Abstract
The D3 dopamine receptor is a therapeutic target for treating various nervous system disorders such as schizophrenia, Parkinson’s disease, depression, and addictive behaviors. The crystal structure of the D3 receptor bound to an antagonist was recently described; however, the structural features that contribute to agonist-induced conformational changes and signaling properties are not well understood. We have previously described the conformation-dependent tolerance and slow response termination (SRT) signaling properties of the D3 receptor and identified the C147 residue in the second intracellular loop (IL2) of the D3 receptor as important for the tolerance property. Interestingly, while IL2 and the C147 residue, in particular, were important for dopamine- and quinpirole-induced tolerance, this residue did not affect the severe tolerance induced by the high affinity, D3 receptor-selective agonist, PD128907. Here, we used D2/D3 receptor chimeras and site-specific D3 receptor mutants to identify another residue, D187, in the second extracellular loop (EC2) of the human D3 receptor that mediates the tolerance property induced by PD128907, quinpirole, pramipexole, and dopamine. Molecular dynamics simulations confirmed the distinct conformation adopted by D3 receptor during tolerance and suggested that in the tolerant D3 receptor the D187 residue in EC2 forms a salt bridge with the H354 residue in EC3. Indeed, site-directed mutation of the H354 residue resulted in loss of PD1287907-induced tolerance. The mapping of specific amino acid residues that contribute to agonist-dependent conformation changes and D3 receptor signaling properties refines the agonist-bound D3 receptor pharmacophore model which will help develop novel D3 receptor agonists.
Keywords: Structure−function, signal transduction, desensitization, functional selectivity, D3 receptor conformation, potassium channels
The D3 dopamine receptor is one of five dopamine receptor subtypes and belongs to the “D2-like” dopamine receptor subfamily that includes the D2 and D4 dopamine receptors. D3 receptor ligands are clinically efficacious in treating Parkinson’s disease, schizophrenia, depression and addictive behaviors, making the D3 receptor an important therapeutic target.1−4 An antagonist-bound crystal structure of D3 dopamine receptor was recently published and identified domains involved in interactions with antagonists.5 Pharmacophore models based on the antagonist-bound D3 receptor structure while successful at identifying novel high affinity D3 receptor antagonists unfortunately are not ideal for identifying novel D3 receptor agonists.6 In the absence of an agonist-bound crystal structure of the D3 receptor, the residues and domains involved in mediating agonist-dependent conformation changes in the D3 receptor are less well understood. Furthermore, with the recent discovery of functionally selective ligands that produce distinct receptor conformation which direct the coupling of the receptor to specific signaling pathways, it has become important to map amino acid residues and structural domains that contribute to diverse agonist-induced conformational changes.7 The mapping of residues involved in the generation of the various receptor conformations induced by different agonists will help refine the agonist-bound receptor pharmacophore models. Such pharmacophore models will facilitate the development of novel functionally selective agonists.
In the case of the D3 dopamine receptor, we have previously identified two agonist-induced signaling properties, tolerance and slow response termination (SRT), which distinguishes the D3 receptor from the closely related D2 receptors.8 The tolerance property of the D3 receptor is the progressive decrease in receptor signaling function upon repeated stimulation by classical agonists, including dopamine. The SRT property is the prolongation of time taken to terminate the signaling function of the D3 receptor, after removal of the agonist. The D3 receptor tolerance property is distinct from classical desensitization as it develops only after the removal of agonist; there is negligible loss of response in the continued presence of agonists. We have shown that the tolerance property is not mediated by D3 receptor internalization, persistent agonist binding or a change in binding affinity;9 however, the D3 receptor does adopt a distinct conformation following agonist-induced tolerance.10 More recently, we have shown that the induction of tolerance and SRT is agonist-dependent.11 Using chimeric D3-D2 receptors and site-directed mutagenesis, we have also shown that the D3 receptor intracellular loop 2 (IL2), and in particular, the C147 residue is important for the D3 receptor tolerance property.8,9 In the studies using chimeric D3-D2 receptors, the substitution of D3 receptor intracellular loops with D2 receptor intracellular loops prevented the induction of the tolerance property; however, reciprocal chimeras in which D2 receptor intracellular loops were substituted with D3 receptor intracellular loops failed to impart the tolerance and SRT properties to the chimeric receptors.8 This suggested that other domains of the D3 receptor contribute to the agonist-induced tolerance and SRT properties. In this paper, we systematically investigated the role of D3 receptor transmembrane 4 (TM4) and extracellular regions in the induction of tolerance and SRT properties. The functional studies were performed using electrophysiological methods that assayed the coupling of human D3 receptors to endogenous G-protein coupled inward rectifier potassium (GIRK) channels in the AtT-20 neuroendocrine cells. The results identified extracellular loop 2 (EC2) and, in particular, the D187 residue in the D3 receptor as important for the agonist-induced tolerance property. These studies were coupled with molecular docking and molecular dynamic (MD) simulations of wild type, mutant, and chimeric D2 and D3 receptors bound to either quinpirole or PD128907. The results show that distinct receptor conformations code for the induction of tolerance properties in the presence of various D3 receptor agonists. The study provides novel structural information and identifies residues that are important for agonist-dependent conformations that determine D3 receptor signaling properties.
Results and Discussion
Our previous studies on D3 receptor, an inhibitor of adenylate cyclase and activator of GIRK channels and mitogen-activated protein kinases (MAPK), have shown that this receptor exhibits tolerance and SRT when presented with different agonists.8,11 In particular, among the different D2/D3 receptor agonist, PD128907 induces the most severe tolerance and SRT11 and is the focus of this paper. The tolerance and SRT properties are not exhibited by the D2 receptor although the two receptors signal through the same pathways and are highly homologous. We have proposed that the distinct D3 receptor tolerance and SRT properties might play an important role in physiological and pathophysiological conditions.9 Under physiological conditions, we have previously proposed that the D3 receptor, with its tolerance and SRT properties, functions as a high pass filter, blocking weak signals and allowing strong signals.12 In pathological conditions, the D3 receptor tolerance and SRT properties are likely to be relevant when there are significant changes in the expression of the D3 receptor due to disease or chronic treatment. For example, studies have shown that in animal models of Parkinson’s disease, there is a significant increase in expression of the D3 receptor in the basal ganglia of animals exhibiting levodopa-induced dyskinesia.13 We have proposed that this increase in D3 receptor expression alters neuronal signaling due to enhanced expression of the tolerance and SRT properties which might contribute to the dyskinetic symptoms.9
Agonist-Specific Structural Determinants of D3 Receptor Tolerance Property
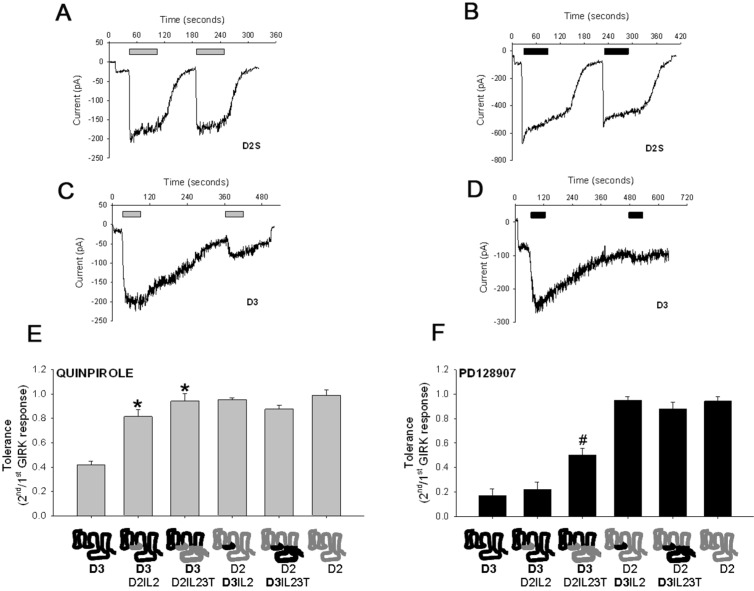
Classical D2/D3 receptor agonists, quinpirole and PD128907, induce tolerance and SRT properties in D3 but not D2 receptor (Figure 1A-D). Of the two agonists, PD128907 induces more significant tolerance and SRT (Figure 1C–F). The acute GIRK response induced by a maximal dose (100 nM) of quinpirole and PD128907 are similar (Supporting Information Table 1); this suggests that the difference in the ability to induce tolerance is not due to differences in the relative efficacy of the two agonists to activate the D3 receptor-GIRK channel signaling pathway. We have previously shown that substitution of D3 receptor IL2 with D2 receptor IL2 (D3D2IL2) abolishes the tolerance induced by quinpirole in the chimeric receptor (Figure 1E).8 However, Figure 1F shows that the D3D2IL2 chimeric receptor still exhibits PD128907-induced tolerance, which is partly attenuated in the D3D2IL23T chimeric receptor. Despite the partial attenuation, substitution of D2 receptor IL2, IL3, and carboxyl tail regions with corresponding D3 receptor regions does not result in the induction of either quinpirole- or PD128907-induced tolerance (Figure 1E and F). This result suggests that the extracellular and/or transmembrane regions of the D3 receptor might be important for the induction of agonist-induced tolerance and SRT properties. The results also suggest that the D3 receptor tolerance induced by the two different agonists is not determined by the same structural domain or by the same series of agonist-induced conformational steps.
Figure 1.
Different structural domains determine the D3 receptor tolerance induced by quinpirole and PD128907. Representative whole cell voltage clamp recordings obtained from AtT-20 cells stably expressing either the human D2S (A and B) or D3 (C and D) dopamine receptors. The cells were held at −65 mV and inward GIRK currents in 30K-ES induced by 100 nM quinpirole (gray; A, C, and E) or 100 nM PD128907 (black; B, D, and F) measured. The agonists were applied for ∼60 s at indicated times. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for quinpirole (E) and PD128907 (F) in AtT-20 cells transiently transfected with either wild type D3 or D2S receptors or various chimeric receptors. Compared to wild type D3 receptor, quinpirole does not induce tolerance in the D3D2IL2 and D3D2IL23T chimeric receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). PD128907-induced tolerance in the D3D2IL23T chimeric receptor is significantly attenuated compared to wild type D3 receptor or the D3D2IL2 chimeric receptor; #P < 0.01, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 10–12 cells).
PD128907-Induced D3 Receptor Tolerance Is Mediated by EC2
Compared to the transmembrane regions and extracellular loops, the extracellular N-terminus region of D2 and D3 receptors is dissimilar with ∼30% identity (Supporting Information Figure 1). To determine if the N-terminal region contributes to the agonist-induced tolerance and SRT properties of the D3 receptor, we constructed chimeric receptors in which we exchanged the N-terminal regions of the D3 and D2 receptors. The results show that chimeric receptor with the N-terminus of D3 receptor on D2 receptor (N-D3 D2S) did not induce quinpirole or PD128907-induced tolerance and SRT (Figure 2A). In addition, the chimeric receptor with the N-terminus of D2 receptor on D3 receptor (N-D2S D3) did not prevent quinpirole or PD128907-induced tolerance (Figure 2B). The results suggest that the N-terminus of D3 receptor is not involved in agonist-induced tolerance and SRT.
Figure 2.
D3 receptor N-terminus and TM4 are not sufficient to impart quinpirole- or PD128907-induced tolerance. Representative whole cell voltage clamp recordings obtained from AtT-20 cells stably expressing either the D2S chimeric receptor with the N-terminus region of the D3 receptor (A) or the D3 chimeric receptor with the N-terminus region of D2S receptor (B). The cells were held at −65 mV and inward GIRK currents in 30K-ES induced by 100 nM quinpirole (gray) or 100 nM PD128907 (black) measured. The agonists were applied for ∼60 s at indicated times. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for quinpirole (C) and PD128907 (D) in AtT-20 cells transiently transfected with either wild type D3 or D2S receptors or various chimeric receptors. Quinpirole- and PD128907-induced tolerance was significantly different only between wild type D3 receptor and all wild type and chimericD2 receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 10–12 cells).
The D3 receptor IL2 is involved in tolerance and is linked to transmembrane 3 (TM3) and transmembrane 4 (TM4). Of these two transmembrane regions, TM4 is less conserved between D2 and D3 receptors (Supporting Information Figure 1). Our recent molecular dynamics studies with D3 receptor homology models showed large shifts in TM4 when a tolerance-inducing (PD128907) versus nontolerance inducing (PBZI) agonist was docked to the D3 receptor;11 therefore, we investigated the role of TM4 in the tolerance and SRT properties of the D3 receptor. Figure 2C and D show that substitution of D2 receptor TM4 with the D3 receptor TM4 in the D2D3IL123T chimeric receptor did not result in quinpirole- or PD128907-induced tolerance. There was also no significant difference in the quinpirole or PD128907-induced acute GIRK response between the D2D3IL123T and D2D3IL123T-TM4 chimeric receptors (Supporting Information Table 1). These results suggest that the addition of TM4 region of D3 receptor is not sufficient to induce the tolerance and SRT properties.
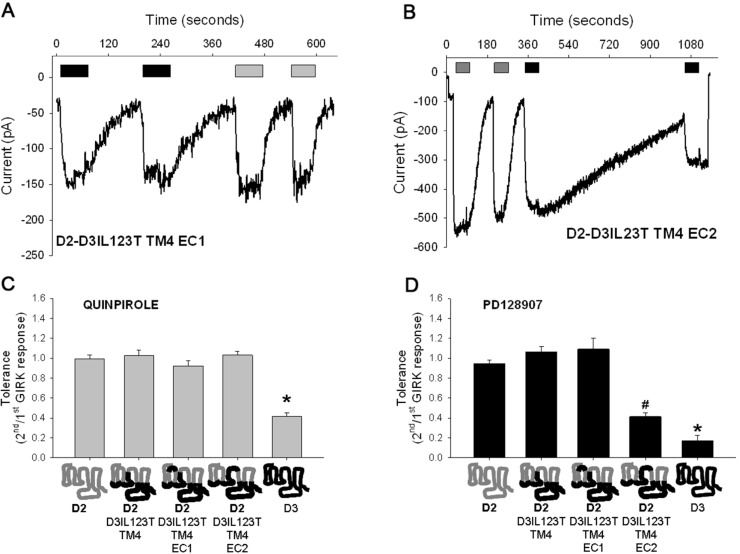
We next investigated if the other extracellular regions of the D3 receptor were necessary for the tolerance and SRT properties. To determine this, we individually substituted EC1 and EC2 of the D2 receptor with EC1 and EC2 of the D3 receptor in the D2D3IL123T-TM4 chimeric receptor. The results in Figure 3 shows that while substitution of D3 receptor EC1 in the chimeric receptor had no effect on quinpirole or PD128907-induced tolerance and SRT (Figure 3A, C, and D), the substitution of D3 receptor EC2 in the chimeric receptor resulted in a significant PD128907-induced tolerance and SRT (Figure 3B and D). Interestingly, this chimeric receptor failed to exhibit quinpirole-induced tolerance and SRT (Figure 3B and C). Results from MD simulations of PD128907-docked models show that unlike the D2D3IL123T-TM4-EC1 chimeric receptor, the D2D3IL123T-TM4-EC2 chimeric receptor has a conformation that is more similar to the wild type D3 receptor than the wild type D2S receptor (Supporting Information Figure 2 and Table 2). The quinpirole- or PD128907-induced acute GIRK response between the D2D3IL123T-TM4-EC1 and D2D3IL123T-TM4-EC2 chimeric receptors was not significantly different (Supporting Information Table 1). These results suggest that the EC2 of D3 receptor is specifically involved in generating a PD128907-induced receptor conformation that causes tolerance and SRT in the chimeric receptor.
Figure 3.
D3 receptor EC2 domain specifically imparts PD128907-induced tolerance to the D2D3IL123T-TM4 chimeric receptor. Representative whole cell voltage clamp recordings in AtT-20 cell transiently transfected with either the D2D3IL123T-TM4-EC1 (A) or D2D3IL123T-TM4-EC2 (B) chimeric receptors. The cells were held at −65 mV and inward GIRK currents in 30K-ES induced by 100 nM quinpirole (gray) or 100 nM PD128907 (black) measured. The agonists were applied for ∼60 s at indicated times. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for quinpirole (C) and PD128907 (D) in AtT-20 cells transiently transfected with either wild type D3 or D2S receptors or various chimeric receptors. Quinpirole- and PD128907-induced tolerance was significantly different between wild type D3 receptor and all wild type and chimeric D2 receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). In addition, PD128907-induced tolerance was significantly different between the D2D3IL12T-TM4-EC2 chimeric receptor and all other receptors shown in panel D; #P < 0.01, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 10–12 cells).
PD128907-Induced D3 Receptor Tolerance Is Specifically Mediated by the D187 Residue in EC2
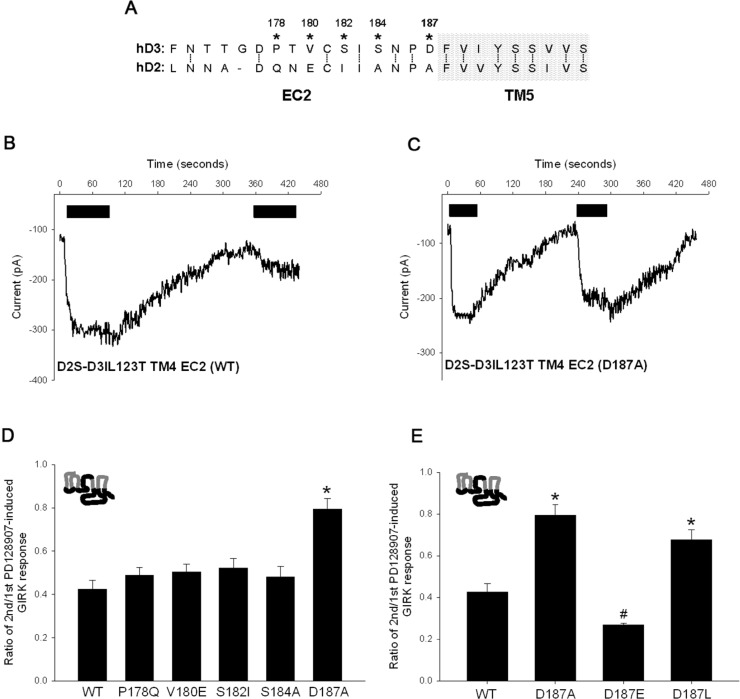
Having identified EC2 of D3 receptor as important for tolerance and SRT, we next used site-directed mutagenesis to mutate amino acid residues that were significantly different between the EC2 of D3 and D2 receptors. There are nine amino acids that are not conserved between the EC2 regions of D3 and D2 receptors (Supporting Information Figure 1). Of these, five are completely distinct whereas four are similar in that they have polar uncharged side chains; for this study we focused on the five amino acid residues in EC2 that were completely different between the D3 and D2 receptors. The EC2 amino acid residues in the chimeric D2D3IL123T-TM4-EC2 receptor were individually mutated to the corresponding D2 receptor amino acid residues (Figure 4A). The results in Figure 4B–D show that only the mutation of the D187 residue in the EC2 of the chimeric receptor to the corresponding D2 receptor alanine residue results in a significant loss of tolerance and a blunted SRT following PD128907 treatment. This result suggests that the D187 residue is centrally involved in the induction of PD128907-induced tolerance property in the chimeric receptor.
Figure 4.
The D187 residue in D3 receptor EC2 is specifically involved in the PD128907-induced tolerance in the D2D3IL123T-TM4-EC2 chimeric receptor. (A) Alignment of amino acid sequences comprising extracellular loop 2 (EC2) and part of the transmembrane 5 (TM5) of the human D2S and D3 receptors. The numbered amino acid residues indicated with asterisks were mutated to corresponding D2 receptor residues in the D2D3IL123T-TM4-EC2 chimeric receptor. Representative whole cell voltage clamp recordings in AtT-20 cell transiently transfected with either the D2D3IL123T-TM4-EC2 (B) or D2D3IL123T-TM4-EC2 (D187A) (C) chimeric/mutant receptors. The cells were held at −65 mV and inward GIRK currents in 30K-ES induced by 100 nM PD128907 (black) measured. The agonists were applied for ∼60 s at indicated times. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for PD128907 in AtT-20 cells transiently transfected with either the D2D3IL123T-TM4-EC2 chimeric receptor (WT) or mutant derivatives in which various residues in the EC2 was mutated to equivalent D2 receptor residues (D). PD128907-induced tolerance was significantly different between the D187A mutant chimeric receptor and all other receptor in panel D; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). (E) PD128907-induced tolerance observed in the D2D3IL12T-TM4-EC2 chimeric receptor was significantly attenuated in the D187A and D187L mutant derivative; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). In contrast, PD128907-induced tolerance observed in the D2D3IL12T-TM4-EC2 chimeric receptor was significantly enhanced in the D187E mutant derivative; #P < 0.01, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 8–14 cells).
Next we determined if a negatively charged residue was important for the induction of tolerance. To test this, we mutated the D187 residue to the similarly sized hydrophobic amino acid, leucine, or an equivalent negatively charged amino acid, glutamate. The result in Figure 4E shows that mutation of the D187 residue to leucine resulted in a significant loss of PD129087-induced tolerance in the D2D3IL123T-TM4-EC2 chimeric receptor. This was consistent with the results obtained with D187A mutation. Interestingly, the mutation of D187 residue to the negatively charged glutamate residue resulted in a significantly enhanced PD128907-induced tolerance in the chimeric receptor (Figure 4E). These results suggest that a negative charge at position 187 in D3 receptor EC2 is critical for the induction of PD128907-induced tolerance in the chimeric receptor.
There was also no significant difference in the PD128907-induced acute GIRK response between the D2D3IL123T-TM4-EC2 chimera and the various mutant chimeric receptors except for the D2D3IL123T-TM4-EC2 (S184A) mutant which showed a significantly higher acute GIRK response compared to the other receptors (Supporting Information Table 3). In addition, we observed that a double mutant construct with the S182I and S184A mutation did not respond to either PD128907 or quinpirole (data not shown). These results suggest that the S182 and S184 residues in the D3 receptor might play an important role in agonist binding.
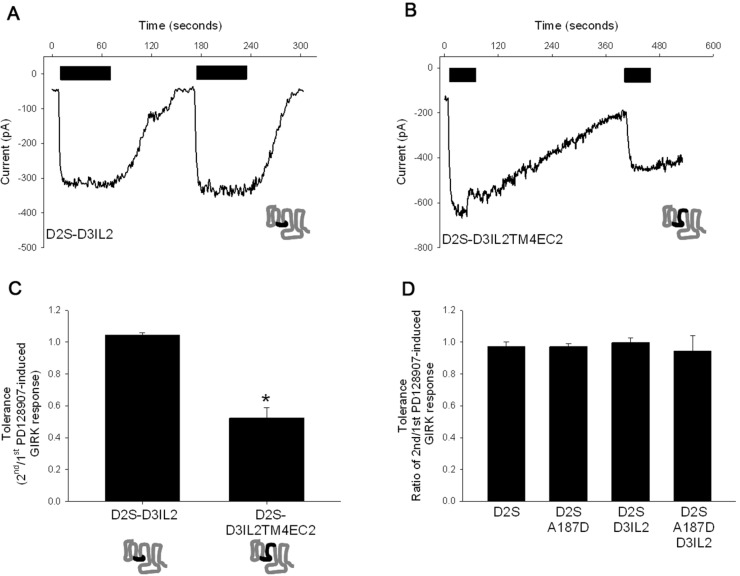
In the above studies, the D3 receptor TM4 and EC2 regions were substituted into a chimeric D2 receptor with all the intracellular regions of the D3 receptor (D2D3IL123T chimera). To determine if all intracellular regions of D3 receptor were necessary, we generated a chimeric receptor in which just the D2 receptor IC2, TM4, and EC2 were substituted with the D3 receptor IC2, TM4, and EC2 (the D2-D3IL2TM4EC2 chimeric receptor; Supporting Information Figure 1). Figure 5A–C shows that the chimeric D2 receptor with the substituted D3 receptor IC2, TM4, and EC2, was sufficient to elicit PD128907-induced tolerance and SRT. We also tested two additional constructs, one in which just the A187 residue in the D2 receptor EC2 was mutated to the negatively charged aspartate (present in the D3 receptor) and another in which the A187D mutation was introduced into the D2-D3IL2 chimeric receptor. The results in Figure 5D show that neither the D2A187D mutant nor the D2A187D-D3IL2 chimeric receptor exhibited PD128907-induced tolerance, suggesting that the point mutation in EC2 and D3IL2 are not sufficient to impart PD128907-induced tolerance and SRT to the chimeric D2 receptor.
Figure 5.
D3 receptor IL2, TM4, and EC2 regions are necessary for mediating PD128907-induced tolerance. Representative whole cell voltage clamp recordings in AtT-20 cell transiently transfected with either the D2D3IL2 (A) or D2D3IL2-TM4-EC2 (B) chimeric receptors. The cells were held at −65 mV and inward GIRK currents in 30K-ES induced by 100 nM PD128907 (black) measured. The agonists were applied for ∼60 s at indicated times. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for PD128907 in AtT-20 cells transiently transfected with either the D2D3IL2 or D2D3IL2-TM4-EC2 chimeric receptors. (C) D2D3IL2-TM4-EC2 chimeric receptor exhibited significant PD128907-induced tolerance compared to the D2D3IL2 chimeric receptor; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). (D) PD128907 did not induce tolerance in wild type D2S receptor, D2S A187D mutant receptor, D2SD3IL2 chimeric receptor or the D2S A187D-D3IL2 mutant/chimeric receptors. The bars represent the mean values ± SEM (n = 6–8 cells).
Differential Role of C147 and D187 Residues in Tolerance-Induced by Different D3 Receptor Agonists
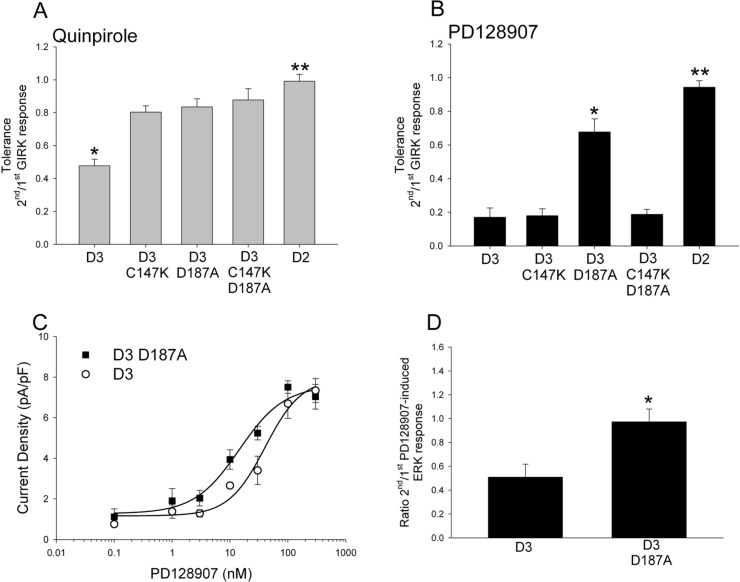
We next determined the effect of mutating the D187 residue in the native non chimeric human D3 receptor and comparing it to the effect of mutating the C147 residue. The cumulative data in Figure 6A and B shows that mutation of the D187 residue to alanine in the intact D3 receptor significantly attenuates tolerance induced by both quinpirole and PD128907; in contrast, the mutation of the C147 residue to lysine in the intact D3 receptor attenuates tolerance induced by quinpirole but not PD128907. We also tested a double mutant D3 receptor in which both the D187 and C147 were mutated to alanine and lysine, respectively. The double mutant did not exhibit quinpirole-induced tolerance but did exhibit PD128907-induced tolerance (Figure 6A and B). This result suggests that tolerance induction is a multistep process and involves different conformations. Interactions of agonists with D187 likely initiates tolerance induction by quinpirole and PD128907, but subsequently the two compounds promote different tolerance-inducing conformations. The results from the double mutant experiment suggest that the PD128907-induced tolerance conformation is stabilized by the C147K mutation such that the D187A mutation does not relieve the tolerance. There was also no significant difference in the quinpirole- or PD128907-induced acute GIRK response between the various mutant D3 receptors (Supporting Information Table 1). We performed a dose–response study to determine if the potency of PD128907 at wild type D3 receptor and D3D187A mutant receptor was significantly different. The results in Figure 6C shows that EC50 was 40.4 ± 14.6 nM and 14.5 ± 4.5 nM for wild type D3 and mutant D3 D187A receptor, respectively. The EC50 values were not statistically different (P = 0.103, Student’s t test), suggesting that the D187A mutation in the EC2 does not significantly affect the potency or efficacy of PD128907 for the D3 receptor.
Figure 6.
PD128907- and quinpirole-induced tolerance is not elicited in the D3 D187A mutant receptor. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for quinpirole (A) and PD128907 (B) in AtT-20 cells transiently transfected with either wild type D2, D3, D3 C147K, D3 D187A, or D3 C147K+D187A mutant receptors. The tolerance induced by quinpirole (A) in wild type D3 receptor is significantly less than in the D2 receptor and D3 C147K, D3 D187A and D3C147K+D187A mutant receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). However, these D3 mutant receptors exhibit quinpirole-induced tolerance when compared to wild type D2 receptor; **P < 0.05, ANOVA (Holm’s-Sidak post hoc test). The tolerance induced by PD128907 (B) in wild type D3 receptor is significantly attenuated only in the D3 D187A mutant receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). However, the D3 D187A mutant receptor exhibits PD128907-induced tolerance when compared to wild type D2 receptor; **P < 0.05, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 10–12 cells). (C) Comparison of PD128907-induced GIRK dose response curves obtained from whole cell voltage clamp recording of AtT-20 cells expressing either wild type D3 receptor (open circle) or D3 D187A mutant receptor (filled square). The PD128907-induced GIRK currents (pA) were normalized for cell size (cell capacitance, pF). The data points were fit with a four-parameter Hill equation. The bars represent the mean values ± SEM (n = 6–8 cells). (D) Cumulative data comparing the mean tolerance of MAPK activation in AtT-20 cells transfected with either wild type human D3 or D3 D187A mutant receptors. Cells were pretreated for 1 min with either vehicle (SES) or 200 nM PD128907, washed 6 times with SES (3 min per wash), and subsequently treated with vehicle (SES) or 200 nM PD128907 for 5 min. The levels of PD128907-induced phosphorylated MAPK proteins were normalized to the levels of total MAPK proteins. Tolerance was determined by dividing the 200 nM PD128907-induced phosphorylation of ERK1/2 at 5 min in cells pretreated with 200 nM quinpirole by the 200 nM PD128907-induced phosphorylation of ERK1/2 at 5 min in cells pretreated with control SES. The wild type D3 receptor expressing cells exhibits significantly reduced response following PD128907 pretreatment compared to D3 D187A mutant expressing cells (*P < 0.05, Student’s t test). The experiments were repeated three independent times.
We have previously shown that the D3 receptor exhibits quinpirole-induced tolerance property when coupling to the MAPK signaling pathway. We next determined if the PD128907-induced tolerance was exhibited in the MAPK signaling pathway and if this tolerance was attenuated in the D187A mutant. Results in Figure 6D shows that PD128907-induced tolerance in the wild type D3 receptor-MAPK signaling pathway is absent in the D187A mutant D3 receptor. The results are consistent with our previous studies that D3 receptor tolerance property is exhibited in multiple signaling pathways and suggests that the D187A mutation abolishes the tolerance property exhibited in the D3 receptor-MAPK signaling pathway.
Quinpirole-induced tolerance is exhibited by the wild type D3 receptor but not by any of the D2-D3 chimeric receptors in which most or all of the transmembrane and extracellular domains are contributed by the D2 receptor, suggesting a unique conformational change adopted by the D3 receptor in the presence of quinpirole (Figures 1E, 2A, 2C, and 3C).10 In the native D3 receptor, the mutation of either the D187 residue in EC2 or the C147 residue in IL2 resulted in the loss of the quinpirole-induced tolerance and tolerance-associated conformation (Figure 6A and Supporting Information Figure 3).9 The results suggest that while the D187 and C147 residues are required for the quinpirole-induced tolerance, they are not sufficient. The quinpirole-induced tolerance is likely mediated by a D3 receptor conformation state that requires cooperation of other TM and EC regions not evaluated in this study.
Structural superpositioning of D3 receptor D187A mutant receptor model activated by PD128907 or quinpirole or D3 receptor C147K mutant receptor model activated by quinpirole shows similar receptor conformations. The average root mean squared deviation between the various conformations was 2 Ǻ (Supporting Information Figure 3). The receptor conformation obtained by superpositioning, most likely represents the D3 receptor conformation associated with attenuated tolerance.
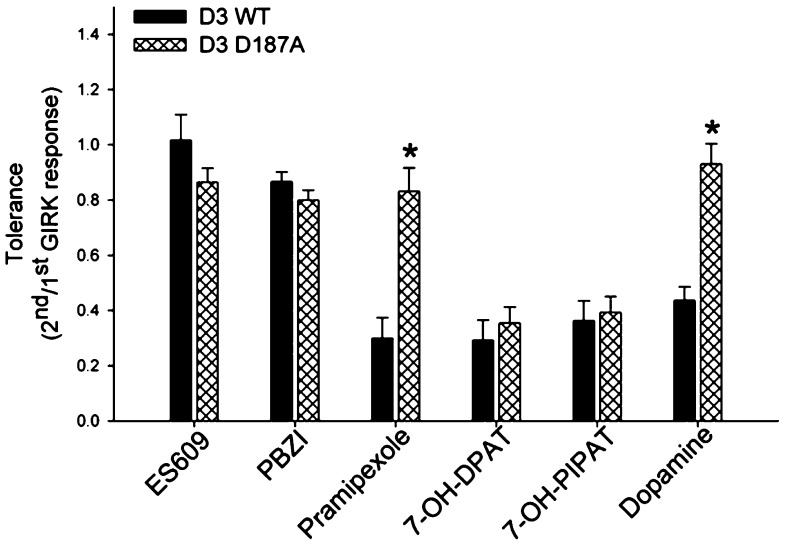
Given the different effects of C147K and D187A mutation on quinpirole- and PD128907-induced tolerance, we compared the effect of six additional D3 receptor agonists to induce tolerance in wild type and mutant D187A D3 receptor (Figure 7). Of the six agonists tested, ES609 and PBZI do not induce D3 receptor tolerance whereas dopamine, pramipexole, 7-OH-DPAT, and 7-OH-PIPAT induce tolerance.11 The results in Figure 7 show that mutating D187 residue to alanine in the D3 receptor prevented the tolerance induced by dopamine and pramipexole but had no effect on tolerance induced by 7-OH-DPAT and 7-OH-PIPAT. The D187A mutation also did not alter the inability of ES609 and PBZI to induce tolerance. These results suggest that while the D187 residue is involved in the tolerance induced by agonists such as quinpirole, PD128907, dopamine and pramipexole, it does not affect the tolerance induced by 7-OH-DPAT and 7-OH-PIPAT. This further suggests multiple conformation changes involving different residues lead to induction of D3 receptor tolerance.
Figure 7.
Dopamine- and pramipexole-induced tolerance is not elicited in the D3 D187A mutant receptor. Tolerance property defined as the ratio of second/first agonist-induced GIRK response was determined for dopamine (100 nM), ES609 (100 nM), cis-8-OH-PBZI (300 nM), pramipexole (300 nM), 7-OH-DPAT (100 nM), and 7-OH-PIPAT (100 nM) in AtT-20 cells transiently transfected with either wild type D3 (black fill) or D3 D187A mutant (cross hatched) receptors. The tolerance induced by dopamine and pramipexole in wild type D3 receptor is significantly attenuated in the D3 D187A mutant receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 5–8 cells).
PD128907-Induced Tolerance Involves Interaction of the D187 Residue in EC2 with the H354 Residue in EC3
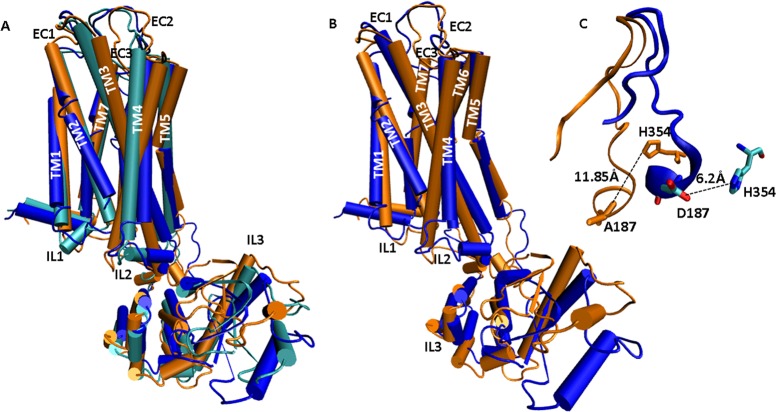
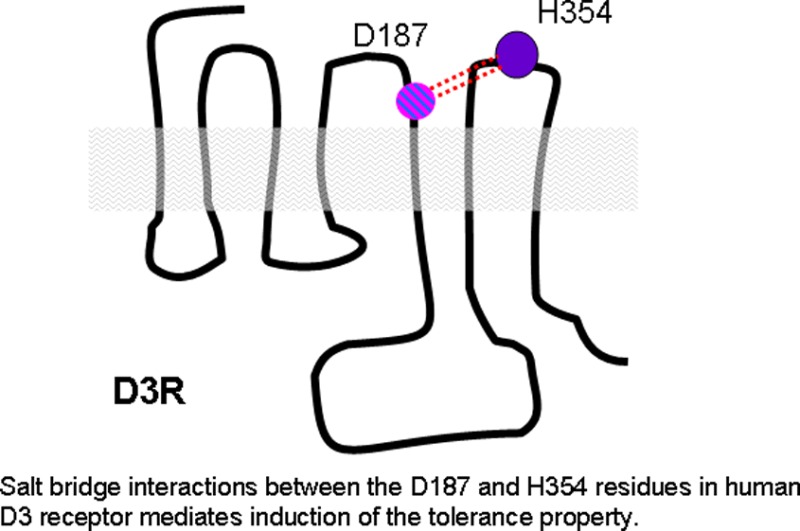
PD128907 induces tolerance in wild type D3 receptor and D3 C147K mutant receptor but not in the D3 D187A mutant receptor (Figure 6B). These functional results were consistent with the MD simulation studies which showed that the PD128907-induced conformation in D3 D187A mutant receptor was significantly different from the PD128907-induced conformation in wild type and D3 C147K mutant receptor (Figure 8A). A quantitative comparison of the PD128907 elicited tolerance-inducing conformation in D3 C147K mutant and the tolerance-attenuated conformation in the D3 D187A mutant shows a root mean squared deviation of 5.7 Å with significant changes in IL2, TM4, EC2, TM5, and IL3 (Figure 8B). While both of these conformations are stable over the time scale of simulations (Supporting Information Figure 4), a strong coupled motion of IL2-TM4-EC2-TM5 seems to induce a conformational change in the D3 D187A mutant receptor that leads to attenuation of tolerance. Structural super positioning of conformations induced in wild type D3 receptor and D187A mutant D3 receptor bound to PD128907 clearly show marked differences in specific regions of the receptor that could be associated with tolerance and SRT (Supporting Information Figure 5). A closer inspection of these conformations suggest a stretch of residues corresponding to the c-terminal end of IL2-TM4-EC2-TM5 and EC3 domains as shown in Supporting Information Figure 5. The IL2-TM4-EC2-TM5 domain motion seems to be coordinated by the motion of EC3 loop. In order to investigate this domain motion, we analyzed the atomic contacts of the D187 residue (which is part of the IL2-TM4-EC2-TM5 domain). The analysis of the atomic contacts of the D187 residue in the simulations showed that in the PD128907-activated D3 wild type receptor and the D3 C147K receptor, the D187 residue forms a salt bridge (∼6.2 Å) with the H354 residue from the EC3 loop (Figure 8C). The charge on the residue seems to play a significant role as these interactions are observed in D187E mutant but are lost in the D187A/L mutant receptors. Quantitatively, when the D187 residue is mutated to an alanine or leucine residue, the salt bridge is disrupted and the distance to H354 increases to ∼11.85 Å (Figure 8C). The D187-H354 interaction is maintained throughout the simulation and during the entire length of the trajectory suggesting a probable role in locking the receptor in a tolerance inducible conformation. Even as the loops move away from each other during the simulation the salt bridge seems to be maintained by a water molecule. In addition, D187 forms strong intraloop hydrogen bonded interactions with the neighboring residues stabilizing the conformation.
Figure 8.
Structural superpositioning of various D3 receptor models represented as cartoon models detailing structural changes involved in tolerance-inducing versus noninducing conformations. (A) Superpositioning of PD128907-activated, wild type D3 receptor (cyan), D3 C147K mutant receptor (blue), and D3 D187A mutant receptor (orange). PD128907 induces tolerance in the wild type D3 and D3 C147K mutant receptors but not in the D3 D187A mutant receptor. (B) Structural superpositioning of PD128907-activated D3-C147K mutant receptor (blue) and D3-D187A mutant receptor (orange) show that a majority of conformational changes are in the domain formed by IL2-TM4-EC2-TM5-IL3. (C) Close-up view of the superposed EC2 loop in PD128907-activated D3-D187A mutant (orange) and D3-C147K mutant (blue) shows that a salt bridge is formed between D187 and H354 which could be an indicator of tolerance-inducing conformations. The residues D187 and H354 from D3-C147K model are shown as licorice sticks and colored atom type; C= cyan, N = blue, O = red. A187 and H354 from D3-D187A model are represented by orange licorice sticks.
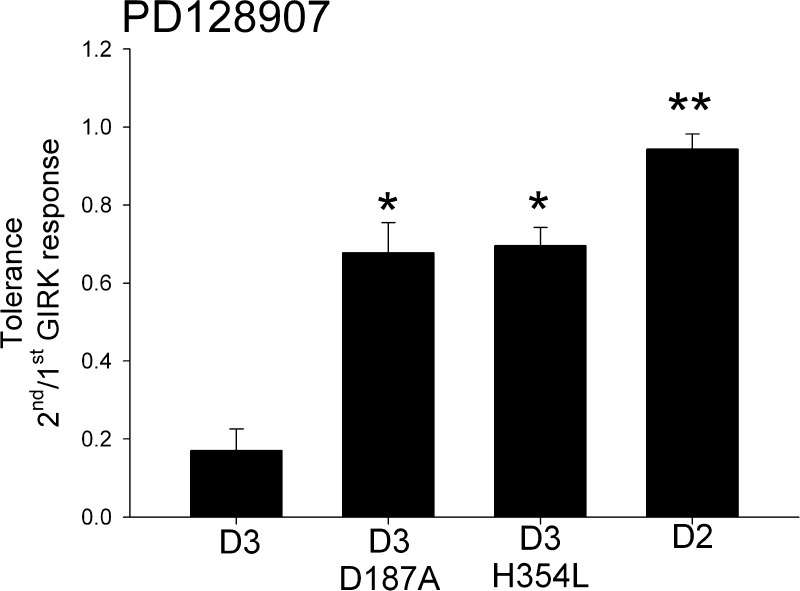
The H354 residue in EC3, but not the D187 residue in EC2, is conserved between the D2 and D3 receptor. To test the role of D187-H354 interaction in PD128907-induced tolerance, we mutated the H354 residue in EC3 to leucine. The H354L mutant D3 receptor failed to elicit quinpirole-induced GIRK response and elicited a significantly reduced PD128907-induced GIRK response (Supporting Information Table 1). In addition, the PD128907-induced GIRK response elicited by the H354L D3 receptor mutant showed attenuated tolerance comparable to the D187A D3 receptor mutant (Figure 9). These results strongly suggest that the H354 residue is important for quinpirole- and PD128907-induced D3 receptor activation. In addition, the results support our D187 and H354 salt bridge model as being important for the induction of D3 receptor tolerance property (Figure 8C).
Figure 9.
Comparison of PD28907-induced tolerance in the GIRK response elicited by the EC2 D187A and EC3 H354L D3 receptor mutants. The tolerance induced by 100 nM PD128907 in wild type D3 receptor is significantly attenuated in the D3 D187A and D3 H354L mutant receptors; *P < 0.01, ANOVA (Holm’s-Sidak post hoc test). However, both mutant receptors exhibit PD128907-induced tolerance when compared to wild type D2 receptor; **P < 0.05, ANOVA (Holm’s-Sidak post hoc test). The bars represent the mean values ± SEM (n = 10 to 15 cells).
Together, these results suggest that the D187 residue in the EC2 plays a more important role in the initial conformational changes than the intracellular C147 residue in mediating the tolerance to different agonists. Taken together with the results from our functional studies which showed a loss of PD128907-induced tolerance in the D187A and D187L mutants (Figure 4D, E and Figure 6B), these results suggest that the D187-H354 salt bridge might be a signature feature of tolerance-inducing agonists at the D3 receptor.
Concluding Remarks
Structure–function studies of the extracellular loops of G-protein coupled receptors (GPCRs) are gaining significance as their role in ligand binding, and ligand-induced conformational changes are being evaluated. Biochemical studies using site-directed mutagenesis studies have demonstrated that residues that make the extracellular loops participate not only in binding of peptide ligands but also small molecule biogenic amines, adenosine, prostanoids, and others.14−18 Some of these studies have demonstrated the role of EC2 in binding ligands and participating in receptor activation.19,20 The crystal structure of bovine rhodopsin revealed that the position of the EC2 loop is within the binding site crevice “capping” the chromophore.21 In addition, the helix movements associated with activation of rhodopsin was shown to be coupled to the displacement of EC2 loop.22 Although the EC2 loops in class-A GPCRs are variable, the bovine rhodopsin structure provided the first template to model EC2 of D2 receptor.23,24 This model provided structural evidence for the role of certain amino acids in EC2 that participated in binding ligands in D2 receptor which were previously established using biochemical methods.18 Our MD simulation studies suggest that the D3 receptor EC2 loop may also participate in stabilizing the extracellular end of the receptor in a preferred conformation that is conducive for binding of certain agonists through interloop interactions with EC3. Indeed, mutation of the D187 residue in EC2 or the H354 residue in EC3 resulted in loss of tolerance. Interestingly, the H354 residue also appears to be important for agonist-induced receptor activation as mutating this residue resulted in loss of response to quinpirole and a significant reduction in response to PD128907. The recent crystallization of the D3 receptor bound to the antagonist, eticlopride, demonstrated both an orthostatic binding site within the TM bundles to which EC2 contributed and the existence of an allosteric binding site outside the TM bundle to which residues of the EC2 also contributed.5
Overall, we propose a model in which D3 receptor agonists such as PD128907 and quinpirole bind and stabilize the D3 receptor in at least two different conformations that induce tolerance in the receptor as evident from our functional studies. This tolerance can be attenuated by a single point mutation of C147K in the IL2 loop for quinpirole, but not PD128907. However, a mutation of D187A in the EC2 loop of D3 receptor leads to attenuation of tolerance in PD128907 and quinpirole. The conformational changes induced in the D3 receptor bound to both these agonists is distinct, providing structural evidence for ligand-induced signaling. This is further supported by double mutant experiments in which a receptor with both D187 and C147 mutated does not elicit quinpirole-induced tolerance but does induce PD128907-induced tolerance. Furthermore, while the D187 residue is involved in the tolerance induced by PD128907, quinpirole, dopamine, and pramipexole, it does not mediate tolerance induced by 7-OH-DPAT and 7-OH-PIPAT. This suggests that induction of D3 receptor tolerance can be mediated by different conformational states that are ligand-dependent. It is interesting that PD128907 and quinpirole both bind and elicit an agonist response at D3 receptors with similar efficacy and their binding mode also maps to same set of residues in the binding pocket, yet they elicit different conformational states. This suggests distinct secondary interactions between the D3 receptor and agonists contribute to the ability to induce different levels of tolerance and SRT. We have recently demonstrated the utility of these ligand-induced conformational changes to design novel D3 selective agonists that do not induce tolerance and SRT in D3 receptors.11 These novel atypical D3 receptor agonists may be beneficial in the clinic due to the suggested involvement of the D3 receptor in Parkinson’s disease, schizophrenia and drug addiction.1−4
Methods
Cell Culture and Transfection
AtT-20 mouse pituitary cells were grown in Ham’s F10 medium supplemented with 10% heat-inactivated horse serum and 5% fetal bovine serum, 2 mM glutamine, and 50 μg/mL gentamicin (Invitrogen, Carlsbad, CA). AtT-20 cells stably expressing either human D2S or D3 receptors were cultured in the above medium, supplemented with 500 μg/mL G418. AtT-20 cells transiently transfected with wild type, mutant, or chimeric receptors were cotransfected with a plasmid containing enhanced green fluorescent protein (EGFP; Clontech, Palo Alto, CA) to identify transfected cells. AtT-20 cells were transfected using the DMRIE-C reagent (Invitrogen). Electrophysiological recordings were made 36–48 h after transfection. For electrophysiological experiments, cells were plated onto glass coverslips which had been previously coated with 40 μg/mL poly-l-lysine (Sigma, St. Louis, MO) and cultured for 24 h prior to transfection.
Generation of Chimeric and Mutant D3R
The chimeric and mutant receptors were generated using a strategy described previously.8 Briefly chimeric receptors and receptors with point mutations were generated by ligating together DNA fragments that were generated using PCR. The PCR was performed with multiple primer pairs that encoded amino acids found at the junction of swapped regions. The cDNA encoding the wild-type and chimeric receptors were subcloned in the pcDNA3 expression plasmid (Invitrogen). The various regions swapped in the different chimeric receptors are shown in Supporting Information Figure 1. All recombinant plasmids were characterized by restriction enzyme analysis and DNA sequencing to verify that no spurious changes were introduced in the entire receptor-coding region. All plasmids used for the transfection studies were purified on two sequential cesium chloride density gradients.
Electrophysiology, Agonists, and Solutions
For electrophysiological experiments, AtT-20 cells were perfused with standard extracellular solution (SES-145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 10 mM HEPES (pH 7.4), and 10 mM glucose). Agonists (dopamine, quinpirole, cis-8-OH-PBZI (Sigma), PD1289207, 7-OH-DPAT, 7-OH-PIPAT, pramipexole (Tocris, Ellisville, MO), and ES609 (Asinex, Durham, NC) were diluted in SES plus 30 mM potassium (30K-ES) to enhance inwardly rectifying currents. Drugs and solutions were applied to cells with a multibarreled micropipet array. Cells were held at −65 mV and whole cell voltage clamp recording performed as described previously.8 The recording glass micropipets were filled with intracellular solution containing 130 mM K-aspartate, 20 mM NaCl, 1 mM MgCl2, 10 mM HEPES (pH 7.4), 10 mM glucose, 0.1 mM GTP, 5 mM Mg-ATP, and 1 mM EGTA.
Data Analysis
Currents were measured using an Axopatch 200B amplifier (Axon Instruments, Union City, CA) and sampled through a Digidata 1322A interface (Axon Instruments) using the pClamp8.0 software (Axon Instruments). Data files were imported into SigmaPlot (SPSS Inc., Chicago, IL) for analysis and display. SigmaPlot was also used to perform analysis of variance and posthoc comparison tests on relevant data. Data were considered statistically different when probability was <0.05.
Detection of ERK Phosphorylation
For the MAPK studies, AtT-20 cells were grown in 12-well plates until 80% confluency and then transfected with plasmids expressing either wild type D3 receptor or D187A mutant D3 receptor as described above. Forty eight hours after transfection, the cells were incubated in Opti-MEM serum-free medium for 3 h prior to treatment. The cells were then pretreated with 200 nM PD128907 or SES for 1 min, followed by 6 SES washes with each wash being of 3 min duration. Subsequently, the cells were treated with SES or 200 nM PD128907 for 5 min and lysed. Equal amount of lysates were run on an SDS-polyacrylamide gel and detected using Western blotting as described previously.9 Activated (phosphorylated) ERK1 and ERK2 were first detected using monoclonal phosphop44/p42 MAPK antibody (1:1000 dilution; Cell Signaling Technology, Danvers, MA). Following the detection of phosphorylated proteins, the membrane was stripped and reprobed to detect total ERK1/ERK2 proteins. Total ERK proteins were detected using a polyclonal antibody (1:1000 dilution; Cell Signaling Technology). Cumulative data was obtained from three independent times with comparable results.
Molecular Modeling and Docking Simulations
The crystal structure of D3 receptor in complex with the antagonist, eticlopride, solved to 2.89 Å was obtained from Protein Data Bank (PDB code: 3PBL).5 The three-dimensional coordinates of D2R, D2-D3 chimeric receptors and D3-D187A, D3-D187E, and D3-D187L mutant receptors were homology modeled using the crystal structure of D3 as a template to the program Modeler.25 The resulting structures were minimized and subject to constrained molecular dynamics simulation to remove steric clashes. Further, PD128907 and quinpirole were docked to the binding pocket of wild type D3, D2 receptors, chimeric receptors and mutant receptor models using GOLD program.26 Fifty independent docking simulations were performed to completely sample the receptor and ligand conformations. The best ranking docked complexes of PD128907 and quinpirole with wild type and mutant D3, D2 receptors were subject to molecular dynamics simulations. All simulations were performed using an explicit 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine lipid bilayer/water model membrane system adopted in the Desmond module27 of the Schrödinger suite program. The model membrane was prealigned to the β2-adrenegic receptor crystal structure template, and default parameters with all-atom OPLS (optimized potentials for liquid simulations) force field were utilized for all the simulations. To completely refine the docked models, a molecular dynamics simulation of 25 ns that included routines for minimization, heating, equilibration, and production was followed.5 We have previously shown that the D3 receptor tolerance develops only after the agonist is washed off;8−11 therefore, in the simulations, the ligand was removed from the production phase of the simulation. Simulation trajectories were analyzed using trajectory analysis modules in VMD program.28
Glossary
Abbreviations
- SRT
slow response termination
- IL
intracellular loop
- EC
extracellular loop
- TM
transmembrane
- MD
molecular dynamics
- OPLS
optimized potentials for liquid simulations
- GPCRs
G-protein coupled receptors
- GIRK
G-protein coupled inward rectifier potassium
- MAPK
mitogen-activated protein kinases
- SES
standard external solution
- 30K-ES
standard external solution with 30 mM potassium
Supporting Information Available
Figure S1: Alignment of the human D2S and D3 receptor primary amino acid sequence. Figure S2: Structural models of D2S, D3 wild type, and D2 D3IL123TM4EC2 chimera. Figure S3: Structural superposition of D3-D187A mutant activated by quinpirole, D3-D187A activated by PD128907, and D3-C147K activated by quinpirole. Figure S4: Root mean squared deviations of PD128907-activated wild type D3 receptor, D3-C147K mutant, and D3-D187A mutant. Figure S5: Root mean squared deviation between PD128907-induced wild type D3 receptor conformation and PD128907-induced D187A mutant D3 receptor conformations. Tables S1 and S3: Acute GIRK-response induced by PD128907 and quinpirole activating wild type, mutant, and chimeric receptors. Table S2: Root mean squared deviation of various chimeric receptors compared to wild type D2 and D3 receptors. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
S.G-M., K.K., and E.V.K. performed the functional experiments, data analysis, and manuscript writing. S.K. performed all the computer simulations, modeling work and analysis, and assisted with manuscript writing. S.G-M, S.K., and E.V.K contributed equal effort.
This work was supported by a grant from the F.M. Kirby Foundation to E.V.K. S.K. acknowledges the support from Department of Microbiology and Immunology, Faculty development funds, Drexel University College of Medicine to set up core computational facilities.
The authors declare no competing financial interest.
Supplementary Material
References
- Newman A. H.; Blaylock B. L.; Nader M. A.; Bergman J.; Sibley D. R.; Skolnick P. (2012) Medication discovery for addiction: translating the dopamine D3 receptor hypothesis. Biochem. Pharmacol. 84(7), 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet A.; Bezard E. (2009) Dopamine receptors and L-dopa-induced dyskinesia. Parkinsonism Relat Disord. Suppl 4, S8–12. [DOI] [PubMed] [Google Scholar]
- Joyce J. N.; Millan M. J. (2007) Dopamine D3 receptor agonists for protection and repair in Parkinson’s disease. Curr. Opin. Pharmacol. 7(1), 100–105. [DOI] [PubMed] [Google Scholar]
- Sokoloff P.; Diaz J.; Le Foll B.; Guillin O.; Leriche L.; Bezard E.; Gross C. (2006) The dopamine D3 receptor: a therapeutic target for the treatment of neuropsychiatric disorders. CNS Neurol. Disord.: Drug Targets 5(1), 25–43. [DOI] [PubMed] [Google Scholar]
- Chien E. Y.; Liu W.; Zhao Q.; Katritch V.; Han G. W.; Hanson M. A.; Shi L.; Newman A. H.; Javitch J. A.; Cherezov V.; Stevens R. C. (2010) Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330, 1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J.; Coleman R. G.; Setola V.; Irwin J. J.; Fan H.; Schlessinger A.; Sali A.; Roth B. L.; Shoichet B. K. (2011) Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat. Chem. Biol. 7, 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J. D.; Clarke W. P.; von Zastrow M.; Nichols D. E.; Kobilka B.; Weinstein H.; Javitch J. A.; Roth B. L.; Christopoulos A.; Sexton P. M.; Miller K. J.; Spedding M.; Mailman R. B. (2007) Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 320(1), 1–13. [DOI] [PubMed] [Google Scholar]
- Kuzhikandathil E. V.; Westrich L.; Bakhos S.; Pasuit J. (2004) Identification and characterization of novel properties of the human D3 dopamine receptor. Mol. Cell. Neurosci. 26, 144–155. [DOI] [PubMed] [Google Scholar]
- Westrich L.; Kuzhikandathil E. V. (2007) The tolerance property of human D3 dopamine receptor is determined by specific amino acid residues in the second cytoplasmic loop. Biochim. Biophys. Acta, Mol. Cell Res. 1773, 1747–1758. [DOI] [PubMed] [Google Scholar]
- Westrich L.; Gil-Mast S.; Kortagere S.; Kuzhikandathil E. V. (2010) Development of tolerance in D3 dopamine receptor signaling is accompanied by distinct changes in receptor conformation. Biochem. Pharmacol. 79, 897–907. [DOI] [PubMed] [Google Scholar]
- Kuzhikandathil E. V.; Kortagere S. (2012) Identification and characterization of a novel class of atypical dopamine receptor agonists. Pharm. Res. 29(8), 2264–2275. [DOI] [PubMed] [Google Scholar]
- Kuzhikandathil E. V.; Oxford G. S. (1999) Activation of human D3 dopamine receptor inhibits P/Q-type calcium channels and secretory activity in AtT-20 cells. J. Neurosci. 19(5), 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezard E.; Ferry S.; Mach U.; Stark H.; Leriche L.; Boraud T.; Gross C.; Sokoloff P. (2003) Attenuation of levodopa-induced dyskinesia by normalizing dopamine D3 receptor function. Nat. Med. 9(6), 762–767. [DOI] [PubMed] [Google Scholar]
- Sakmar T. P. (2002) Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr. Opin. Cell Biol. 14, 189–195. [DOI] [PubMed] [Google Scholar]
- Conner M.; Hawtin S. R.; Simms J.; Wootten D.; Lawson Z.; Conner A. C.; Parslow R. A.; Wheatley M. (2007) Systematic analysis of the entire second extracellular loop of the V(1a) vasopressin receptor: key residues, conserved throughout a G-protein-coupled receptor family, identified. J. Biol. Chem. 282, 17405–17412. [DOI] [PubMed] [Google Scholar]
- Fu W.; Shen J.; Luo X.; Zhu W.; Cheng J.; Yu K.; Briggs J. M.; Jin G.; Chen K.; Jiang H. (2007) Dopamine D1 receptor agonist and D2 receptor antagonist effects of the natural product (−)-stepholidine: molecular modeling and dynamics simulations. Biophys. J. 93, 1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds K. A.; Katritch V.; Abagyan R. (2009) Identifying conformational changes of the beta(2) adrenoceptor that enable accurate prediction of ligand/receptor interactions and screening for GPCR modulators. J. Comput.-Aided Mol. Des. 23, 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.; Javitch J. A. (2004) The second extracellular loop of the dopamine D2 receptor lines the binding-site crevice. Proc. Nat. Acad. Sci. U.S.A. 101, 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal H.; Jagannathan R.; Bhat M. B.; Karnik S. S. (2010) Ligand-specific conformation of extracellular loop-2 in the angiotensin II type 1 receptor. J. Biol. Chem. 285, 16341–16350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K.; Imamoto Y.; Yamashita T.; Shichida Y. (2010) Functional analysis of the second extracellular loop of rhodopsin by characterizing split variants. Photochem. Photobiol. Sci. 9, 1490–1497. [DOI] [PubMed] [Google Scholar]
- Palczewski K.; Kumasaka T.; Hori T.; Behnke C. A.; Motoshima H.; Fox B. A.; Trong I. L.; Teller D. C.; Okada T.; Stenkamp R. E.; Yamamoto M.; Miyano M. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- Ahuja S.; Eilers M.; Hirshfeld A.; Yan E. C.; Ziliox M.; Sakmar T. P.; Sheves M.; Smith S. O. (2009) 6-s-cis Conformation and polar binding pocket of the retinal chromophore in the photoactivated state of rhodopsin. J. Am. Chem. Soc. 131, 15160–15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler E. L.; Hassan S. A.; Kortagere S.; Weinstein H. (2006) Ab initio computational modeling of loops in G-protein-coupled receptors: lessons from the crystal structure of rhodopsin. Proteins 64, 673–690. [DOI] [PubMed] [Google Scholar]
- Kortagere S.; Roy A.; Mehler E. L. (2006) Ab initio computational modeling of long loops in G-protein coupled receptors. J. Comput.-Aided Mol. Des. 20, 427–436. [DOI] [PubMed] [Google Scholar]
- Sali A.; Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815. [DOI] [PubMed] [Google Scholar]
- Jones G.; Willett P.; Glen R. C. (1995) Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 245, 43–53. [DOI] [PubMed] [Google Scholar]
- Kevin J., Bowers E. C., Huafeng X., Dror R. O., Eastwood M. P., Gregersen B. A., Klepeis J. L., Kolossvary I., Moraes M. A., Sacerdoti F. D., Salmon J. K., Shan Y., and Shaw D. E. (2006) Scalable Algorithms for Molecular Dynamics Simulations on Commodity Clusters. Proceedings of the ACM/IEEE Conference on Supercomputing, Tampa, FL, November 11–17 (SC06). [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graphics 14(27–28), 33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.