Abstract

We have used protein engineering to expand the palette of genetically encoded calcium ion (Ca2+) indicators to include orange and improved red fluorescent variants, and validated the latter for combined use with optogenetic activation by channelrhodopsin-2 (ChR2). These indicators feature intensiometric signal changes that are 1.7- to 9.7-fold improved relatively to the progenitor Ca2+ indicator, R-GECO1. In the course of this work, we discovered a photoactivation phenomenon in red fluorescent Ca2+ indicators that, if not appreciated and accounted for, can cause false-positive artifacts in Ca2+ imaging traces during optogenetic activation with ChR2. We demonstrate, in both a beta cell line and slice culture of developing mouse neocortex, that these artifacts can be avoided by using an appropriately low intensity of blue light for ChR2 activation.
Keywords: Ca2+ imaging, fluorescent proteins, Ca2+ indicators, channelrhodopsin, photoactivation
We have previously reported the engineering of a spectral palette of fluorescent protein (FP)-based Ca2+ indicators that enable imaging of Ca2+ dynamics in single cells with up to three spectrally distinct colors.1 As the longest wavelength member of this series, the red fluorescent R-GECO1 (excitation at 561 nm and emission 589 nm with Ca2+), provides the advantages of diminished autofluorescence background and deeper tissue penetration associated with imaging with longer wavelength light.
In addition to benefiting from the inherent advantages of imaging at longer wavelengths, red-shifted Ca2+ indicators can also enable monitoring of intracellular Ca2+ concentration during excitation or activation of a second chromophore in the violet to green wavelength range.2,3 An example of such a chromophore is the cation-selective light activated channel channelrhodopsin-2 (ChR2) from Chlamydomonas reinhardtii.4 When expressed in neurons, activation of ChR2 causes membrane depolarization5 which is accompanied by a rapid rise in intracellular Ca2+, due to its partial permeability to Ca2+ (ref (4)) and the action of voltage gated Ca2+ channels.6,7 Accordingly, ChR2 is the prototypical example of an optogenetic actuator that can be used to manipulate the activity of neurons with visible light. However, the action spectrum of ChR2 extends from the ultraviolet through to ∼550 nm,4,8,9 restricting the ability of researchers to image neuronal activation with popular genetically encoded (e.g., cameleons10 and G-CaMP11) or synthetic dye-based indicators.7 Substantially red-shifted ChR2 homologues with diminished excitation at ∼400 nm have been reported,8,12,13 though even these variants retain strong excitation throughout the visible region of the spectrum.14 As we have elaborated in a recent literature review, the challenges imposed by the relative dearth of color choices in terms of both optogenetic actuators and genetically encoded Ca2+ indicators has severely limited the number of reports in which researchers have attempted to visualize Ca2+ dynamics following optogenetic neuron activation.15
Results and Discussion
In an effort to expand the number of color choices of Ca2+ indicator available to researchers, we took inspiration from the far-red DsRed-derived mPlum variant16 and attempted to similarly engineer variants of R-GECO1 with further red-shifted emission. However, introduction of various combinations of the key mutations of mPlum (V16E, F65I, L124V, and I161M by DsRed numbering) into R-GECO1 resulted in nonfluorescent proteins or indicators with a drastically diminished Ca2+ response relative to R-GECO1. Of those variants that retained a Ca2+-dependent signal, only the F65I/I161M variant retained a signal change of at least 3× (compare to 16× for R-GECO1) and had a red-shifted emission peak (597 nm with Ca2+). Reasoning that alternative substitutions at key positions might further red-shifted fluorescence, we created a genetic library in which the two residues (16 and 65) that are most important to the red shift of mPlum17 were fully randomized. We performed ratiometric fluorescence-based screening of this library expressed in the periplasm of bacterial colonies,1 and picked those variants with the highest ratio of emission through a 660–700 nm bandpass filter over emission through a 573–648 nm bandpass filter (excitation through a 510–560 nm bandpass filter). This approach led to the identification of the red-shifted V16T/F65I variant (emission maximum at 606 nm with Ca2+) with a 9.5× intensity change. Further directed evolution with library creation by error-prone PCR and ratiometric screening ultimately led to the production of a variant designated as carmine (i.e., a deep red color) GECO 1 (CAR-GECO1) with fluorescence excitation and emission peaks at 560 and 609 nm, respectively, in the Ca2+-bound state (Figure 1a,b). CAR-GECO1 has 6 mutations relative to R-GECO1 (Figure 1d, Supporting Information Figure 1 and Table 1), an intensity change of 27×, a Kd for Ca2+ of 490 nM (Supporting Information Figure 2, and Tables 2 and 3), and a 2-photon cross section of 45 GM at 1052 nm (Figure 1c). The increased Stokes shift of CAR-GECO1 relative to R-GECO1 suggests that, like with mPlum, directed evolution produced a dynamic Stokes shift in the new variant.17
Figure 1.

Characterization of improved indicators. (a) Excitation spectra of O-GECO1 (orange), R-GECO1.2 (red) and CAR-GECO1 (dark red) with (solid line) and without (dashed line) Ca2+. Inset: 25× y-axis zoom. (b) Emission spectra represented as in (a). (c) Two-photon excitation spectra of O-GECO1, R-GECO1, and CAR-GECO1, colored as in (a, b). (d–f) Models of (d) CAR-GECO1; (e) R-GECO1.2; and (f) O-GECO1, showing location of substitutions relative to R-GECO1. The structural model used in (d–f) is based on PDB IDs 3EVR(36) and 2H5Q(28) and was previously described.1
Screening of the same library (i.e., randomization of positions 16 and 65) also led to the discovery of R-GECO1.1 with the V16L and F65L mutations, a 27× intensity change, and essentially the same excitation and emission maxima as R-GECO1. Further directed evolution with library creation by error-prone PCR led to the identification of R-GECO1.2 which has 8 mutations relative to R-GECO1 (Figure 1e, Supporting Information Figure 1 and Table 1), a 2-fold improved intensity change (33×) (Figure 1a,b) relative to R-GECO1, and a Kd for Ca2+ of 1200 nM (Supporting Information Figure 2, and Tables 2 and 3).
Similar to the strategy used to identify red-shifted variants, our efforts to generate an orange fluorescent variant were inspired by the mutations present in the orange DsRed-variant mOrange.18 Accordingly, we introduced the K163 mutation and created a gene library in which the codons for 4 positions (M66, F83, I197, and Q213 by DsRed numbering) were randomized to codon subsets encoding amino acids S/T/A/L/I/V, L/Y/F, K/R/I/V/G/E, and L/Q, respectively. Screening of this library led to the identification of an orange fluorescent (excitation at 545 nm and emission 565 nm with Ca2+) GECO variant, O-GECO0.1, with a 44× intensity change. Further directed evolution ultimately produced O-GECO1 with nine mutations relative to R-GECO1 (Figure 1f, Supporting Information Figure 1 and Table 1), a fluorescence response of 146× (Figure 1a,b), a Kd for Ca2+ of 1500 nM (Supporting Information Figure 2, and Tables 2 and 3), and a 2-photon cross-section of 85 GM at 1048 nm (Figure 1c). The large in vitro response of O-GECO1 is similar to that of the latest generation G-CaMP variants, G-CaMP5F (162×) and G-CaMP5H (158×).19
To demonstrate the utility of the new Ca2+ indicators, we performed systematic characterization of the dynamic response in HeLa cells treated with histamine followed by ionomycin/Ca2+ and ionomycin/EGTA (Supporting Information Figure 3 and Table 4). Although there is a great deal of cell-to-cell heterogeneity in such experiments, averaging the results from many individual cells revealed that the improved intensity changes observed in vitro were preserved in cells. Of the three indicators, O-GECO1 gave the best performance in cells, exhibiting maximum fluorescence intensity changes of 41 ± 10-fold (N = 49 cells) following treatment with histamine (Supporting Information Video 1). Under identical conditions, R-GECO1.2 and CAR-GECO1 had histamine-induced fluorescence changes of 11.2 ± 2.5- and 10.5 ± 1.8-fold, respectively.
As the red-shifted excitation spectra of R-GECO1, CAR-GECO1, and R-GECO1.2 enable these Ca2+ indicators to be efficiently excited at wavelengths greater than 550 nm, we expected that they could be used to monitor Ca2+ concentration changes associated with ChR2 activation without causing unintentional additional activation of ChR2. To test this idea, we undertook a series of Ca2+ imaging experiments with a pancreatic islet insulinoma beta cell line, INS-1, transfected with a plasmid encoding either cytoplasmic R-GECO1.2 (i.e., no targeting sequence) or R-GECO1.2 anchored to the plasma membrane with the N-terminal peptide sequence of the Src kinase Lyn (Lyn-R-GECO1.2).20 For optogenetic manipulation experiments, these cells were cotransfected with a second plasmid encoding ChR2(T159C)-EGFP.21 Control experiments with cells transfected with only the Ca2+ indicator revealed that Lyn-R-GECO1.2 exhibited a slightly higher fluorescence intensity increase than R-GECO1.2 in response to KCl stimulated membrane depolarization and elevation of cytoplasmic and subplasmalemmal Ca2+ (Supporting Information Figure 4a–c) and was notably more sensitive than Fluo-3 and Lck-G-CaMP3 (Supporting Information Figure 4d,e).
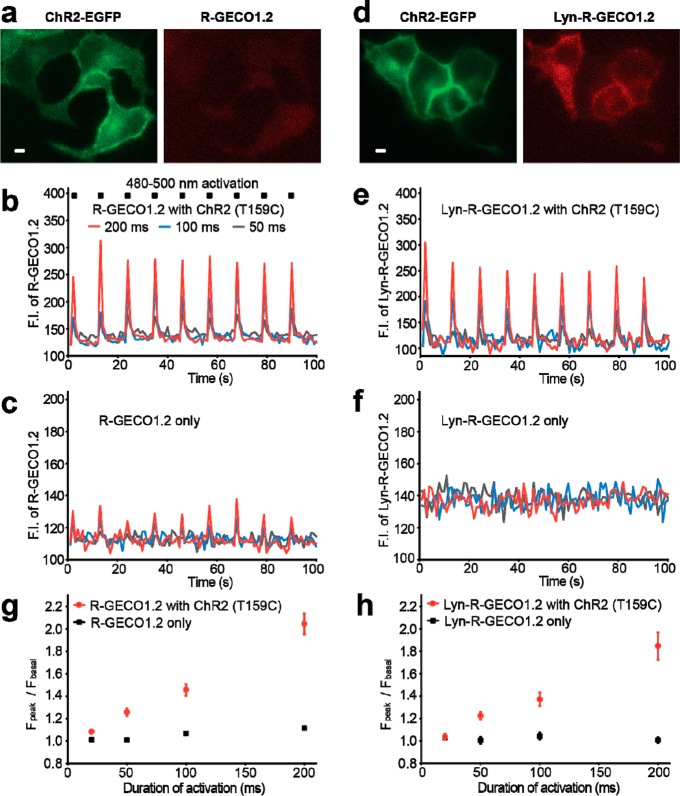
When we imaged INS-1 cells cotransfected with plasmids encoding R-GECO1.2 (or Lyn-R-GECO1.2) and ChR2(T159C)-EGFP (Figure 2a,d), we found that we were able to achieve robust apparent increases in cytoplasmic and subplasmalemmal Ca2+ using brief episodes (20–200 ms) of blue light illumination (480–500 nm at 135 mW/cm2) to activate ChR2(T159C)-EGFP (Figure 2b,e,g,h). However, control experiments with cells expressing only R-GECO1.2 also revealed transient increases in red fluorescence during blue light illumination lasting 100 ms or longer (Figure 2c). This ChR2-independent increase in red fluorescence amounted to as much as 12% of the total fluorescence signal for 200 ms illumination (Figure 2c,g). For reasons that remain unclear, the magnitude of the ChR2-independent fluorescent response was diminished in cells expressing Lyn-R-GECO1.2, and essentially indistinguishable from noise even for 200 ms illumination (Figure 2f,h). Notably, the blue light was turned off during acquisition of the red fluorescence image, so these transient increases were not due to additional excitation power. A similar effect was observed when using R-GECO1 in combination with 440 nm activation of the Zebrafish blue opsin22 coupled to the Gq signaling pathway (Supporting Information Figure 5). Brief (5 ms) pulses of 440 nm blue light at high power (33 W/cm2) consistently produced an approximate 20% increase in R-GECO1 fluorescence, even in those that were not transfected with the opsin. However, upon illuminating the cell with a train of 3 or more pulses of 440 nm light, a threshold in opsin activation was crossed and a significantly larger (80%), more persistent (10–30 s), and more heterogeneous R-GECO1 response was observed due to a Gq driven release of Ca2+ from intracellular stores. Akerboom et al. have recently reported similar observations of R-GECO1 photoactivation.3
Figure 2.
Imaging ChR2-induced Ca2+ transients in INS-1 cells. (a, d) Example fluorescence images of INS-1 cells expressing ChR2(T159C)-EGFP and either (a) R-GECO1.2 or (d) Lyn-R-GECO1.2. Scale bar = 2 μm. (b, e) ChR2-dependent fluorescence vs time traces for cells expressing ChR2(T159C)-EGFP and either (b) R-GECO1.2 or (e) Lyn-R-GECO1.2, during intervals of blue light irradiation (480–500 nm at 135 mW/cm2) for 50–200 ms. (c, f) ChR2-independent fluorescence vs time traces for cells expressing only (c) R-GECO1.2 or (f) Lyn-R-GECO1.2, during intervals of blue light irradiation as described for (b, e). (g, h) Average signal enhancements of R-GECO1.2 or Lyn-R-GECO1.2 upon increasing doses of blue light illumination in cells expressing (g) R-GECO1.2 only (n = 22) or R-GECO1.2 with ChR2(T159C)-EGFP (n = 33); or (h) Lyn-R-GECO1.2 (n = 18) or Lyn-R-GECO1.2 with ChR2(T159C)-EGFP (n = 24).
To determine if a similar combination of optogenetic activation and Ca2+ imaging could be achieved in intact brain tissue, we attempted to use CAR-GECO1 and ChR2(T159C)-EGFP to manipulate and image intracellular Ca2+ in mouse neocortical slice culture. In utero electroporation of mice embryos at embryonic day 13 with either a vector encoding CAR-GECO1 or vectors for both CAR-GECO1 and ChR2(T159C)-EGFP, was performed. Confocal fluorescence imaging of the transfected neocortical neurons that had been born at the ventricular surface on the day of electroporation was performed 6 days later (Figure 3a,d). Similar to our results in beta cells, Ca2+ elevations in cells transfected with only CAR-GECO1 showed transient increases in red fluorescence during confocal laser scanning with a 36 mW 405 nm laser at 100% power (90 μJ/μm2) for 1 s (Figure 3b). At 5% power levels (4.5 μJ/μm2, for 1 s), these increases in red fluorescence decreased to a level where they were practically indistinguishable from noise (Figure 3c). Cotransfection with plasmids for both CAR-GECO1 and ChR2(T159C)-EGFP, followed by confocal laser scanning with the 405 nm laser at 5% power, resulted in a robust CAR-GECO1 fluorescence response due to intracellular Ca2+ elevations (Figure 3f). No activation of ChR2(T159C)-EGFP was observed when transfected cells were illuminated with light from a 10 mW 561 nm laser at 50% power (12.5 μJ/μm2) for 1 s (Supporting Information Figure 6). Taken together, our imaging results in beta cells and neurons empirically demonstrate that, while this photoactivation phenomenon certainly complicates the use of red fluorescent Ca2+ indicators for use in combination with ChR2(T159C)-EGFP, it by no means prevents it. By appropriately adjusting the dosage or the duration of the light used for ChR2(T159C)-EGFP activation, it is feasible to perform optogenetic activation and Ca2+ imaging concurrently.
Figure 3.
Confocal imaging of ChR2(T159C)-induced Ca2+ elevations in mouse neocortical slice culture. Fluorescence images and response to photoactivation light for neocortial neurons transfected with only CAR-GECO1 (a–c) or cotransfected with CAR-GECO1 and ChR2(T159C)-EGFP (d–f). Scale bar = 50 μm. (b, c) Fluorescence vs time traces for cells transfected with only CAR-GECO1, during intervals of illumination at region of interest 1 (ROI-1) with a 405 nm laser at either 100% (90 μJ/μm2) (b) or 5% (4.5 μJ/μm2) (c) power. Control cells at ROI-2 and 3 were not illuminated. (e, f) Identical experimental conditions to (b, c) using tissue that has been cotransfected with CAR-GECO1 and ChR2(T159C)-EGFP. Based on the colocalization of green and red fluorescence, the neuron being photoactivated (at ROI-1) is expressing both CAR-GECO1 and ChR2(T159C)-EGFP.
In an effort to obtain further insight into the mechanism of the GECO photoactivation, we undertook an in vitro characterization of the effect of intense violet light illumination on the fluorescence intensity of R-GECO1, R-GECO1.2, CAR-GECO1, O-GECO1, and RCaMP1.07.2 Consistent with our cell imaging results, we found that all five purified proteins exhibited substantial transient increases in the proportion of proteins in an anionic red fluorescent form upon illumination with light from a 405 nm laser at 1200 mW/cm2 in the absence of Ca2+. At physiological pH (7.2), we observed absorbance increases of 631 ± 36% for CAR-GECO1, 434 ± 22% for R-GECO1.2, 256 ± 12% for R-GECO1, 291 ± 6% for RCaMP1.07, and 249 ± 52% for O-GECO1 (Figure 4a–c and Supporting Information Figure 7a, b). The recovery to the initial state proceeded rapidly once the 405 nm laser was turned off, and did not appear to be accelerated by illumination with longer wavelength visible light. With the exception of O-GECO1, all the rising portions (τon) of the intensity versus time curves were well fit as monoexponential functions with τon values ranging from 0.22 ± 0.02 s to 0.31 ± 0.02 s (Supporting Information Table 5 and Figure 8). O-GECO1 was best fit as a biexponential function with τon values of 0.11 ± 0.04 s (major component) and 5.4 ± 1.2 s (minor component). In contrast, the falling portions (τoff) for most indicators were best fit as biexponential functions, with the exception of R-GECO1.2 (τoff = 0.54 ± 0.05 s), which lacked the slow minor component (τoff2 ∼ 1.6–4.2 s) that was shared by the other GECOs.
Figure 4.
Reversible photoactivation of Ca2+ indicators during 405 nm illumination. Solutions of purified Ca2+ indicators were illuminated with a violet light laser (405 nm, 150 mW) for 5 s intervals. (a–c) Absorbance changes for the fluorescent (anionic) form of the Ca2+-free state during 5 s violet light pulses (black solid line) for (a) CAR-GECO1 at 560 nm; (b) R-GECO1.2 at 570 nm; (c) O-GECO1 at 545 nm. (d–f) Transient absorbance spectra (dashed lines) acquired immediately after the onset of illumination for (d) CAR-GECO1; (e) R-GECO1.2; (f) O-GECO1. (g–i) Transient absorbance spectra (dashed lines) acquired immediately after the end of illumination for (g) CAR-GECO1; (h) R-GECO1.2; (i) O-GECO1.
Inspection of the transient absorbance spectra revealed that the new species produced by photoactivation had a peak absorbance (λmax = 545 nm) that was blue-shifted relative to R-GECO1 (and 1.2), CAR-GECO1, and RCaMP1.07, and essentially identical to that of O-GECO1 (Figure 4d–f and Supporting Information Figure 7c, d). In all cases, formation of the new species was associated with a decrease in the proportion of the neutral, nonfluorescent form of the chromophore (Figure 4d–f and Supporting Information Figure 7c, d). Excitation at the transiently formed 545 nm peak during photoactivation did not give rise to significant fluorescence, yet 405 nm illumination itself generated a slightly red-shifted emission peak (λmax = 605 nm). For the red fluorescent GECOs, the fast increase in the 545 nm peak was followed by a slower increase in a longer wavelength absorbing species, effectively indistinguishable from the normal red fluorescent state (Figure 4d–f and Supporting Information Figure 7c, d). Upon turning the photoactivation light off, the intensity of the 545 nm peak decreased faster than that of the red fluorescent state (Figure 4g–i and Supporting Information Figure 7e, f). The fast formation and disappearance of the 545 nm intermediate species may explain why τon for O-GECO1 and τoff for most of the indicators were best fit as biexponential functions. For all five proteins, we observed that photoactivation became most significant at pH ∼7 and diminished at higher and lower pH values (Figure 5a–c and Supporting Information Figure 7g, h). The photoactivation effect was greatly diminished for the Ca2+-bound states of all Ca2+ indicators (Figure 5d–f, note the y-scale difference relative to Figure 5a–c; and Supporting Information Figure 7i, j, note the y-scale difference relative to Figure 7g, h), presumably because of the much smaller fraction of the protein in the neutral ground state. We observed a proportional decrease in photoactivation as the power of the 405 nm laser was decreased from 1200 to 1.2 mW/cm2 (Supporting Information Figure 9). At laser intensities in the range of 12 mW/cm2 or less (<1% laser power), intensity changes for the dim Ca2+-free state were limited to less than 50%.
Figure 5.

pH dependence of photoactivation of Ca2+ indicators. Change of absorbance in the Ca2+-free state (a–c) and Ca2+-bound state (d–f) with violet light (405 nm) illumination as a function of pH for: (a, d) CAR-GECO1; (b, e) R-GECO1.2; (c, f) O-GECO1.
The above results have led us to propose a model that captures the key features of the photoactivation phenomenon (Figure 6). Specifically, illumination shifts the chromophore ground state equilibrium toward a nonfluorescent anionic form (assumed to be anionic due to its long-wavelength absorbance) that has a blue-shifted absorbance relative to the red fluorescent form that predominates at high pH and in the Ca2+-bound state. This blue-shifted species then undergoes a slower conversion to the typical red fluorescent form. When illumination is turned off, this process is reversed and both the blue-shifted form (faster) and the red fluorescent form (slower) convert back to the neutral ground state form. The fact that the photoactivated form is not identical to the high pH anionic form rules out the simplest photoactivation mechanism in which violet/blue light illumination reversibly shifts the equilibrium between the protonation states of the chromophore toward the anionic form.23 Rather, we must turn to possible mechanisms in which the photoactivated form has an alternate structure, conformation, or microenvironment. One possibility is that violet or blue light illumination induces a Z to E isomerization of the chromophore, with the E isomer stabilized in an anionic state with a higher energy (blue-shifted) absorbance. Precedent indicates that, in some cases, the E isomers of FP chromophore are neutral and nonfluorescent,24−26 while in other cases they are anionic and fluorescent.27 Precedents also indicate that the time constants for Z to E recovery in the dark are much slower than those observed with the R-GECOs, and are typically in the range of minutes25 to hours.26
Figure 6.

Proposed mechanism of photoactivation in red fluorescent GECOs.
An alternative photoactivation mechanism, which lacks precedent but could also explain the observed results, is that blue light illumination induces the formation of an mOrange-type chromophore structure with a third ring. In mOrange (and O-GECO1), a threonine in the first position of the chromophore-forming tripeptide forms a dihydrooxazole ring due to attack of the side chain hydroxyl group on the carbonyl moiety of the preceding residue.28 This cyclization decreases the conjugation of the (otherwise red fluorescent) chromophore, resulting in a blue-shifted absorption and emission. To test the possibility that photoactivation induces a similar cyclization to form a 6-member sulfonium-containing ring (Supporting Information Figure 10a) in R-GECO1, we substituted the methionine at the first position of the chromophore (position 223; Supporting Information Figure 10a) with glutamine. It is unlikely that the glutamine side chain could participate in an analogous cyclization reaction. The resulting R-GECO-M223Q variant retained its response to Ca2+ and could be photoactivated (Supporting Information Figure 10b–d). We conclude that photoactivation does not involve the formation of a 6-member sulfonium-containing ring, leaving Z to E isomerization as the most reasonable explanation for the photoactivation phenomenon.
In conclusion, O-GECO1, R-GECO1.2, and CAR-GECO1 are engineered Ca2+ indicators that undergo intensiometric changes of 146×, 33×, and 27×, respectively. CAR-GECO1 retains a Kd that is similar to that of R-GECO1, and is most sensitive to Ca2+ changes in the range of 100–1000 nM. In contrast, R-GECO1.2 and O-GECO1 have lower affinity for Ca2+ and are most sensitive to Ca2+ changes in the range of 500–3000 nM. These indicators enable the detection of transient elevation in intracellular Ca2+ concentration with improved sensitivity. However, due to an inherent ability to be photoactivated, care must be taken when these indicators are used in conjunction with wavelengths of light that correspond to the absorbance profile of the protonated form of the chromophore (from ∼350 to ∼500 nm). Fortunately, using decreased duration or intensities of violet/blue activation light can circumvent this problem. If high power optogenetic activation with violet/blue light is unavoidable, a delay of several seconds will be required to allow the GECO fluorescence to recover to baseline. Under such circumstances, R-GECO1.2 would be the indicator of choice due to its fast τoff, recovering to approximately 5% of baseline within 2.9 s. Tethering of the Ca2+ indictor to the inner leaflet of plasma membrane (i.e., Lyn-R-GECO1.2) also helps to mitigate the problem of photoactivation, yet this approach may be limited to the subplasmalemmal microenvironment and may not be applicable to other cellular compartments. Yet another alternative would be to use the recently described RCaMP red fluorescent Ca2+ indicators that exhibit smaller Ca2+-dependent changes in fluorescence, but do not undergo photoactivation.3 We anticipate that these new tools will stimulate further efforts to combine the use of genetically encoded Ca2+ indicators and optogenetic tools for the simultaneous activation and imaging of excitable cells in different tissues including brains.
Methods
Engineering of Ca2+ Indicators
R-GECO1 in pTorPE1 was used as the initial template for all engineering. Mutations of V16E, F65I, L124V, and I161M were introduced by using primers mplum_V16E, mplum_F65I, mplum_L124V, and mplum_I161M and the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). Oligonucleotides used in this work are listed in Supporting Information Table 6. Primers V16NNK and F65NNK were used to randomize position 16 and 65 in a single reaction. Random mutagenesis was performed by error-prone PCR amplification29 using primers FW_XbaI-6His and RV_GCaMP-Stop-HindIII. For constructing the library of potentially orange-fluorescent GECO variants, primers oy_M66DYA, oy_F83TWH, oy_K163M, oy_I197RDA, and oy_Q213CWG were used. In this library, the codon for position 66 was DYA, the codon for position 83 was TWH, the codon for position 163 was ATG, the codon for position 197 was RDA, and the codon for position 213 was CWG, where D = A or G or T, Y = C or T, W = A or T, H = A or C or T, and R = A or G. R-GECO1 with M223Q was constructed using the mutagenic primerQC_R-GECO1_M66Q.
Screening of Gene Libraries
The procedure for screening libraries of O-GECO1 and R-GECO1.2 variants was identical to the procedure previously described for screening of R-GECO libraries.1 The procedure for screening CAR-GECO1 libraries was essentially identical, except for the use of ratiometric imaging of the E. coli colonies. Colonies expressing CAR-GECO variants, grown on 10 cm Petri dishes, were illuminated by light through an 535/50 nm excitation filter, and fluorescence emission was captured through both a 610/75 nm filter and a 680/40 nm emission filter. Colonies with ratios of 680/40 nm to 610/75 nm emission intensities that were in the highest 0.1% were picked and cultured individually. Bacteria expressing either O-GECO1 or R-GECO1.2 or CAR-GECO1 variants were cultured in liquid LB medium (with 0.0016% l-arabinose and 100 μg/mL ampicillin) for 36 h at 28 °C. Subsequent in vitro testing of picked variants was performed as described previously.1
Characterization of Improved Variants
For detailed characterization of O-GECO1, R-GECO1.2, and CAR-GECO1, proteins were expressed and purified as previously described.1 Spectral measurements were performed in solutions containing 10 mM EGTA or 10 mM EGTACa, 30 mM MOPS, 100 mM KCl, pH 7.2. For determination of fluorescence quantum yield, mOrange was used as a standard for O-GECO1 and mCherry for R-GECO1.2 and CAR-GECO1. Procedures for measurement of fluorescence quantum yield, extinction coefficient, pKa, Kd for Ca2+, and Ca2+-association kinetics have been described previously.1 Two-photon absorption spectra and cross sections were measured using a previously described method and optical setup.30 Briefly, a femtosecond Optical Parametric Amplifier (TOPAS-C, Light Conversion) was used for two-photon excitation of fluorescence. Fluorescence signal was collected from the first millimeter of the sample solution, contained in 1 cm optical cuvette. Two-photon absorption spectral profiles were measured relative to Styryl 9 M in chloroform in the 1090–1200 nm region and relative to Rhodamine B in methanol in the 780–1080 nm region. Two-photon cross sections were obtained relative to Rhodamine B in methanol at 1050 and 1114 nm in independent experiments. The concentration of chromophores was determined using peak extinction coefficients (from Supporting Information Table 2) and the Lambert–Beer law.
Assembling RCaMP1.07
RCaMP1.07 was assembled by overlap PCR using primers pBad_XhoI_Fw, fix_K47V_T49V_Rv, fix_K47V_T49V_Fw, CaMP_extend_Rv, assemble_fw1, assemble_fw2, connect_fw12, assemble_Rv (Supporting Information Table 6) with R-GECO1 in pTorPE as the template. PCR products were purified by gel electrophoresis in 1% agarose gel (Agarose S, Nippon Gene Co.). Gel extractions were carried out with the GeneJET gel extraction kit (Fermentas). Digestion was performed with the XhoI and HindIII restriction enzymes, and then the major DNA fragment was purified as described for PCR products. The purified product was then ligated into a pBAD/His B plasmid (Invitrogen). The ligation product was transformed into DH10B E. coli by electroporation and transformed bacteria were grown on agar plates with 400 μg/mL ampicillin overnight. Colonies were picked and cultured in LB media with 100 μg/mL ampicillin overnight. Plasmid DNA purification was performed to obtain DNA plasmids inserted regions were sequenced to verify their identity.
In Vitro Characterization of Photoactivation
For characterization of photoactivation in GECOs and RCaMP1.07, purified proteins were diluted to an absorbance value in the range of 0.1–0.5 in a buffered solution with a pH value ranging from 3 to 11. Buffered solutions were prepared as previously described.1 Kinetics experiments were performed using a UV–visible spectrometer (Agilent 8453 spectrophotometer). Purified proteins samples in buffered pH solutions were illuminated by a 150 mW (1200 mW/cm2) 405 nm laser (Changchun New Industries Optoelectronics Tech. Co., Ltd.) three times with a duration of 5 s each time during a kinetic measurement. When measuring photoactivation with different light source power, neutral density filters (2000a UVND, Chroma) were used while the rest of experimental conditions were kept the same.
Construction of Mammalian Expression Plasmids
To construct mammalian expression plasmids, pTorPE containing O-GECO1, R-GECO1.2, or CAR-GECO1 were used as the initial templates. PCR amplification of the GECO gene was performed using primers GCaMP_FW_BamH1 and GCaMP_RV_EcoR1. PCR products were purified by gel electrophoresis in 1% agarose gel (Agarose S, Nippon Gene Co.). Gel extractions were carried out with the GeneJET gel extraction kit (Fermentas). The resulting products were subjected to digestion with the BamHI and EcoRI restriction enzymes, and purified as described above for PCR products. The resulting DNA fragments were ligated with a modified pcDNA3 plasmid1 that had previously been digested with the same two enzymes. To construct the plasmid for expression of CAR-GECO1 in brain slices, the pEFx vector (constructed by replacement of the CMV-enhancer and -actin promoter of pCAGGS with the human elongation factor 1α (EF1α) promoter)31 was digested with BamHI and EcoRI and ligated with the CAR-GECO1 gene fragment cut from the pcDNA3 plasmid with the same enzymes. The pEFx vector enabled us to achieve high levels of CAR-GECO1 expression in brain slices. DNA sequencing was used to verify the inserted gene sequences, and large scale plasmid preparations to produce sufficient plasmid DNA for mammalian cell transfections was carried out as previously described.28
To generate Lyn-R-GECO1.2, R-GECO1.2 plasmid was digested with Kpn1 and BamH1. Oligos containing Lyn sequence (sense or antisense) and Kpn1 or BamH1 restriction sites were ligated with the digested plasmid. To construct ChR2(T159C)-EGFP, ChR2(T159C) was PCR-amplified from vector pCI-Synapsin-ChR2(T159C) using primers containing EcoR1 and BamH1 restriction sites. The amplified product was then ligated with EGFP-N1 that has been digested with EcoR1 and BamH1.
HeLa Cell Culture and Imaging
HeLa cells (40–60% confluent) on collagen-coated 35 mm glass bottom dishes (Mastumami) were transfected with 1 μg of plasmid DNA and 4 μL of Lipofectamine 2000 (Life technologies) according to the manufacturer’s instructions. After 2 h incubation, the media was exchanged to Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and the cells were incubated for an additional 24 h at 37 °C in a CO2 incubator. Immediately prior to imaging, cells were washed twice with Hank’s balanced salt solution (HBSS), and then 1 mL of 20 mM HEPES buffered HBSS (HHBSS) was added.
Cell imaging was performed with an inverted microscope (Eclipse Ti-E, Nikon) equipped with a digital CCD camera (ORCA-R2, Hamamatsu Photonics K.K.), a microscanning stage (MSS-BT110, Chuo Precision Industrial Co. Ltd.), and an incubator system (Chamlide IC, Live Cell instrument). The AquaCosmos software package (Hamamatsu Photonics K.K.) was used for automated microscope and camera control. For determination of the dynamic range of new indicators in live cells, cells were imaged with a 40× oil objective lens (NA 1.3), excitation light intensity was decreased to 1% with a neutral density filter (ND100), and a 0.6× relay lens was attached before CCD camera. We used the following combinations of excitation filters, dichroic mirrors, and emission filters, respectively: 534/20, 552, and 572/28 nm for O-GECO1; and 562/40, 593, and 624/40 nm for R-GECO1.2 and CAR-GECO1. All filters were purchased from Semrock and had part numbers FF01-534/20-25, FF552-Di01-25×36, FF01-572/28-25, FF01-562/40-25, FF593-Di02-25×36, and FF01-624/40-25. Imaging was performed at room temperature.
For imaging of histamine-induced Ca2+ dynamics, 500 ms exposure images (2 × 2 binning) were acquired every 5 s for 15 min. Approximately 30 s after the start of the experiment, histamine (500 μM in PBS, 10 μL) was added to a final concentration of 5 μM. After 15 min of imaging, cells were washed twice with HBSS and then incubated for 10 min in 1 mL HHBSS to allow histamine-induced oscillations to subside. Cells were then once again imaged as described above, with exposures every 10 s for a duration of 5 min. Approximately 30 s after the start of the imaging, 1 mL of 2 mM CaCl2, 10 μM ionomycin in Ca2+- and Mg2+-free HHBSS (HHBSS(−)) was added to the dish via peristaltic pump (AC-2110, ATTO). After 5 min of imaging, cells were washed three times with HHBSS(−), 1 mL of 1 mM EGTA and 5 μM ionomycin in HHBSS(−) was added to the media, and cells were imaged once again with exposures every 5 s for a duration of 5 min.
Ca2+ Imaging of R-GECO1.2 and ChR2 Activation in INS-1 cells
INS-1(832/13) cells were seeded on 35-mm glass bottom imaging dishes (MatTek) and transfected 24 h later with R-GECO1.2 with or without ChR2(T159C)-EGFP by Metafectene (Biontex) according to the manufacturer’s instructions. Cells were then cultured for additional 24 h before imaging. Photoactivation and imaging were carried out on an inverted wide-field fluorescence microscope (Axiovert 200, Carl Zeiss) through a 63× oil objective (NA 1.25). Fluorescence excitation was controlled by a Lambda DG4 exciter (Sutter Instrument, Novato CA) equipped with a 175 W Xenon lamp (PerkinElmer, PE175BF). To activate ChR2(T159C)-EGFP, or to image Fluo-3 or G-CaMP, a band-pass filter (S490/20x, Chroma) was used to provide excitation light at the intensity of 135 mW/cm2 (measured immediately above the objective lens using a LI-190 quantum sensor and a LI-250A light meter (LI-COR Biosciences)). Fluorescence emissions from R-GECO1.2 (605 ± 26 nm), Fluo-3 or G-CaMP (525 ± 18 nm) were collected using an EMCCD camera (iXon3 897, Andor).
Light Driven Gq Activation
The hybrid Gq/t α subunit was a chimeric Gq subunit in which the last C-terminal 18 amino acids of Gq were replaced with Gt. This is a commonly used strategy for converting receptor specificity of a particular α subunit.32 To create the Gq/t hybrid, a series of PCR primers were designed to amplify the coding region of Gq and replace the C-terminal 18 amino acids of Gq with Gt. The final PCR product was cloned in the CMV expression plasmid pUB2.1 (Addgene plasmid 40728) using an InFusion joining reaction (Clontech, Mountain View, CA). HEK 293 cells were cotransfected with CMV expression plasmids carrying the Zebrafish blue opsin, the hybrid Gq/t α subunit, and the R-GECO1 calcium sensor using Lipofectamine 2000 (Life Technologies, Grand Island, NY) following the manufacturer’s recommended protocol. Time lapse imaging was done with an Olympus IX70 inverted microscope fitted with computer controlled filter wheel and shutter (Sutter Instrument, Novato, CA) in the excitation path. A Retiga camera (Surrey, BC, Canada) and a 40× objective lens were used to collect the images. An Exfo light source (Mississauga, Ontario, Canada) with a metal halide lamp provided the excitation light. A 440/20 excitation filter was used to activate the Zebrafish blue opsin, and a 556/20 excitation filter was used to excite the R-GECO1. The dichroic and emission filters were from Semrock (Rochester, NY) “Pinkel” set (GFP/DsRed-2X-A-000).
Ca2+ Imaging with ChR2 Activation in Mice Brain Slices
For imaging of cultured mice brain slices, pallium tissue of day 13 embryos of ICR strain mice was electroporated with plasmids (pEFx/CAR-GECO1 and pEF-1/ChR2(T159C)-EGFP) using a CUY21 electroporator (Nepagene) and an applied voltage of 30 V as previously described.33,34 Cerebral walls were isolated from the electroporated embryos 6 days later and were sliced (300 μm), embedded in collagen gel,35 and imaged in artificial cerebrospinal fluid containing NaCl (125 mM), KCl (5 mM), NaH2PO4 (1.25 mM), MgSO4 (1 mM), CaCl2 (2 mM), NaHCO3 (25 mM), and glucose (20 mM) at pH 7.4. All images were obtained with confocal upright microscope (A1R, Nikon) equipped with a 25× water immersion lens (NA 1.10, Nikon).34 Light from a 36 mW 405 nm diode laser was used to activate ChR2(T159C)-EGFP, and CAR-GECO1 was imaged with excitation with a 10 mW 561 nm laser and collection of fluorescence through a 570–620 nm bandpass filter. Expression of ChR2(T159C)-EGFP was confirmed by EGFP florescence excited by a 488 nm laser.
For imaging neocortical neurons in mice brain slices, images were first acquired with 561 nm laser excitation at 10% power (2.5 μJ/μm2) for 1 min. Samples were then stimulated for 1 s, by 405 nm laser at 100% (90 μJ/μm2) or 5% (4.5 μJ/μm2) power, or by 561 nm laser with 50% power (12.5 μJ/μm2). Following stimulation, images were acquired with 561 nm laser excitation at 10% power (2.5 μJ/μm2) for 15 s. This 1 s stimulation and 15 s acquisition process were repeated 7 more times. Finally, the samples were imaged by 561 nm laser with 10% power (2.5 μJ/μm2) for 1 min.
Acknowledgments
The authors thank Dr. Thomas Oertner for providing the plasmid encoding ChR2(T159C), Akira Sakakibara for the EF2 vector, Pamela Raymond for the Zebrafish blue opsin, Wayne Moffat and Joel Weiner for technical support. Technical support was provided by the Nikon Imaging Center at Hokkaido University and the University of Alberta MBSU. Plasmid requests will be handled through Addgene and covered under the Uniform Biological Material Transfer Agreement.
Supporting Information Available
Additional figures, tables, and video. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
J.W. performed directed evolution and the majority of the characterization of O-GECO1, R-GECO1.2 and CAR-GECO1. L.L. performed imaging and optogenetic activation in INS-1 cells. T. Matsuda and Y.-F.C. performed imaging and optogenetic activation in neurons. Y.Z. performed kinetic characterization. A.R. and M.D. acquired the 2-photon spectra. S.A. imaged Ca2+ dynamics in HeLa cells. Y.A. developed the microscope for optogenetic activation and imaging. K.M. and T.E.H. performed imaging and optogenetic activation with the short wavelength opsin. K.S. performed electroporation of mouse embryos. T. Miyata prepared brain slices from transfected mouse embryo and processed imaging data. R.E.C., W.-H.L, and T.N. directed research, devised experiments, and wrote the manuscript.
This work was supported by Alberta Innovates (Y.Z.); the U.S. National Institute of General Medical Sciences grant R01 GM086198 (M.D.); R01 GM077593 (W.-H.L); JSPS, CIHR (NHG 99085 and MOP 123514) and NSERC (R.E.C.); PRESTO from JST, FIRST, and MEXT Grant-in-Aid for Scientific Research on Innovative Areas 23115003 (T.N.), and 3111502 (T.M.). R.E.C. holds a Tier II CRC in Bioanalytical Chemistry.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Zhao Y.; Araki S.; Wu J.; Teramoto T.; Chang Y. F.; Nakano M.; Abdelfattah A. S.; Fujiwara M.; Ishihara T.; Nagai T.; Campbell R. E. (2011) An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura M.; Sasaki T.; Kobayashi C.; Ikegaya Y.; Nakai J. (2012) An improved genetically encoded red fluorescent Ca2+ indicator for detecting optically evoked action potentials. PLoS One 7, e39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerboom J.; Calderón N. C.; Tian L.; Wabnig S.; Prigge M.; Tolö J.; Gordus A.; Orger M. B.; Severi K. E.; Macklin J. J.; Patel R.; Pulver S. R.; Wardill T. J.; Fischer E.; Schüler C.; Chen T. -W.; Sarkisyan K. S.; Marvin J. S.; Bargmann C. I.; Kim D. S.; Kügler S.; Lagnado L.; Hegemann P.; Gottschalk A.; Schreiter E. R.; Looger L. L. (2013) Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G.; Szellas T.; Huhn W.; Kateriya S.; Adeishvili N.; Berthold P.; Ollig D.; Hegemann P.; Bamberg E. (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. U.S.A. 100, 13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden E. S.; Zhang F.; Bamberg E.; Nagel G.; Deisseroth K. (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Zhang Y. P.; Oertner T. G. (2007) Optical induction of synaptic plasticity using a light-sensitive channel. Nat. Methods 4, 139–141. [DOI] [PubMed] [Google Scholar]
- Prigge M.; Rösler A.; Hegemann P. (2010) Fast, repetitive light-activation of CaV3.2 using channelrhodopsin 2. Channels (Austin) 4, 241–247. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Prigge M.; Beyrière F.; Tsunoda S. P.; Mattis J.; Yizhar O.; Hegemann P.; Deisseroth K. (2008) Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat. Neurosci. 11, 631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Y.; Lin M. Z.; Steinbach P.; Tsien R. Y. (2009) Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys. J. 96, 1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki A.; Llopis J.; Heim R.; McCaffery J. M.; Adams J. A.; Ikura M.; Tsien R. Y. (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 388, 882–887. [DOI] [PubMed] [Google Scholar]
- Nakai J.; Ohkura M.; Imoto K. (2001) A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nat. Biotechnol. 19, 137–141. [DOI] [PubMed] [Google Scholar]
- Govorunova E. G.; Spudich E. N.; Lane C. E.; Sineshchekov O. A.; Spudich J. L. (2011) New channelrhodopsin with a red-shifted spectrum and rapid kinetics from Mesostigma viride. mBio 2 e00115–e00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge M.; Schneider F.; Tsunoda S. P.; Shilyansky C.; Wietek J.; Deisseroth K.; Hegemann P. (2012) Color-tuned channelrhodopsins for multiwavelength optogenetics. J. Biol. Chem. 287, 31804–31812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Y. (2011) A user’s guide to channelrhodopsin variants: features, limitations and future developments. Exp. Physiol. 96, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford S. C.; Wu J.; Zhao Y.; Campbell R. E.; Knöpfel T. (2013) Optogenetic reporters. Biol. Cell 105, 14–29. [DOI] [PubMed] [Google Scholar]
- Wang L.; Jackson W. C.; Steinbach P. A.; Tsien R. Y. (2004) Evolution of new nonantibody proteins via iterative somatic hypermutation. Proc. Natl. Acad. Sci. U.S.A. 101, 16745–16749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbyad P.; Childs W.; Shi X.; Boxer S. G. (2007) Dynamic Stokes shift in green fluorescent protein variants. Proc. Natl. Acad. Sci. U.S.A. 104, 20189–20194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N. C.; Campbell R. E.; Steinbach P. A.; Giepmans B. N.; Palmer A. E.; Tsien R. Y. (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Akerboom J.; Chen T. W.; Wardill T. J.; Tian L.; Marvin J. S.; Mutlu S.; Calderón N. C.; Esposti F.; Borghuis B. G.; Sun X. R.; Gordus A.; Orger M. B.; Portugues R.; Engert F.; Macklin J. J.; Filosa A.; Aggarwal A.; Kerr R. A.; Takagi R.; Kracun S.; Shigetomi E.; Khakh B. S.; Baier H.; Lagnado L.; Wang S. S.; Bargmann C. I.; Kimmel B. E.; Jayaraman V.; Svoboda K.; Kim D. S.; Schreiter E. R.; Looger L. L. (2012) Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. J. Neurosci. 32, 13819–13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh M. D. (1999) Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta 1451, 1–16. [DOI] [PubMed] [Google Scholar]
- Berndt A.; Schoenenberger P.; Mattis J.; Tye K. M.; Deisseroth K.; Hegemann P.; Oertner T. G. (2011) High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. U.S.A. 108, 7595–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihtelic T. S.; Doro C. J.; Hyde D. R. (1999) Cloning and characterization of six zebrafish photoreceptor opsin cDNAs and immunolocalization of their corresponding proteins. Vis. Neurosci. 16, 571–585. [DOI] [PubMed] [Google Scholar]
- Jung G.; Mais S.; Zumbusch A.; Brauchle C. (2000) The Role of Dark States in the Photodynamics of the Green Fluorescent Protein Examined with Two-Color Fluorescence Excitation Spectroscopy. J. Phys. Chem. A 104, 873–877. [Google Scholar]
- Andresen M.; Stiel A. C.; Trowitzsch S.; Weber G.; Eggeling C.; Wahl M. C.; Hell S. W.; Jakobs S. (2007) Structural basis for reversible photoswitching in Dronpa. Proc. Natl. Acad. Sci. U.S.A. 104, 13005–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J. N.; Ai H. W.; Campbell R. E.; Remington S. J. (2007) Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc. Natl. Acad. Sci. U.S.A. 104, 6672–6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam V.; Lelimousin M.; Boehme S.; Desfonds G.; Nienhaus K.; Field M. J.; Wiedenmann J.; McSweeney S.; Nienhaus G. U.; Bourgeois D. (2008) Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. U.S.A. 105, 18343–18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J.; Wilmann P. G.; Beddoe T.; Oakley A. J.; Devenish R. J.; Prescott M.; Rossjohn J. (2003) The 2.0-angstrom crystal structure of eqFP611, a far red fluorescent protein from the sea anemone Entacmaea quadricolor. J. Biol. Chem. 278, 44626–44631. [DOI] [PubMed] [Google Scholar]
- Shu X.; Shaner N. C.; Yarbrough C. A.; Tsien R. Y.; Remington S. J. (2006) Novel chromophores and buried charges control color in mFruits. Biochemistry 45, 9639–9647. [DOI] [PubMed] [Google Scholar]
- Cirino P. C., Mayer K. M., and Umeno D. (2003) Generating Mutant Libraries Using Error-Prone PCR. In Directed Evolution Library Creation (Arnold F. H., and Georgiou G., Eds.), pp 3–9, Humana Press, Totowa, New Jersey. [DOI] [PubMed] [Google Scholar]
- Makarov N. S.; Drobizhev M.; Rebane A. (2008) Two-photon absorption standards in the 550–1600 nm excitation wavelength range. Opt. Express 16, 4029–4047. [DOI] [PubMed] [Google Scholar]
- Minobe S.; Sakakibara A.; Ohdachi T.; Kanda R.; Kimura M.; Nakatani S.; Tadokoro R.; Ochiai W.; Nishizawa Y.; Mizoguchi A.; Kawauchi T.; Miyata T. (2009) Rac is involved in the interkinetic nuclear migration of cortical progenitor cells. Neurosci. Res. 63, 294–301. [DOI] [PubMed] [Google Scholar]
- Conklin B. R.; Farfel Z.; Lustig K. D.; Julius D.; Bourne H. R. (1993) Substitution of three amino acids switches receptor specificity of Gq alpha to that of Gi alpha. Nature 363, 274–276. [DOI] [PubMed] [Google Scholar]
- Saito T.; Nakatsuji N. (2001) Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240, 237–246. [DOI] [PubMed] [Google Scholar]
- Tabata H.; Nakajima K. (2008) Labeling embryonic mouse central nervous system cells by in utero electroporation. Dev. Growth Differ. 50, 507–511. [DOI] [PubMed] [Google Scholar]
- Miyata T.; Kawaguchi A.; Okano H.; Ogawa M. (2001) Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron 31, 727–741. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Shui B.; Kotlikoff M. I.; Sondermann H. (2008) Structural basis for calcium sensing by GCaMP2. Structure 16, 1817–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.