotic treatment ranged from 4 to 29 d (mean: 12.25±7.5 d).
Table 1.
Demographic Information for Twelve Bronchoalveolar Lavage Samples
| BAL 1 | BAL 2 | BAL 4 | BAL 6 | BAL 7 | BAL 8 | BAL 9 | BAL 10 | BAL 11 | BAL 13 | BAL 14 | BAL 15 | Mean (range) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (years) | 64 | 52 | 18 | 56 | 56 | 24 | 60 | 32 | 54 | 18 | 49 | 64 | 45.6 (18–64) |
| Hospital LOS (days) | 17 | 45 | 24 | 61 | 17 | 28 | 10 | 34 | 20 | 73 | 80 | 4 | 34.4 (4–80) |
| Length of stay in ICU | 14 | 36 | 16 | 56 | 14 | 18 | 10 | 19 | 18 | 21 | 19 | 4 | 20.4 (4–56) |
| Ventilator days | 8 | 19 | 14 | 8 | 9 | 15 | 10 | 17 | 15 | 9 | 10 | 4 | 11.5 (4–19) |
| Total days of Abx therapy | 8 | 29 | 9 | 9 | 5 | 19 | 4 | 16 | 9 | 15 | 19 | 4 | 12.3 (4–29) |
Abx=antibiotic; BAL=bronchoalveolar lavage; ICU=intensive care unit; LOS=length of stay.
FIG. 1.
Mean number of bacterial species identified in bronchoalveolar lavage (BAL) samples by 16s rRNA pyrosequencing, according to the use or non-use of antibiotics (ABX) before collection of the BAL sample.
Table 2 presents the flora identified in BAL specimens by standard culture methods. Culture failed to grow an organism or the organisms found were considered “normal respiratory tract flora” in three BAL samples.
Table 2.
Bacterial Identification Results for Twelve Bronchoalveolar Lavage (BAL) Samples by Standard Culture Methods and Culture-Independent Pyrosequencinga–c
| BAL sample no. | Culture results (cfu) | Species by 454 pyrosequencing (% sequences) | Phylum by 454 pyrosequencing(% sequences) | Antibiotic treatment |
|---|---|---|---|---|
| 1 | Klebsiella pneumoniae (1) |
Staphylococcus aureus (99.6) Streptococcus spp. (0.2) |
Firmicutes (99.94) | Ticarcillin–clavulanic acid and piperacillin–tazobactam |
| 2 |
Klebsiella oxytoca (100,000) Haemophilus influenzae (100,000) Respiratory tract flora (20,000) |
Streptococcus spp. (41.3) Streptococcus anginosus (15.5) |
Firmicutes (72.7) Proteobacteria (20.8) |
|
| 4 | Haemophilus influenzae (50,000) |
Haemophilus spp. (58.22) Haemophis influenza (36.7) |
Proteobacteria (97.7) | |
| 6 | None |
Mycoplasma spp. (89.6) Prevotella spp. (3.3) |
Tenericutes (89.8) | |
| 7 | Respiratory tract flora (25,000) |
Klebsiella spp. (35.2) Stenotrophomonas maltophilia (28.8) |
Proteobacteria (97.4) | |
| 8 |
Enterobacter cloacae (100,000) Pseudomonas aeruginosa (20,000) |
Methylomicrobium spp. (8.97) Staphylococcus aureus (7.6) |
Proteobacteria (46.1) Firmicutes (26.6) |
Piperacillin–tazobactam and vancomycin |
| 9 |
Klebsiella pneumoniae (100,000) Stenotrophomonas maltophilia (100,000) |
Haemophilus influenza (42.2) Haemophilus spp. (32.7) |
Proteobacteria (77.7) Firmicutes (9.3) |
Piperacillin–tazobactam and vancomycin |
| 10 | Staphylococcus aureus (7,000) | Haemophilus influenza (17.9) Haemophilus spp. (14.5) | Proteobacteria (48.3) Firmicutes (30.1) |
Sulfamethoxazole–trimethoprim |
| 11 |
Streptococcus pneumoniae (50,000) Haemophilus influenzae (100,000) |
Staphylococcus aureus (13.5) Ruminococcus spp. (7.5) |
Firmicutes (44.8) Proteobacteria (37.6) |
|
| 13 | Enterobacter aerogenes (100,000) | Haemophilus influenza (39.2) Haemophilus spp. (26.9) | Proteobacteria (76.3) Tenericutes (19.7) |
Vancomycin |
| 14 | None |
Mycoplasma spp. (69.0) Klebsiella spp. (9.5) |
Tenericutes (69.1) Proteobacteria (30.9) | Vancomycin, ticarcillin–clavulanic acid, and piperacillin–tazobactam |
| 15 | Staphylococcus aureus (50,000) |
Pseudomonas spp. (91.6) Pseudomonas aeruginosa (6.4) |
Proteobacteria (98.4) |
Estimated number of colony-forming units (cfu) is listed in parentheses for standard culture.
Percent of pyrosequences identified in each sample is denoted by the appropriate taxonomic designation.
Antibiotic column denotes type of antibiotic treatment (if any) that was given before BAL.
Analysis with 454 pyrosequencing yielded 47,789 sequences with an average of 3,982 sequence readings per BAL sample. Pyrosequencing identified a mean of 60.5 species per BAL sample (range: 15–128±37 species), an average of 43.4 genera (range: 12–101±30 genera), and 7.3 phyla (range: 3–14±3 phyla). Table 2 lists the two most predominant bacterial species identified in each BAL sample as determined by the relative percentages of sequences identified in the sample. The Fisher exact test was used to test the concordance between the pyrosequencing and culture methods of identifying bacterial species. Detection of bacterial species with the two methods was discordant (p=0.009). Overall, the major species identified are listed in Table 3, with the predominant phyla identified through 16s rRNA sequencing being Proteobacteria (52.8%; chiefly represented by gram-negative organisms such as Escherichia and Salmonella), Firmicutes (24.5%; chiefly consisting of gram-positive organisms such as Streptococcus, Staphylococcus, and Clostridium), and Tenericutes (15.7%; such as Mycoplasma). Diversity indices for each sample are listed in Table 4.
Table 3.
Fifteen Most Common Bacterial Species Identified in Fifteen Bronchoalveolar Lavage (BAL)Samples by 16s rRNA Pyrosequencinga,b
| Species | BAL 1 | BAL 2 | BAL 4 | BAL 6 | BAL 7 | BAL 8 | BAL 9 | BAL 10 | BAL 11 | BAL 13 | BAL 14 | BAL 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staphylococcus aureus | 99.58 | 7.75 | 0.05 | 0.03 | 0 | 7.64 | 0 | 7.61 | 13.50 | 0 | 0 | 0 |
| Mycoplasma spp. | 0 | 0 | 0 | 89.64 | 0 | 0 | 6.84 | 1.35 | 0.30 | 19.67 | 68.96 | 0 |
| Haemophilus influenzae | 0 | 0 | 36.72 | 0 | 0 | 0 | 42.24 | 17.96 | 0.30 | 39.19 | 0 | 0.05 |
| Haemophilus spp. | 0 | 0 | 58.22 | 0.03 | 0 | 0 | 32.71 | 14.48 | 0.56 | 26.87 | 0 | 0.05 |
| Pseudomonas spp. | 0 | 7.44 | 0 | 0 | 0.02 | 1.91 | 0 | 0.83 | 4.25 | 0.00 | 0.02 | 91.56 |
| Klebsiella spp. | 0 | 3.15 | 1.71 | 2.31 | 35.16 | 0.54 | 1.54 | 3.17 | 1.19 | 9.47 | 29.95 | 0.05 |
| Streptococcus spp. | 0.15 | 41.32 | 0.23 | 0.68 | 0.35 | 2.33 | 3.45 | 3.30 | 5.97 | 4.01 | 0 | 0 |
| Stenotrophomonas maltophilia | 0.01 | 0.52 | 0 | 0.08 | 28.81 | 0.17 | 0 | 0.83 | 0 | 0 | 0 | 0.07 |
| Stenotrophomonas spp. | 0 | 0.36 | 0.02 | 0 | 19.92 | 0.58 | 0 | 1.17 | 1.76 | 0 | 0.00 | 0.12 |
| Prevotella spp. | 0 | 1.03 | 0.16 | 3.27 | 0.31 | 0.91 | 5.29 | 4.57 | 0.63 | 0.01 | 0.15 | 0.02 |
| Ruminococcus spp. | 0 | 0 | 0.00 | 0.11 | 0.02 | 2.74 | 0 | 5.35 | 7.27 | 0.01 | 0 | 0.10 |
| Streptococcus anginosus | 0.08 | 15.50 | 0.02 | 0 | 0 | 0.96 | 0.04 | 0.96 | 0.93 | 0 | 0 | 0 |
| Escherichia spp. | 0 | 0.77 | 0.02 | 0 | 6.13 | 0.17 | 0 | 1.26 | 0.76 | 0 | 0 | 0 |
| Methylomicrobium spp. | 0 | 0 | 0 | 0 | 0 | 8.97 | 0 | 0 | 2.65 | 0 | 0 | 0 |
| Pseudomonas aeruginosa | 0 | 0.52 | 0 | 0 | 0 | 0 | 0 | 0 | 0.27 | 0 | 0 | 6.40 |
Prevalence was determined by total number of sequences identified across all samples, with the most prevalent species listed in descending order of prevalence.
Representation of percent sequences identified in each particular BAL specimen is denoted for the 15 most prevalent species.
Table 4.
Diversity Indices for Each Bronchoalveolar Lavage Samplea
| Sample ID | BAL 1 | BAL 2 | BAL 4 | BAL 6 | BAL 7 | BAL 8 | BAL 9 | BAL 10 | BAL 11 | BAL 13 | BAL 14 | BAL 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sOTU | 35 | 116 | 73 | 69 | 76 | 150 | 110 | 148 | 193 | 98 | 43 | 30 |
| Shannon | 0.91 | 4.0 | 2.2 | 1.7 | 2.7 | 6.2 | 2.9 | 5.17 | 6.5 | 3.3 | 2.0 | 0.8 |
| chao1 | 65 | 160.6 | 104.2 | 127.0 | 82.3 | 226.5 | 169.7 | 254.6 | 240.9 | 170.7 | 43.1 | 37.4 |
Alpha diversity measurements made with Quantitative Insights into Microbial Ecology (QIIME) software were made on a rarified set of 1,590 sequences from each sample over 10 iterations and averaged. The 3% divergence species operational taxonomic units (sOTUs) were calculated to determine the predicted numbers of OTUs. Shannon and chao1 indices were calculated for each sample and are listed accordingly.
BAL=bronchoalveolar lavage.
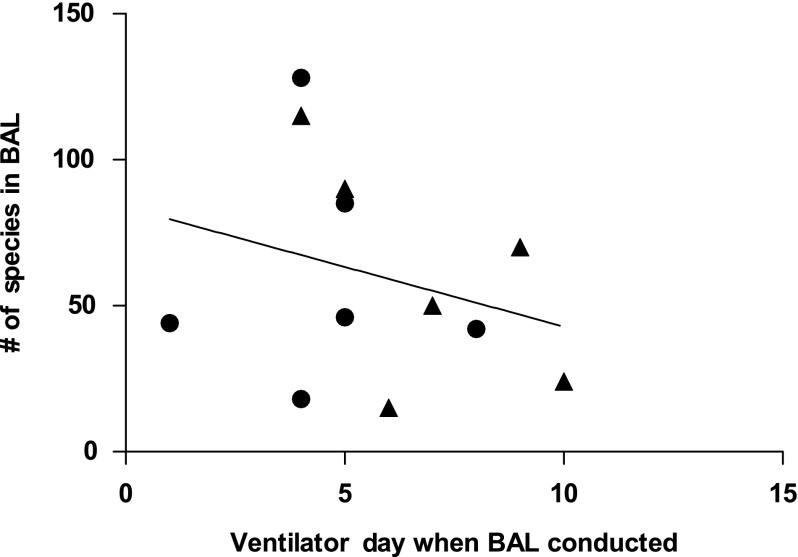
Figure 2 shows the correlation between the day of ventilation on which a BAL sample was collected and the number of species in the sample identified by 16S rRNA sequencing (r=−0.274, p=0.388; r2=0.075). The correlation between the number of species identified by 16S rRNA sequencing and total number of days of ventilation (Fig. 3) was significant (r=0.7435, p=0.0056; r2=0.5528).
FIG. 2.
Number of bacterial species identified in bronchoalveolar lavage (BAL) sample relative to the number of days for which the patient was on a ventilator before collection of the sample. Each point represents an individual BAL sample. Closed circles represent patients who were not receiving antibiotics at the time of sample collection. Closed triangles represent patients who were receiving antibiotic treatment at the time of sample collection.
FIG. 3.
Plot of linear regression of number of bacterial species identified in bronchoalveolar lavage (BAL) samples versus days of ventilation. Each closed circle represents a single BAL sample from one patient. Closed circles represent individuals who were not receiving antibiotics at the time of BAL-sample collection. The solid triangles represent individuals who were receiving antibiotic treatment at the time of BAL-sample collection.
Discussion
Traditional culture methods are considered the gold standard for identifying bacteria in the clinical setting. However, many patients showing signs of an infection have results of bacterial culture that remain negative even though their symptoms resolve frequently with antibiotic treatment. Therefore, a method of identifying bacteria that is both independent of and more sensitive than culture is desirable. In the present study, we used a culture-independent, DNA-based method to identify bacterial populations in BAL samples from mechanically ventilated trauma patients. With this, we identified a broad and diverse range of bacteria that showed poor concordance with the results of culture. The organisms identified with the DNA-based method were chiefly those commonly encountered in clinical practice, with a mixture of gram-positive and gram-negative organisms and Mycoplasma. The DNA-based method was also able to identify multiple species simultaneously within the same BAL sample. These findings are concordant with reports published previously that the pulmonary tract is not sterile [32]. These results are surprising and may have a significant effect on clinical care.
When comparing the bacteria identified in a single specific BAL sample through culture and the culture-independent method, low levels of concordance were observed. This lack of concordance is not unfounded, with previous studies having demonstrated little concordance between bacteria identified through traditional methods of culture in antemortem and post-mortem clinical samples [33,34]. Of the 12 BAL samples examined in the present study, both culture and the NGS method identified the same predominant species (Haemophilus sp.) in only a single sample. The three samples that were culture-negative or denoted as containing normal respiratory tract flora were identified as containing predominantly Mycoplasma and Klebsiella. Although some of the species of these two genera are described as atypical pathogens for pneumonia [35], other studies have identified “atypical pathogens” as the predominant species in respiratory tract infections [15]. These findings are similar to previously published data for comparisons of traditional culture and culture-independent methods [6]. Zhou et al. [15], in their examination of sputum samples from patients with lower respiratory tract infections, found statistically significant differences in the two methods' sensitivity in detecting bacteria. A potential explanation for the disparity in the results with the two methods may be the ability of DNA-based methods to identify bacteria that are difficult to culture or dead, and therefore undetectable via culture. Additionally, there may be a bias in culture-based methods toward bacteria that grow more readily than and outcompete bacteria that do not grow well in culture [6], whereas these latter bacteria may survive or thrive quite well in vivo.
Although the standard method of culture primarily identifies one or two species of bacteria in a particular specimen, our findings suggest that the respiratory tract is colonized simultaneously by multiple bacterial species. In a small number of BAL samples, we identified more than 450 species of bacteria that varied both in their presence and prevalence in these samples. Interestingly, in several samples that were culture-negative, we identified organisms (Mycoplasma) that are relevant clinically and difficult to culture. Previous studies have suggested that Mycoplasma is present in a substantial percentage (∼40%) of patients with suspected ventilator-acquired pneumonia [36].
When we stratified our data according to use or non-use of antibiotic therapy at the time of BAL, we found no significant difference in the mean number of bacteria in BAL samples. There was also no correlation of the number of days of mechanical ventilation a subject had had before the BAL procedure with the number of bacterial species identified by 454 pyrosequencing.
A major barrier to recovery from trauma is the total number of days on which a patient receives mechanical ventilation. When correlating the number of bacterial species identified in the BAL samples in our study with the total number of days on which the patient from whom the sample came had required mechanical ventilation, we found a significant correlation between these two variables. We surmise that continued mechanical ventilation probably permits the expansion and selection of particular phyla of organisms rather than facilitating such expansion as a generalized effect for all microbes. Although some of the patients who had BAL were treated with antibiotics after their BAL specimens were collected, patients with increased multi-species colonization had mechanical ventilation for a longer period than those with fewer species identified by 16S rRNA sequencing. Additionally, the inability of standard culture methods to identify a potential pathogen for a perceived respiratory infection in a mechanically ventilated patient from whom the culture specimen was taken may hinder the patient's recovery by impeding effective antibiotic therapy. Recent data suggest that timely and correct antibiotic use in sepsis improves outcomes significantly [37]. The use of culture-independent methods such as 16S rRNA pyrosequencing for identifying bacteria in BAL samples may provide an effective alternative for such identification in samples that are culture-negative or that yield inconclusive results with standard culture, and can be much more rapid and specific.
The present pilot study had several limitations, primarily in its small number of BAL samples. However, the findings of the study are surprising, and should be confirmed in a larger study with targets for analysis identified by the present work. Additionally, the use of the 16S rRNA sequencing to identify bacterial taxa currently precludes establishment of the antibiotic sensitivity pattern of a putative pathogen. A further limitation of 16S rRNA sequencing is the inability to absolutely quantify bacterial counts in a sample. Methods are being developed to assist in such quantification, but were not used in the present study. The existing data derived from NGS also suffer from a lack of standardization across studies of lung microbiota. However, as NGS advances, it is inevitable that a scientific consensus of sampling methodology, analysis, and data presentation across studies of pulmonary microbiota will occur [30].
The findings in the present study permit the conclusion that the respiratory tract is occupied by a community of multiple bacterial species. This multi-species colonization may not only affect recovery from traumatic injury but may increase the susceptibility to respiratory infection. More sensitive, accurate, and timely identification of the microbial organisms present in the respiratory tract may hold promise for improving the clinical management of mechanically ventilated patients.
In conclusion, our study, albeit done with a relatively small number of samples, indicates that NGS can identify numerous bacteria in BAL fluid samples from mechanically ventilated trauma patients in whom such bacterial identification is clinically indicated. The concordance between our results with this method and those with standard culture techniques was disparate, a finding that is currently of unknown clinical relevance. The predominant organisms identified through NGS technology are generally those encountered typically in clinical practice based upon our experiences.
Acknowledgments
This study was supported in part by grant UL1 RR024982 from the Clinical and Translational Science Award (CTSA) program of the National Center for Research Resources, National Institutes of Health.
Author Disclosure Statement
Scot Dowd is an employee of Molecular Research LP, and received a salary from the diagnostic laboratory of Molecular Research, LP, Shallowater, TX. The remaining authors have no competing financial interests related to this study.
References
- 1.Eagye KJ. Nicolau DP. Kuti JL. Impact of superinfection on hospital length of stay and costs in patients with ventilator-associated pneumonia. Semin Respir Crit Care Med. 2009;30:116–123. doi: 10.1055/s-0028-1119815. [DOI] [PubMed] [Google Scholar]
- 2.Boyce JM. Potter-Bynoe G. Dziobek L. Solomon SL. Nosocomial pneumonia in Medicare patients. Hospital costs and reimbursement patterns under the prospective payment system. Arch Intern Med. 1991;151:1109–1114. doi: 10.1001/archinte.151.6.1109. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich ES. Demmler M. Schulgen G, et al. Nosocomial pneumonia: A cost-of-illness analysis. Infection. 2002;30:61–67. doi: 10.1007/s15010-002-1083-8. [DOI] [PubMed] [Google Scholar]
- 4.Labelle AJ. Arnold H. Reichley RM, et al. A comparison of culture-positive and culture-negative health-care-associated pneumonia. Chest. 2010;137:1130–1137. doi: 10.1378/chest.09-1652. [DOI] [PubMed] [Google Scholar]
- 5.Caliendo AM. Multiplex PCR and emerging technologies for the detection of respiratory pathogens. Clin Infect Dis. 2011;52:S326–S330. doi: 10.1093/cid/cir047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowd SE. Sun Y. Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamady M. Walker JJ. Harris JK, et al. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE. Backhed F. Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koren O. Spor A. Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA. 2011;108:4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley RE. Lozupone CA. Hamady M, et al. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fierer N. Hamady M. Lauber CL. Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolcott RD. Gontcharova V. Sun Y, et al. Bacterial diversity in surgical site infections: Not just aerobic cocci any more. J Wound Care. 2009;18:317–323. doi: 10.12968/jowc.2009.18.8.43630. [DOI] [PubMed] [Google Scholar]
- 13.Hilty M. Burke C. Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erb-Downward JR. Thompson DL. Han MK, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Y. Lin P. Li Q, et al. Analysis of the microbiota of sputum samples from patients with lower respiratory tract infections. Acta Biochim Biophys Sin (Shanghai) 2010;42:754–761. doi: 10.1093/abbs/gmq081. [DOI] [PubMed] [Google Scholar]
- 16.Pugin J. Clinical signs and scores for the diagnosis of ventilator-associated pneumonia. Minerva Anestesiol. 2002;68:261–265. [PubMed] [Google Scholar]
- 17.Pugin J. Auckenthaler R. Mili N, et al. Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic “blind” bronchoalveolar lavage fluid. Am Rev Respir Dis. 1991;143:1121–1129. doi: 10.1164/ajrccm/143.5_Pt_1.1121. [DOI] [PubMed] [Google Scholar]
- 18.Minei JP. Nathens AB. West M, et al. Inflammation and the host response to injury, a large-scale collaborative project: Patient-oriented research core–standard operating procedures for clinical care. II. Guidelines for prevention, diagnosis and treatment of ventilator-associated pneumonia (VAP) in the trauma patient. J Trauma. 2006;60:1106–1113. doi: 10.1097/01.ta.0000220424.34835.f1. [DOI] [PubMed] [Google Scholar]
- 19.Edrington TS. Dowd SE. Farrow RF, et al. Development of colonic microflora as assessed by pyrosequencing in dairy calves fed waste milk. J Dairy Sci. 2012;95:4519–4525. doi: 10.3168/jds.2011-5119. [DOI] [PubMed] [Google Scholar]
- 20.Hume ME. Hernandez CA. Barbosa NA, et al. Molecular identification and characterization of ileal and cecal fungus communities in broilers given probiotics, specific essential oil blends, and under mixed Eimeria infection. Foodborne Pathog Dis. 2012;9:853–860. doi: 10.1089/fpd.2011.1093. [DOI] [PubMed] [Google Scholar]
- 21.Suchodolski JS. Dowd SE. Wilke V, et al. 16S rRNA Gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS One. 2012;7:e39333. doi: 10.1371/journal.pone.0039333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy-Szakal D. Ross MC. Dowd SE, et al. Maternal micronutrients can modify colonic mucosal microbiota maturation in murine offspring. Gut Microbes. 2012;3:426–433. doi: 10.4161/gmic.20697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooda S. Boler BM. Serao MC, et al. 454 pyrosequencing reveals a shift in fecal microbiota of healthy adult men consuming polydextrose or soluble corn fiber. J Nutrition. 2012;142:1259–1265. doi: 10.3945/jn.112.158766. [DOI] [PubMed] [Google Scholar]
- 24.Dowd SE. Zaragoza J. Rodriguez JR, et al. Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST) BMC Bioinformatics. 2005;6:93. doi: 10.1186/1471-2105-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole JR. Wang Q. Cardenas E, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dowd SE. Wolcott RD. Sun Y, et al. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP) PLoS One. 2008;3:e3326. doi: 10.1371/journal.pone.0003326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Y. Wolcott RD. Dowd SE. Tag-encoded FLX amplicon pyrosequencing for the elucidation of microbial and functional gene diversity in any environment. Methods Mol Biol. 2011;733:129–141. doi: 10.1007/978-1-61779-089-8_9. [DOI] [PubMed] [Google Scholar]
- 28.Kuczynski J. Stombaugh J. Walters WA, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011 Dec;Chapter 10(Unit 10.7) doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon CE. Weaver W. Urbana, II: University of Illinois Press; 1949. The mathematical theory of communication. [Google Scholar]
- 30.Chao A. Non-parametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
- 31.Thomson RB. Specimen collection, transport, and processing: Bacteriology. Manual of Clinical Microbiology. In: Murray PR, editor; Baron EJ, editor; Jorgensen JH, et al., editors. Washington, DC: ASM Press; 2007. pp. 309–310. [Google Scholar]
- 32.Beck JM. Young VB. Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160:258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatachalam V. Hendley JO. Willson DF. The diagnostic dilemma of ventilator-associated pneumonia in critically ill children. Pediatr Crit Care Med. 2011;12:286–296. doi: 10.1097/PCC.0b013e3181fe2ffb. [DOI] [PubMed] [Google Scholar]
- 34.Gomez R. Murray CK. Hospenthal DR, et al. Causes of mortality by autopsy findings of combat casualties and civilian patients admitted to a burn unit. J Am Coll Surg. 2009;208:348–354. doi: 10.1016/j.jamcollsurg.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal J. Awasthi S. Rajput A, et al. Atypical bacterial pathogens in community-acquired pneumonia in children: A hospital-based study. Trop Doct. 2009;39:109–111. doi: 10.1258/td.2008.080248. [DOI] [PubMed] [Google Scholar]
- 36.Muir MT. Cohn SM. Louden C, et al. Novel toxin assays implicate Mycoplasma pneumoniae in prolonged ventilator course and hypoxemia. Chest. 2011;139:305–310. doi: 10.1378/chest.10-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar A. Ellis P. Arabi Y, et al. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest. 2009;136:1237–1248. doi: 10.1378/chest.09-0087. [DOI] [PubMed] [Google Scholar]