Abstract
Polymorphisms in CYP2D6 and CYP2C19 affect the efficacy and safety of tricyclics, with some drugs being affected by CYP2D6 only, and others by both polymorphic enzymes. Amitriptyline, clomipramine, doxepin, imipramine, and trimipramine are demethylated by CYP2C19 to pharmacologically active metabolites. These drugs and their metabolites, along with desipramine and nortriptyline, undergo hydroxylation by CYP2D6 to less active metabolites. Evidence from published literature is presented for CYP2D6 and CYP2C19 genotype–directed dosing of tricyclic antidepressants.
The use of tricyclics to treat psychological disorders has declined in part because of the occurrence of undesirable side effects. Although tricyclics are still used to treat depression,1 their main therapeutic use is often for pain management.2,3 Interindividual differences in side effects and treatment response have been associated with variability of tricyclic plasma concentrations.4,5 Because both enzymes influence plasma concentrations, the effectiveness and tolerability of tricyclics are affected by CYP2D6 metabolism and partially by CYP2C19 metabolism.4 The purpose of this guideline is to provide information regarding how to use existing CYP2D6 and/or CYP2C19 genotyping test results to guide dosing of tricyclics for psychological disorders and pain management, focusing particularly on amitriptyline and nortriptyline.
Optimal therapeutic plasma concentrations for the tricyclics have been defined.6 Poor or ultrarapid metabolizers of CYP2D6 and CYP2C19 may have tricyclic plasma concentrations outside the recommended therapeutic range, thereby increasing the risk of treatment failure or side effects.7,8,9,10 Therefore, this guideline takes into consideration both clinical outcomes and observed tricyclic plasma concentrations based on genotype/phenotype characteristics. Detailed guidelines for use of other laboratory tests including therapeutic drug monitoring of tricyclics are beyond the scope of this article. The Clinical Pharmacogenetics Implementation Consortium (CPIC) of the National Institutes of Health's Pharmacogenomics Research Network develops peer-reviewed gene–drug guidelines that are published and updated periodically at http://www.pharmgkb.org based on new developments in the field.
Focused Literature Review
A systematic literature review focused on CYP2D6 and CYP2C19 genotyping and its relevance to gene-based dosing of tricyclics was conducted (see Supplementary Data online). This guideline was developed based on interpretation of the literature by the authors and experts in the field.
Genes: CYP2D6 and CYP2C19
CYP2D6 background
The CYP2D6 gene is highly polymorphic.11 More than 100 known allelic variants and subvariants have been identified, and there are substantial ethnic differences in observed allele frequencies (Supplementary Table S1 online). The most commonly reported alleles are categorized into functional groups as follows: functional (e.g., CYP2D6*1 and *2), reduced function (e.g., CYP2D6*9, *10, and *41), and nonfunctional (e.g., CYP2D6*3–*6).11,12 Because CYP2D6 is subject to deletions or duplications, most clinical laboratories also report copy number. Deletions are indicated by the CYP2D6*5 allele, and gene duplications are denoted by an “xN” following the allele (e.g., CYP2D6*1xN, where xN represents the number of CYPD6 gene copies).
CYP2C19 background
Similar to CYP2D6, the CYP2C19 gene is highly polymorphic; more than 30 known allelic variants and subvariants have been identified.13 Although there are ethnic differences in allele frequencies (Supplementary Table S2 online), the majority of patients carry a CYP2C19*1, *2, or *17 allele.13 CYP2C19*1 is the wild-type allele encoding a fully functional enzyme, and CYP2C19*2 is the most common loss-of-function allele. Multiple CYP2C19 loss-of-function alleles have been identified (e.g., CYP2C19*3–*8), but their allele frequencies are <1%, with the exception of CYP2C19*3, which has a frequency of 2–15% in Asians.13
The CYP2C19*17 allele results in enhanced gene transcription purportedly leading to increased metabolic activity.14 However, the clinical importance of the CYP2C19*17 allele is a matter of debate. Some studies indicate that CYP2C19*17 enhances clopidogrel response and increases the probability of therapeutic failure for proton pump inhibitors or antidepressants due to altered drug plasma concentrations, but there are conflicting clinical data.13,14,15,16,17,18,19
Genetic test interpretation
Clinical laboratories usually test for the more frequently observed CYP2D6 and CYP2C19 genetic variants and translate the results into star-allele (*) nomenclature. Each star-allele, or haplotype, is defined by a specific combination of single-nucleotide polymorphisms and/or other genetic variants within the gene locus of either CYP2D6 or CYP2C19.11,13 Genetic test results are reported as the summary of inherited maternal and paternal star-alleles referred to as a diplotype (e.g., CYP2D6*1/*2 and CYP2C19*1/*1). See Supplementary Tables S3–S6 online for the more frequently observed alleles and their functional status.
Scoring systems have been developed in an attempt to provide a uniform approach to quantitate the predicted functional status of CYP2D6 alleles as follows: 1 for functional, 0.5 for reduced function, and 0 for nonfunctional alleles (Supplementary Table S4 online).11,12 The activity value for each allele of the diplotype is totaled to provide a CYP2D6 activity score. If CYP2D6 gene duplications are detected, the activity value of the duplicated allele is multiplied by the number of duplications present before calculating the activity score. The CYP2D6 activity score is used to assign phenotype in this guideline as follows: patients with an activity score of 0 are classified as poor metabolizers, those with a score of 0.5 are intermediate metabolizers, those with a score from 1.0 to 2.0 are extensive metabolizers, and those with a score >2.0 are classified as ultrarapid metabolizers (Table 1, Supplementary Tables S7 and S8 online). Extensive metabolizers are considered to have normal CYP2D6 enzyme activity.
Table 1. Assignment of likely phenotypes based on diplotypes.
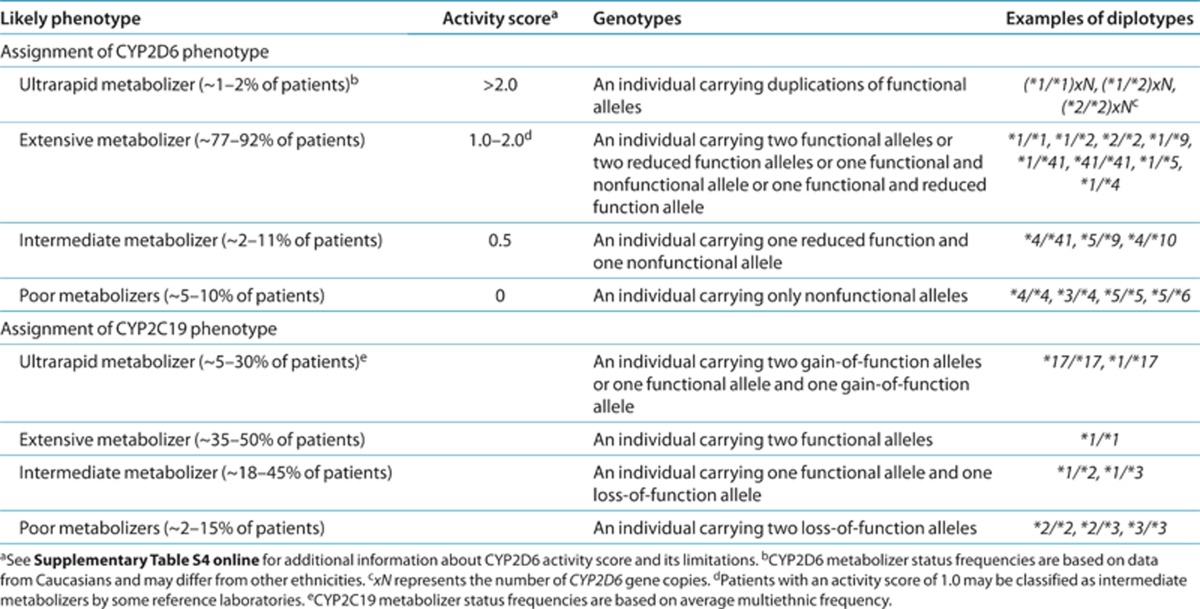
There is a lack of consensus with regard to whether patients with a CYP2D6 activity score of 1.0 should be assigned an extensive or intermediate phenotype.11 Pharmacokinetic data suggest that patients with an activity score of 1.0 have a higher CYP2D6 metabolic capacity as compared with patients with an activity score of 0.5 but less CYP2D6 enzyme activity as compared with patients with an activity score of 2.0.12 Patients with one functional and one nonfunctional CYP2D6 allele do not have significantly different nortriptyline plasma concentrations as compared with patients with two functional CYP2D6 alleles,20 although another study reported conflicting data.7 Herein, we classified patients with a CYP2D6 activity score of 1.0 as extensive metabolizers, which is analogous to the CPIC guideline for codeine.11
The predicted CYP2C19 phenotype based on the diplotype (Table 1 and Supplementary Table S9 online) is consistent with the CPIC guideline for clopidogrel.13 Patients with two functional alleles are categorized as extensive metabolizers, which is considered normal CYP2C19 enzyme activity. Individuals carrying one or two loss-of-function alleles are considered intermediate and poor metabolizers, respectively. Patients with two gain-of-function alleles are classified as ultrarapid metabolizers. The predicted phenotype for a patient carrying the CYP2C19*17 gain-of-function allele in combination with a loss-of-function allele is a matter of debate. Limited data suggest that CYP2C19*17 may not compensate for the CYP2C19*2 allele.15,21
Reference laboratories use varying methods to assign phenotypes. Before pharmacotherapy modifications are made based on this guideline, it is advisable to determine a patient's phenotype as described above.
Available genetic test options
CYP2D6 and CYP2C19 genotyping is available from several reference laboratories (see Supplementary Data online and http://www.pharmgkb.org).
Incidental findings
Independent of drug metabolism and response, there are currently no diseases or conditions that have been convincingly linked to variants in the CYP2D6 or CYP2C19 genes.11,13 Reports describing an association between CYP2D6 ultrarapid metabolizers and suicidality, and CYP2C19 polymorphisms and depressive symptoms are available.22,23,24 These associations are poorly understood and may be explained by alterations in either drug or endogenous substrate metabolism.
Drugs: Amitriptyline and Nortriptyline
Background
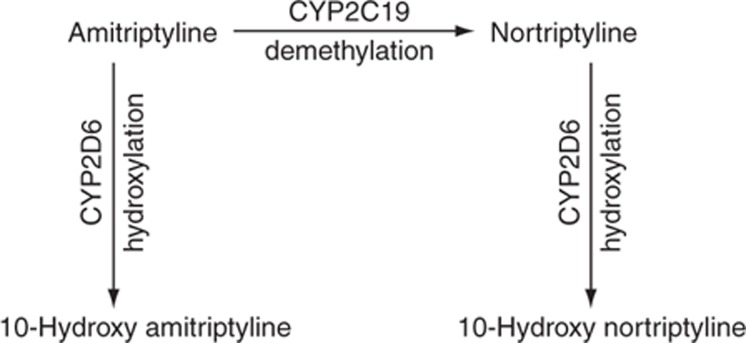
Tricyclics are mixed serotonin and norepinephrine reuptake inhibitors used to treat several disease states including depression, obsessive–compulsive disorder, and neuropathic pain in addition to migraine prophylaxis. The tricyclics have similar but distinct chemical structures referred to as tertiary and secondary amines. The pharmacological properties of the tertiary and secondary amines differ, with tertiary amines having a more pronounced serotonergic effect and secondary amines having a greater noradrenergic effect (Supplementary Table S10 online).25,26 The tertiary amines (e.g., amitriptyline) are mainly metabolized by CYP2C19 to desmethyl metabolites (Figure 1), also referred to as secondary amines (e.g., nortriptyline). It should be noted that the desmethyl metabolites, nortriptyline as well as desipramine, are antidepressant drugs themselves with distinct clinical features that differ from the parent drugs amitriptyline and imipramine. Both the tertiary and secondary amines are metabolized by CYP2D6 to less active hydroxy metabolites (Figure 1, Supplementary Table S10 online). CYP2C19 impacts the ratio of tertiary to secondary amine plasma concentrations but may have less influence on overall drug clearance than CYP2D6.27 However, CYP2C19 metabolism may modulate antidepressant activity and side effects through the pharmacological actions of the tertiary amines. Serotonin reuptake inhibition is expected be more pronounced in CYP2C19 poor metabolizers due to the decreased conversion of tertiary to secondary amines.26
Figure 1.
Major metabolic pathway of amitriptyline and nortriptyline.
Patients may be predisposed to treatment failure or adverse effects due to polymorphisms in CYP2D6 and CYP2C19 altering drug clearance or the ratio of parent drug to metabolites, respectively. Tricyclics are associated with multiple adverse effects, which can cause patients to fail therapy. Common adverse effects include anticholinergic, central nervous system, and cardiac effects. Tertiary and secondary amines along with their metabolites have unique side-effect profiles as detailed in Supplementary Table S11 online.
Both amitriptyline and nortriptyline are used as model drugs for this guideline because the majority of pharmacogenomic studies have focused on these two drugs. However, the results of these studies may apply to other tricyclics because these drugs have comparable pharmacokinetic properties.4,8 Tricyclics are well absorbed from the gastrointestinal tract, and the average extent of first-pass metabolism is ~50%, although the average first-pass metabolism of doxepin may be closer to 70%.4 The clearance of tricyclics is mostly a linear process, but saturation of the hydroxylation pathway may occur at higher plasma concentrations for certain tricyclics, including imipramine and desipramine.4,28 In addition, extrapolated dose adjustments based on metabolizer status are similar across the tricyclic class.8 Because some studies investigating the influence of CYP2D6 and/or CYP2C19 genotype/phenotype on the pharmacokinetics of tricyclics used a single dose, it should be noted that tricyclic metabolism is thought to be similar after single and multiple dosing.5
Linking genetic variability to variability in drug-related phenotypes
Psychiatric disorders such as depression have a 30–50% failure rate with initial treatment, which may, in part, be attributed to adverse effects or altered plasma concentrations.9,29 There is substantial evidence linking CYP2D6 and CYP2C19 genotypes to phenotypic variability in tricyclic side-effect and pharmacokinetic profiles. Modifying pharmacotherapy for patients who have CYP2D6 or CYP2C19 genomic variants that affect drug efficacy and safety could potentially improve clinical outcomes and reduce the failure rate of initial treatment. The application of a grading system to the evidence linking CYP2D6 and CYP2C19 genotypic variations to phenotypic variability in response to amitriptyline or nortriptyline indicates a high quality of evidence in the majority of cases (Supplementary Tables S12 and S13 online). This body of evidence, rather than randomized clinical trials, provides the basis for amitriptyline and nortriptyline dosing recommendations in Tables 2 and 3. Because the tricyclics have comparable pharmacokinetic properties, it may be reasonable to apply this guideline to other tricyclics, including clomipramine, desipramine, doxepin, imipramine, and trimipramine (Supplementary Tables S14–S18 online), with the acknowledgment that there are fewer data supporting dose adjustments for these drugs than for amitriptyline or nortriptyline.
Table 2. Dosing recommendations for amitriptyline and nortriptyline based on CYP2D6 phenotype.
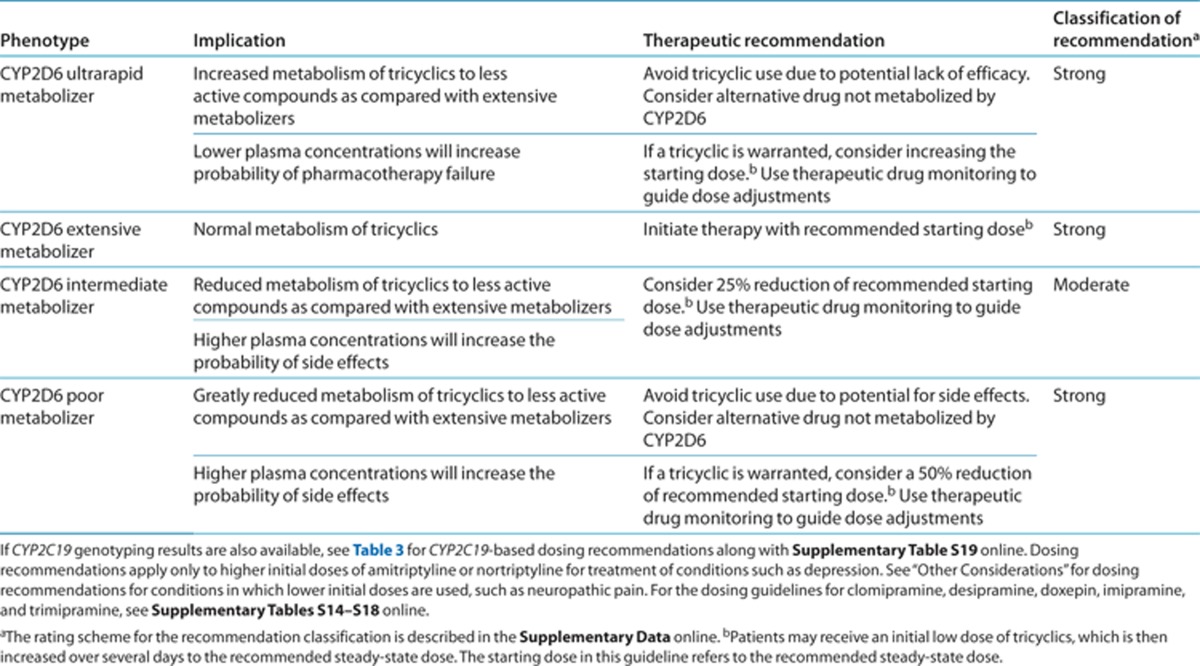
Therapeutic recommendations
CYP2D6 dosing recommendations. For neuropathic pain treatment, in which lower initial doses of tricyclics are used, gene-based dosing recommendations are found in the “Other considerations” section. Table 2 summarizes the gene-based dosing recommendations for amitriptyline and nortriptyline based on CYP2D6 phenotype for situations requiring a higher initial dose, such as depression treatment. The recommended starting dose of amitriptyline or nortriptyline does not need adjustment based on genotype for CYP2D6 extensive metabolizers. A 25% reduction of the recommended dose may be considered for CYP2D6 intermediate metabolizers.30 Because patients with a CYP2D6 activity score of 1.0 are inconsistently categorized as intermediate or extensive metabolizers in the literature, these are difficult to evaluate, resulting in a moderate recommendation classification.
CYP2D6 ultrarapid metabolizers have a higher probability of failing amitriptyline or nortriptyline pharmacotherapy due to subtherapeutic plasma concentrations, and therefore alternative agents are preferred. There are documented cases of CYP2D6 ultrarapid metabolizers receiving large doses of nortriptyline to achieve therapeutic concentrations.10 However, very high plasma concentrations of the nortriptyline hydroxy metabolite were present, which may increase the risk for cardiotoxicity. If a tricyclic is warranted, there are insufficient data in the literature to calculate a starting dose for a patient with CYP2D6 ultrarapid metabolizer status, and therapeutic drug monitoring is strongly recommended. Adverse effects are more likely in CYP2D6 poor metabolizers due to elevated tricyclic plasma concentrations,31 therefore, alternative agents are preferred. If a tricyclic is warranted, consider a 50% reduction of the usual dose; therapeutic drug monitoring is strongly recommended.
CYP2C19 dosing recommendations. Dosing recommendations for neuropathic pain treatment with amitriptyline are discussed in the “Other considerations” section. Table 3 summarizes the gene-based dosing recommendations for CYP2C19 and amitriptyline when higher initial starting doses are warranted. The usual starting dose of amitriptyline may be used in CYP2C19 extensive and intermediate metabolizers. Although CYP2C19 intermediate metabolizers would be expected to have a modest increase in the ratio of amitriptyline to nortriptyline plasma concentrations, the evidence does not indicate that CYP2C19 intermediate metabolizers should receive an alternative dose.
Table 3. Dosing recommendations of amitriptyline based on CYP2C19 phenotype.
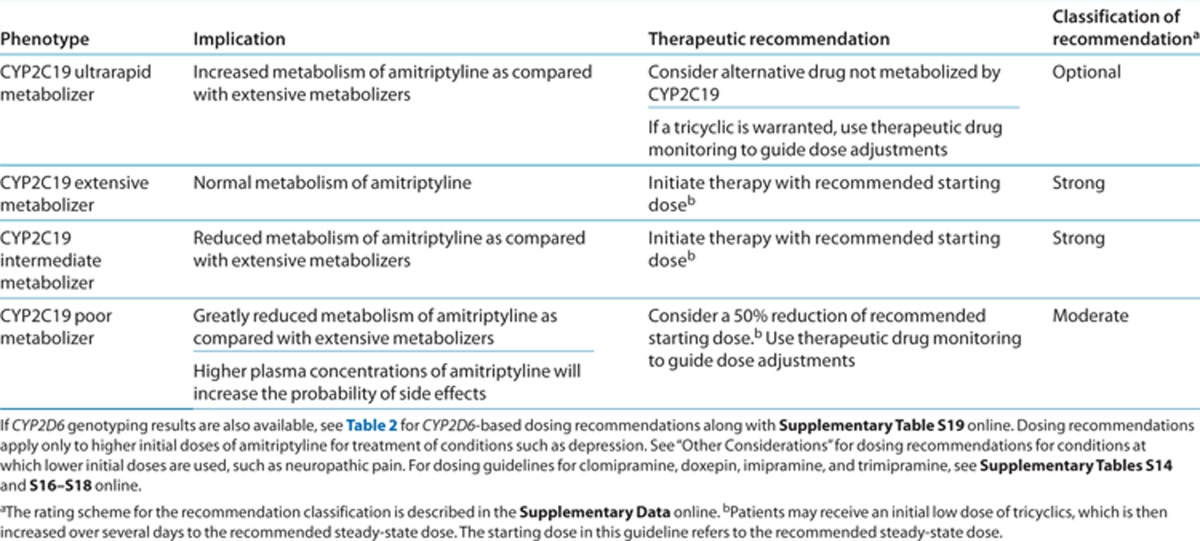
Patients taking amitriptyline who are CYP2C19 ultrarapid metabolizers may be at risk of having altered plasma concentrations or adverse events. Although the CYP2C19*17 allele did not alter the sum of amitriptyline plus nortriptyline plasma concentrations, it was associated with higher nortriptyline plasma concentrations, possibly increasing the risk of adverse events.15 For patients taking amitriptyline, extrapolated pharmacokinetic data suggest that CYP2C19 ultrarapid metabolizers may need a dose increase.8 Due to the need for further studies investigating the clinical importance of the CYP2C19*17 allele and the possibility of altered tricyclic concentrations, we recommend consideration of an alternative tricyclic or other drug not affected by CYP2C19. Because the clinical importance of CYP2C19*17 is currently poorly understood, this recommendation is classified as optional. If amitriptyline is administered to a CYP2C19 ultrarapid metabolizer, therapeutic drug monitoring is recommended.
CYP2C19 poor metabolizers are expected to have a greater ratio of amitriptyline to nortriptyline plasma concentrations.32 The elevated amitriptyline plasma concentrations may increase the chance of a patient experiencing side effects. Consider a 50% reduction of the usual amitriptyline starting dose along with therapeutic drug monitoring.8
CYP2D6 and CYP2C19 combined dosing recommendations. Although specific combinations of CYP2D6 and CYP2C19 alleles are likely to result in additive effects on the pharmacokinetic properties of tricyclics, little information is available on how to adjust initial doses based on combined genotype information. Patients carrying at least one CYP2D6 nonfunctional allele and two CYP2C19 functional alleles had an increased risk of experiencing side effects when administered amitriptyline, whereas patients with at least one CYP2C19 loss-of-function allele and two CYP2D6 functional alleles had a low risk of experiencing side effects.10,33 Because there is only sparse clinical evidence for an additive effect of CYP2D6 and CYP2C19 on tricyclic dosing, the recommendations are classified as optional (Supplementary Table S19 online).
Other considerations
Gene-based dosing recommendations for neuropathic pain treatment. Amitriptyline is often used at lower dosages (e.g., 0.1 mg/kg/day in pediatric patients) for treatment of neuropathic pain than when used for depressive disorders.2,3 Because of the lower dosage, it is less likely that CYP2D6 or CYP2C19 poor or intermediate metabolizers will experience adverse effects due to supratherapeutic plasma concentrations of amitriptyline.34 Therefore, we recommend no dose modifications for poor or intermediate metabolizers when prescribed amitriptyline at a lower dose for treatment of neuropathic pain, but these patients should be monitored closely for side effects. If larger doses of amitriptyline are warranted, we recommend following the gene-based dosing guidelines presented in Tables 2 and 3.
Providing dose recommendations for CYP2C19 ultrarapid metabolizers when amitriptyline is prescribed at lower doses for neuropathic pain treatment is difficult. On the basis of predicted and observed pharmacokinetic data, CYP2D6 ultrarapid metabolizers are at risk of failing amitriptyline therapy for neuropathic pain, and thus alternative agents such as gabapentin should be considered.35 Although little information is available on how to adjust initial amitriptyline doses based on combined CYP2D6 and CYP2C19 genetic results when treating neuropathic pain, caution should be used when patients have a combination of poor or ultrarapid phenotypes (e.g., a CYP2D6 poor metabolizer also having CYP2C19 ultrarapid or poor metabolism).
Consideration of drug interactions and patient characteristics. Patients treated for psychiatric disorders often require multiple medications, which can influence tricyclic plasma concentrations, side effects, and therapeutic failure.4 For example, patients taking amitriptyline in combination with a potent CYP2D6 inhibitor, such as fluoxetine, may have dramatic increases in plasma concentrations.36 It has been suggested that patients taking strong CYP2D6 inhibitors should be treated similarly to CYP2D6 poor metabolizers.11 In addition, patients with increased age, liver disease, and reduced renal function may require reduced doses of tricyclics.4,37,38 Drug–drug interactions along with patient characteristics should be considered in addition to the gene-based dosing recommendations presented herein.
Minor metabolic pathways of tricyclics. Other cytochrome P450 enzymes, including CYP3A4 and CYP1A2, metabolize tricyclics to a lesser extent.4,34,39,40 There is currently no strong evidence supporting gene-based dosing recommendations for other cytochrome P450 enzymes that metabolize tricyclics.
Potential benefits and risks for the patient
For patients who have existing CYP2D6 and/or CYP2C19 genotyping test results, the potential benefit is identifying those patients who are at an elevated risk of experiencing side effects or therapeutic failure. For those patients, dose adjustments can be made or an alternative agent selected. A limitation inherent to most commercially available genotyping tests is that rare or de novo variants are not detected. In addition, some alleles are not well characterized, resulting in uncertainty when predicting the phenotype for some genetic test results. Genotyping is reliable when performed in qualified reference laboratories, but, as with any laboratory test, an error can occur. Any errors in genotyping or phenotype prediction, along with the presence of a rare genomic variant not tested for, could potentially affect the patient lifelong.
Caveats: appropriate use and/or potential misuse of genetic tests
The application of genotype-based dosing is most appropriate when initiating therapy with a tricyclic. Obtaining a pharmacogenetic test after months of drug therapy may be less helpful in some instances, given that the drug dose may have already been adjusted based on plasma concentrations, response, or side effects. Similar to all diagnostic tests, genetic tests are one of several pieces of clinical information that should be considered before initiating drug therapy.
Disclaimer
CPIC guidelines reflect expert consensus based on clinical evidence and peer-reviewed literature available at the time they are written; they are intended only to assist clinicians in decision making and to identify questions for further research. New evidence may have emerged since the time a guideline was submitted for publication. Guidelines are limited in scope and are not applicable to interventions or diseases not specifically identified. Guidelines do not account for all individual variations among patients and cannot be considered inclusive of all proper methods of care or exclusive of other treatments. It remains the responsibility of the health-care provider to determine the best course of treatment for a patient. Adherence to any guideline is voluntary, with the ultimate determination regarding its application to be made solely by the clinician and the patient. CPIC assumes no responsibility for any injury to persons or damage to persons or property arising out of or related to any use of CPIC's guidelines, or for any errors or omissions.
Author Contributions
All the authors contributed to the development of the gene-based dosing guidelines and writing of the manuscript and supplementary material.
Acknowledgments
We acknowledge the critical input of M. Relling, T. Klein, K. Crews, and members of the Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network and funding by the National Institutes of Health (NIH). J.K.H. is supported by NIH/National Institute of General Medical Sciences (NIGMS) Pharmacogenomics Research Network (U01 GM92666 and U01 HL105198) and by ALSAC. C.F.T. and K.S. are supported by NIH/NIGMS (R24 GM61374). E.D.K. is supported by R01 GM63674 and R01 DA14211. T.C.S. is supported by R01 GM088076 and the Agency for Healthcare Research and Quality (R01 HS19818-01). V.L.E. is supported by the NIMH (R01 MH082784). D.J.M. is supported by a New Investigator Salary Award from the Canadian Institutes of Health Research, a New Investigator Fellowship Award from the Ontario Mental Health Foundation, and an Early Researcher Award by the Ministry of Research and Innovation of Ontario. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Agency for Healthcare Research and Quality.
The authors declared no conflict of interest.
Footnotes
SUPPLEMENTARY MATERIAL is linked to the online version of the paper at http://www.nature.com/cpt
Supplementary Material
References
- American Psychiatric Association, A.P. Practice Guideline for the Treatment of Patients With Major Depressive Disorder 3rd edn. (American Psychiatric Publishing, Arlington, VA, 2010); [Google Scholar]
- Watson C.P. The treatment of neuropathic pain: antidepressants and opioids. Clin. J. Pain. 2000;16:S49–S55. doi: 10.1097/00002508-200006001-00009. [DOI] [PubMed] [Google Scholar]
- Laird B., Colvin L., Fallon M. Management of cancer pain: basic principles and neuropathic cancer pain. Eur. J. Cancer. 2008;44:1078–1082. doi: 10.1016/j.ejca.2008.03.022. [DOI] [PubMed] [Google Scholar]
- Rudorfer M.V., Potter W.Z. Metabolism of tricyclic antidepressants. Cell. Mol. Neurobiol. 1999;19:373–409. doi: 10.1023/a:1006949816036. [DOI] [PubMed] [Google Scholar]
- Potter W.Z., Zavadil A.P., 3rd, Kopin I.J., Goodwin F.K. Single-dose kinetics predict steady-state concentrations on imipramine and desipramine. Arch. Gen. Psychiatry. 1980;37:314–320. doi: 10.1001/archpsyc.1980.01780160084010. [DOI] [PubMed] [Google Scholar]
- Hiemke C., et al. AGNP consensus guidelines for therapeutic drug monitoring in psychiatry: update 2011. Pharmacopsychiatry. 2011;44:195–235. doi: 10.1055/s-0031-1286287. [DOI] [PubMed] [Google Scholar]
- Dalén P., Dahl M.L., Bernal Ruiz M.L., Nordin J., Bertilsson L. 10-Hydroxylation of nortriptyline in white persons with 0, 1, 2, 3, and 13 functional CYP2D6 genes. Clin. Pharmacol. Ther. 1998;63:444–452. doi: 10.1016/S0009-9236(98)90040-6. [DOI] [PubMed] [Google Scholar]
- Stingl J.C., Brockmöller J., Viviani R. Mol. Psychiatry. e-pub ahead of print 8 May 2012; 2012. Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J., Seeringer A. Clinical implications of pharmacogenetics of cytochrome P450 drug metabolizing enzymes. Biochim. Biophys. Acta. 2007;1770:489–494. doi: 10.1016/j.bbagen.2006.09.019. [DOI] [PubMed] [Google Scholar]
- Bertilsson L., Aberg-Wistedt A., Gustafsson L.L., Nordin C. Extremely rapid hydroxylation of debrisoquine: a case report with implication for treatment with nortriptyline and other tricyclic antidepressants. Ther. Drug Monit. 1985;7:478–480. [PubMed] [Google Scholar]
- Crews K.R., et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin. Pharmacol. Ther. 2012;91:321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaedigk A., Simon S.D., Pearce R.E., Bradford L.D., Kennedy M.J., Leeder J.S. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin. Pharmacol. Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- Scott S.A., et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin. Pharmacol. Ther. 2011;90:328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S.C., et al. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin. Pharmacol. Ther. 2006;79:103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- de Vos A., van der Weide J., Loovers H.M. Association between CYP2C19*17 and metabolism of amitriptyline, citalopram and clomipramine in Dutch hospitalized patients. Pharmacogenomics J. 2011;11:359–367. doi: 10.1038/tpj.2010.39. [DOI] [PubMed] [Google Scholar]
- Rudberg I., Mohebi B., Hermann M., Refsum H., Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Clin. Pharmacol. Ther. 2008;83:322–327. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- Li-Wan-Po A., Girard T., Farndon P., Cooley C., Lithgow J. Pharmacogenetics of CYP2C19: functional and clinical implications of a new variant CYP2C19*17. Br. J. Clin. Pharmacol. 2010;69:222–230. doi: 10.1111/j.1365-2125.2009.03578.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson Rosenborg S., et al. Kinetics of omeprazole and escitalopram in relation to the CYP2C19*17 allele in healthy subjects. Eur. J. Clin. Pharmacol. 2008;64:1175–1179. doi: 10.1007/s00228-008-0529-z. [DOI] [PubMed] [Google Scholar]
- Schenk P.W., et al. The CYP2C19*17 genotype is associated with lower imipramine plasma concentrations in a large group of depressed patients. Pharmacogenomics J. 2010;10:219–225. doi: 10.1038/tpj.2009.50. [DOI] [PubMed] [Google Scholar]
- Dahl M.L., Bertilsson L., Nordin C. Steady-state plasma levels of nortriptyline and its 10-hydroxy metabolite: relationship to the CYP2D6 genotype. Psychopharmacology (Berl.) 1996;123:315–319. doi: 10.1007/BF02246640. [DOI] [PubMed] [Google Scholar]
- Sibbing D., et al. Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J. Thromb. Haemost. 2010;8:1685–1693. doi: 10.1111/j.1538-7836.2010.03921.x. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J., Lang U., Stamm T., Sander T., Gallinat J. Association of CYP2D6 genotypes and personality traits in healthy individuals. J. Clin. Psychopharmacol. 2006;26:440–442. doi: 10.1097/01.jcp.0000229484.52955.22. [DOI] [PubMed] [Google Scholar]
- Zackrisson A.L., Lindblom B., Ahlner J. High frequency of occurrence of CYP2D6 gene duplication/multiduplication indicating ultrarapid metabolism among suicide cases. Clin. Pharmacol. Ther. 2010;88:354–359. doi: 10.1038/clpt.2009.216. [DOI] [PubMed] [Google Scholar]
- Sim S.C., et al. Association between CYP2C19 polymorphism and depressive symptoms. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:1160–1166. doi: 10.1002/ajmg.b.31081. [DOI] [PubMed] [Google Scholar]
- Gillman P.K. Tricyclic antidepressant pharmacology and therapeutic drug interactions updated. Br. J. Pharmacol. 2007;151:737–748. doi: 10.1038/sj.bjp.0707253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K., Jerling M., Böttiger Y., Yasuda S., Morita S., Bertilsson L. Pronounced differences in the dispositon of clomipramine between Japanese and Swedish patients. J. Clin. Psychopharmacol. 1999;19:393–400. doi: 10.1097/00004714-199910000-00002. [DOI] [PubMed] [Google Scholar]
- Jiang Z.P., et al. The role of CYP2C19 in amitriptyline N-demethylation in Chinese subjects. Eur. J. Clin. Pharmacol. 2002;58:109–113. doi: 10.1007/s00228-002-0445-6. [DOI] [PubMed] [Google Scholar]
- Cooke R.G., Warsh J.J., Stancer H.C., Reed K.L., Persad E. The nonlinear kinetics of desipramine and 2-hydroxydesipramine in plasma. Clin. Pharmacol. Ther. 1984;36:343–349. doi: 10.1038/clpt.1984.185. [DOI] [PubMed] [Google Scholar]
- Thase M.E. Achieving remission and managing relapse in depression. J. Clin. Psychiatry. 2003;64(suppl. 18):3–7. [PubMed] [Google Scholar]
- Swen J.J., et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- Bertilsson L., Mellström B., Sjökvist F., Mårtenson B., Asberg M. Slow hydroxylation of nortriptyline and concomitant poor debrisoquine hydroxylation: clinical implications. Lancet. 1981;1:560–561. doi: 10.1016/s0140-6736(81)92894-4. [DOI] [PubMed] [Google Scholar]
- Shimoda K., et al. The impact of CYP2C19 and CYP2D6 genotypes on metabolism of amitriptyline in Japanese psychiatric patients. J. Clin. Psychopharmacol. 2002;22:371–378. doi: 10.1097/00004714-200208000-00007. [DOI] [PubMed] [Google Scholar]
- Steimer W., et al. Amitriptyline or not, that is the question: pharmacogenetic testing of CYP2D6 and CYP2C19 identifies patients with low or high risk for side effects in amitriptyline therapy. Clin. Chem. 2005;51:376–385. doi: 10.1373/clinchem.2004.041327. [DOI] [PubMed] [Google Scholar]
- Halling J., Weihe P., Brosen K. The CYP2D6 polymorphism in relation to the metabolism of amitriptyline and nortriptyline in the Faroese population. Br. J. Clin. Pharmacol. 2008;65:134–138. doi: 10.1111/j.1365-2125.2007.02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin R.H., et al. Pharmacologic management of neuropathic pain: evidence-based recommendations. Pain. 2007;132:237–251. doi: 10.1016/j.pain.2007.08.033. [DOI] [PubMed] [Google Scholar]
- el-Yazigi A., Chaleby K., Gad A., Raines D.A. Steady-state kinetics of fluoxetine and amitriptyline in patients treated with a combination of these drugs as compared with those treated with amitriptyline alone. J. Clin. Pharmacol. 1995;35:17–21. doi: 10.1002/j.1552-4604.1995.tb04740.x. [DOI] [PubMed] [Google Scholar]
- Nelson J.C., Mazure C., Jatlow P.I. Clinical implications of the pharmacokinetics of tricyclic antidepressants. Psychopharmacol. Ser. 1989;7:219–227. doi: 10.1007/978-3-642-74430-3_24. [DOI] [PubMed] [Google Scholar]
- Young R.C., Alexopoulos G.S., Dhar A.K., Kutt H. Plasma 10-hydroxynortriptyline and renal function in elderly depressives. Biol. Psychiatry. 1987;22:1283–1287. doi: 10.1016/0006-3223(87)90039-4. [DOI] [PubMed] [Google Scholar]
- Ketter T.A., et al. The emerging role of cytochrome P450 3A in psychopharmacology. J. Clin. Psychopharmacol. 1995;15:387–398. doi: 10.1097/00004714-199512000-00002. [DOI] [PubMed] [Google Scholar]
- Shen W.W. Cytochrome P450 monooxygenases and interactions of psychotropic drugs: a five-year update. Int. J. Psychiatry Med. 1995;25:277–290. doi: 10.2190/29NP-2XPN-X0ME-MQWU. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.