Abstract
When resectable, invasive pancreatic ductal adenocarcinoma (PDAC) is most commonly treated with surgery and radiochemotherapy. Given the intricate local anatomy and locoregional mode of dissemination, achieving clean surgical margins can be a significant challenge. On the basis of observations that cathepsin E (CTSE) is overexpressed in PDAC and that an United States Food and Drug Administration (FDA)-approved protease inhibitor has high affinity for CTSE, we have developed a CTSE optical imaging agent [ritonavir tetramethyl-BODIPY (RIT-TMB)] for potential intraoperative use. We show nanomolar affinity [half maximal inhibitory concentration (IC50) of 39.9 ± 1.2 nM] against CTSE of the RIT-TMB in biochemical assays and intracellular accumulation and target-to-background ratios that allow specific delineation of individual cancer cells. This approach should be useful for more refined surgical staging, planning, and resection with curative intent.
Introduction
The incidence of pancreatic ductal adenocarcinoma (PDAC) has increased during the past several decades and now ranks as the fourth leading cause of cancer death in the United States. Patients usually present late in the disease process with symptoms of weight loss, pain, and/or jaundice. While improvements in imaging technology have generally increased the spatial resolution of scans as well as interventional capabilities, these technologies rarely allow detection of the small lesions without anatomic abnormalities or precursor lesions (pancreatic intraepithelial neoplasia). 18F-fluorodeoxyglucose-positron emission tomography imaging is generally considered less sensitive since many cancers do not show increased glucose uptake [1,2] or are simply too small, i.e., below the detection threshold. As a result, patients often present with advanced stage disease when the decision to surgically resect is not straightforward. Moreover, the incidence of recurrent disease following surgical resection remains high, which underscores the need for improved detection of residual disease. Unfortunately, there are currently no reliable intraoperative imaging approaches to delineate microscopically invading cancers or to distinguish malignancy from inflammation for more accurate mapping and detection of cancer spread. In short, there is an urgent need for molecularly targeted imaging agents to improve the detection of this highly lethal disease.
Cathepsin E (CTSE) has emerged as a promising PDAC target from different lines of research. The Hanahan group demonstrated overexpression of CTSE in the ripTag model and subsequently confirmed high levels in the K-ras mouse model. Here, it was shown that 3.5% of the total protein within PDAC lysates consisted of CTSE protein (unpublished). Second, a meta-analysis by the Ramaswamy laboratory, employing whole-genome transcriptional arrays to analyze more than 500 samples (including primary human tumors, cell line screens, mouse model analysis, and metasearch), identified CTSE as a prime target. Finally, there are literature reports pointing to CTSE as a target both recently [3–6] and even dating back to the early 1990s [7–11]. There are several features that make CTSE a unique imaging target: 1) it exists only intracellularly and is not secreted, 2) it exists in a pro-form and is not proteolytically active (thus is not efficiently targeted by activity-based probes or substrates except for perhaps a small fraction of active enzyme), and 3) enzyme activity in live cells is not inhibitable by classic pan-cathepsin inhibitors. Furthermore, unlike other cathepsins (B, D, S, K) that can show high stromal levels [12–16], CTSE overexpression appears to be unique to PDAC tissue. Additionally, CTSE is expressed in the earliest forms of pancreatic intraepithelial neoplasia [3,6]. Interestingly, the United States Food and Drug Administration (FDA)-approved human immunodeficiency virus (HIV) protease inhibitor ritonavir (RIT; Figure 1B) has been shown to have a reasonable coaffinity for CTSE [17]. We hypothesized that this small molecule drug scaffold could be modified to yield high affinity imaging agents for intraoperative optical imaging and with appropriate pharmacokinetics for in vivo use.
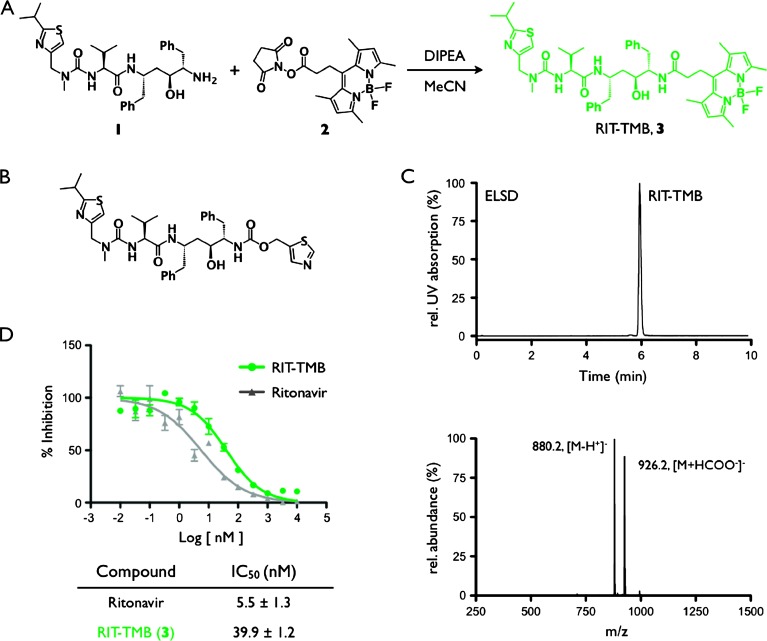
Figure 1.
RIT-TMB synthesis and characterization. (A) Synthesis of RIT-TMB (3). (B) Chemical structure of RIT, P2′ thiazole on the right side of the molecule. (C) HPLC chromatogram and LC-MS analysis of purified RIT-TMB. (D) Binding affinity data for RIT parent compound and RIT-TMB against human CTSE.
In this research, we systemically analyzed different RIT variants and conjugation points, linkers to modulate hydrophilicity, imaging reporters, and labeling strategies. These compounds were screened against CTSE protein using surface plasmon resonance (SPR) analysis. We discovered that one particular compound [RIT-tetramethyl-BODIPY (TMB)] had low nM affinity, was cell permeable, and was fluorescent. Here, we show that this compound is selective for PDAC in human tissue samples and allows in vivo imaging of PDAC in mouse models.
Materials and Methods
General
RIT was purchased from Selleck Chemicals (Houston, TX), (S)-N-((2S,4S,5S)-5-amino-4-hydroxy-1,6-diphenylhexan-2-yl)-2-(3-((2-isopropylthiazol-4-yl)methyl)-3-methylureido)-3-methylbutanamide (desthiazolylmethyloxycarbonyl, 1) and 2S,3S,5S-5-amino-2-[N-[[(5-thiozolyl)methoxy]carbonyl]amino]-1,6-diphenyl-3-hydroxyhexane, 4, were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and BODIPY 493/503 succinimidyl ester (TMB-NHS) was purchased from Invitrogen (Grand Island, NY). Proton and carbon nuclear magnetic resonance (1H and 13C NMR) spectra were recorded on a Varian AS-400 (400 MHz) spectrometer. Chemical shifts are reported in parts per million downfield from tetramethylsilane. Data are reported as follows: chemical shift (κ), integration, multiplicity (bs, broad singlet; s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet), and coupling constants in Hz. Liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) analysis and high-performance liquid chromatography (HPLC) purifications were performed on a Waters (Milford, MA) LC-MS System. For LC-ESI-MS analyses, a Waters XTerra C18 5-µm column was used. Preparative HPLC runs for synthetic intermediates used an Atlantis Prep T3 OBD 5-µM column [eluents: 0.1% trifluoroacetic acid (TFA) (vol/vol) in water and MeCN; gradient: 0 to 1.5 minutes, 5% to 100% B; 1.5 to 2.0 minutes, 100% B]. Nu/Nu mice [Cox-7; Massachusetts General Hospital (MGH), Boston, MA] were housed and handled according to MGH Institutional Animal Care and Use Committee guidelines. Mice undergoing surgical procedures or imaging were anesthetized by isoflurane inhalation at a flow rate of 2% isoflurane and 2 L/min oxygen, which was reduced to 1 L/min oxygen for extended anesthesia during imaging. Mice implanted with cells in window chambers and orthotopically in the pancreas were administered antibiotics in their drinking water. Anesthetized mice were monitored at all times.
Homology Modeling and Docking Simulations
A homology model of CTSE was obtained as previously described [18] using the SWISS-MODEL Workspace server (SWISS-MODEL Workspace, http://swissmodel.expasy.org) [19]. The model was templated on a crystal structure of pepsin (PEP4). RIT and RIT-TMB were docked into the resulting CTSE homology model. Autodock Vina version 1.1.2 [20] was used for all docking simulations after preparing files using Autodock Tools 1.5.6rc3 [21]. All bonds were treated as rotatable with the exception of amide and ring bonds. Docking simulations were conducted to generate 20 structures at an exhaustiveness of 25.
RIT-NH-Ac (5) and Ac-NH-RIT (6) Synthesis and Characterization
(S)-N-((2S,4S,5S)-5-amino-4-hydroxy-1,6-diphenylhexan-2-yl)-2-(3-((2-isopropylthiazol-4-yl)methyl)-3-methylureido)-3-methylbutanamide, 1 (10 mg, 0.017 mmol), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) (50 mg, 0.261 mmol), triethylamine (600 µl, 4.32 mmol), and glacial acetic acid (50 µl, 0.870 mmol) were combined in dichloromethane (2 ml) and stirred at room temperature for 1 hour. Solvent was removed under reduced pressure and the resulting crude mixture was redissolved in 400 µl of acetonitrile water (1:4). The desired (S)-N-((2S,4S,5S)-5-acetamido-4-hydroxy-1,6-diphenylhexan-2-yl)-2-(3-((2-isopropylthiazol-4-yl)methyl)-3-methylureido)-3-methylbutanamide (RIT-NH-Ac, 5) was isolated by HPLC purification in 34.1% yield and 99% purity; 1H NMR (400 MHz, DMSO-d6) δ 7.65 (d, J = 8.5 Hz, 1H), 7.48 - 7.41 (m, 1H), 7.26 - 7.03 (m, 11H), 5.99 (d, J = 8.6 Hz, 1H), 4.50 - 4.34 (m, 2H), 4.09 (d, J = 7.2 Hz, 1H), 3.97 (q, J = 9.1, 8.6 Hz, 1H), 3.94 - 3.86 (m, 1H), 3.61 - 3.53 (m, 1H), 3.22 (p, J = 6.9 Hz, 1H), 2.90 - 2.81 (m, 3H), 2.74 - 2.53 (m, 4H), 1.90 - 1.80 (m, 1H), 1.66 (s, 3H), 1.39 (d, J = 8.2 Hz, 2H), 1.28 (d, J = 6.9 Hz, 6H), 0.72 (d, J = 6.7 Hz, 6H). 2S,3S,5S-5-amino-2-[N-[[(5-thiozolyl)methoxy]carbonyl]amino]-1,6-diphenyl-3-hydroxyhexane, 4 (10 mg, 0.024 mmol), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) (65 mg, 0.339 mmol), triethylamine (790 µl, 5.68 mmol), and glacial acetic acid (65 µl, 1.14 mmol) were combined in dichloromethane (2 ml) and stirred at room temperature for 1 hour. Solvent was removed under reduced pressure and the resulting crude mixture was redissolved in 400 µl of acetonitrile water (1:4). 2S,3S,5S-5-acetamido-2-[N-[[(5-thiozolyl)methoxy]carbonyl]amino]-1,6-diphenyl-3-hydroxyhexane, 6, was isolated by HPLC purification in 30.9% yield and 99% purity; 1H NMR (400 MHz, DMSO-d6) δ 1H NMR (400 MHz, DMSO-d6) δ 9.04 (d, J = 0.8 Hz, 1H), 7.85 (q, J = 0.8 Hz, 1H), 7.57 (d, J = 8.7 Hz, 1H), 7.24 - 7.06 (m, 12H), 6.87 (d, J = 9.3 Hz, 1H), 5.20 - 5.09 (m, 2H), 4.14 - 4.05 (m, 1H), 3.86 - 3.77 (m, 1H), 3.51 (td, J = 6.9, 2.0 Hz, 1H), 2.74 - 2.59 (m, 4H), 1.63 (s, 3H), 1.42 (m, 2H).
RIT-TMB (3) Synthesis and Characterization
(S)-N-((2S,4S,5S)-5-Amino-4-hydroxy-1,6-diphenylhexan-2-yl)-2-(3-((2-isopropylthiazol-4-yl)methyl)-3-methylureido)-3-methylbutanamide, 1 (6.9 mg, 0.012 mmol) and BODIPY 493/503 succinimidyl ester, 2 (2 mg, 0.012 mmol) were added to a 2-ml vial. To the same vial, dry acetonitrile (1 ml) and diisopropylethyl amine (6.3 µl, 0.036 mmol, 3 eq) were added. The reaction mixture was stirred for 14 hours at room temperature and was purified by preparative HPLC (20 ml/min, 10 minutes, 5% acetonitrile in water to 95% acetonitrile in water; see specifics above). The fractions were collected and the solvent was removed by rotary evaporation to yield 11.1 mg of orange solid (63% yield) in 99% purity; 1H NMR (400 MHz, methanol-d4) δ 8.27 (bs, 1H), 7.84 (d, J = 8.7 Hz, 1H), 7.68 (d, J = 9.4 Hz, 1H), 7.24 (d, J = 4.4 Hz, 4H), 7.19 - 7.12 (m, 6H), 7.07 (ddt, J = 5.4, 4.3, 3.6 Hz, 1H), 6.17 - 6.09 (m, 3H), 4.59 - 4.41 (m, 2H), 4.35 - 4.22 (m, 2H), 4.00 - 3.89 (m, 1H), 3.74 (td, J = 6.9, 1.9 Hz, 1H), 3.36 - 3.20 (m, 1H), 3.21 - 2.93 (m, 5H), 2.89 - 2.62 (m, 4H), 2.43 (s, 6H), 2.40 - 2.25 (m, 2H), 1.94 (dq, J = 13.7, 7.0 Hz, 1H), 1.69 - 1.53 (m, 2H), 1.35 (d, J = 6.9 Hz, 7H), 1.28 (s, 1H), 0.84 (dd, J = 6.7, 4.8 Hz, 11H); 13C NMR (100 MHz, methanol-d4) δ 177.7, 171.6, 170.1, 157.9, 153.7, 153.3, 146.3, 141.5, 139.9, 139.2, 131.0, 129.8, 129.5, 128.5, 128.3, 128.2, 128.0, 126.2, 126.1, 122.1, 114.5, 70.2, 69.2, 60.0, 48.7, 47.4, (39.095), 38.0, 36.5, 34.9, 24.5, 23.3, 23.2, 19.9, 18.7, 16.4, 14.5; LC-ESI-MS m/z (%) ES-: observed, 880.2 [M - H+]-1 (100); calculated, 880.5 [M - H+]-1.
SPR Measurements
SPR measurements were performed on a Biacore T100 instrument using a Sensor Chip CM5 (carboxymethylated dextran matrix immobilized on a gold surface). Immobilization reagents (amine coupling kit) were purchased from GE Healthcare (Pitttsburg, PA). Control software and data evaluation software supplied with the instrument were employed for setting up experiments and analyzing data.
Immobilization of CTSE to the sensor surface. Carboxyl groups on the dextran matrix of flow cell 2 were activated with a 1:1 solution of EDC/NHS (600 second injection at 10 µl/min) followed by a solution of CTSE [20 µg/ml in acetate buffer (pH 4.5), 750 second injection at 5 µl/min]. The remaining NHS-ester groups on the sensor surface were quenched with ethanolamine (420-second injection at 10 µl/min). This resulted in a CTSE immobilization level of 2980 response units. The reference flow cell 1 (blank immobilization) was prepared by activating carboxyl groups with a 1:1 solution of EDC/NHS (420-second injection at 10 µl/min) and quenched with a solution of ethanolamine (420-second injection at 10 µl/min).
Binding measurements between RIT-NH-Ac/Ac-NH-RIT and immobilized CTS. Binding was measured at 25°C using sodium acetate buffer solution (pH 3.5) containing 2% DMSO as running and dilution buffer. Using the single-cycle kinetics method, the analyte was injected in duplicate at increasing concentrations from 7.8 to 2000 nM in 1:4 dilution in a single cycle without regeneration. The resulting binding curves were double reference subtracted and fitted to a 1:1 Langmuir binding model for analyses.
Biochemical Assays
We used a commercially available assay to determine CTSE inhibition. In short, recombinant human CTSE (10 µg, 1294-AS; R&D Systems, Minneapolis, MN) was diluted with 100 µl of 25 mM MES and 150 mM NaCl (pH 6) buffer. From the resulting solution, 10 µl was added to 990 µl assay buffer (0.1 M NaOAc, 0.1 M NaCl, pH 3.5). This solution was incubated for 30 minutes at room temperature. It was then added to 4 ml of assay buffer. Separately, the fluorogenic peptide substrate (Mca-PLGL-Dpa-AR-NH2, R&D Systems ES001) was diluted 1:299 in assay buffer. The assay was performed in a 96-well, black, flat-bottomed plate. To each well, the designated dilution of RIT-TMB, RIT, RIT-NH-Ac, and Ac-NH-RIT was added. Control samples did not contain inhibitor and background measurement samples did not contain CTSE. All reaction mixtures were adjusted to a final volume of 100 µl. To each well, 49.5 µl of CTSE was then added. The inhibitor was incubated with the CTSE for 3 minutes at room temperature. Then, 49.5 µl of fluorogenic substrate was added to each well. The resulting concentrations of the inhibitor ranged from 0.01 nM to 10 µM. All experiments were performed in triplicate. This mixture was incubated for 7 minutes at room temperature. The fluorescence of each well was measured using a Tecan Safire 2 microplate reader (Tecan Group, Mannedorf, Switzerland). Half maximal inhibitory concentration (IC50) values were calculated from raw fluorescence data using Prism (GraphPad Software, La Jolla, CA).
Cell Culture, Cloning, and Generation of Fluorescent Cell Lines
MIA PaCa-2 (PaCa-2) and PANC-1 human pancreatic adenocarcinoma cells (ATCC, Manassas, VA) were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 1% glutamine/penicillin/streptomycin (Mediatech, Herndon, VA). AsPC1 (human), Panc02, and AH367 pancreatic carcinoma cell lines were grown in RPMI supplemented with 10% FBS and 1% glutamine/penicillin/streptomycin (Mediatech). Panc02 were kindly provided by Dr Jeffrey Schlom, Chief of the Laboratory of Tumor Immunology and Biology at the National Cancer Institute. AH367 were kindly provided by Dr Nabeel Bardeesy at MGH. All cells were grown in a 37°C humidified incubator with 5% CO2. Human CTSE (Human Mammalian Gene Collection-verified FL cDNA, clone ID 5174814) in a pSPORT6 vector (Thermo Scientific Open Biosystems, Waltham, MA) was amplified using the Platinum PCR SuperMix High Fidelity (Invitrogen/Life Technologies) and gel purified using the Qiaex gel purification kit (Qiagen, Venlo, The Netherlands), then cloned into a BamH1/EcoR1-digested pLVX-mCherry-N1 plasmid using the In Fusion Cloning Kit (Clontech, Mountain View, CA). Ligated DNA was transformed into TOP10 cells using the manufacturer recommended rapid transformation protocol (Invitrogen/Life Technologies). Plasmid DNA was extracted from ampicillin-resistant colonies grown first on Luria-Bertani agar plates and then in Luria-Bertani broth, and plasmids were verified with DNA sequencing (Genewiz, South Plainfield, NJ).
Lentiviral particles were generated by transfecting the HEK293T cell line with packaging vectors (pMDL gag/pol, pRSV Rev, VSV-g; obtained from Addgene, Cambridge, MA) and pLVX-CTSE-mCherry for approximately 3 days, at which point the media were collected and filtered through a 0.45-µm cellulose acetate syringe filter, aliquoted into cryotubes, and snap frozen in liquid nitrogen. Cells (1 x 105 cells/well) were seeded in a 12-well plate and transduced with lentivirus in a 1:1 ratio with growth medium with 5 µg/ml polybrene. After 72 hours, cells were trypsinized and split into dishes with growth medium containing 3 µg/ml puromycin. For imaging experiments, the brightest cells were sorted using the FACS Aria II (BD Biosciences). Sorting was performed at the Harvard Stem Cell Institute-Center for Regenerative Medicine Flow Cytometry Core facility at MGH.
Western Blot
Lysate was collected from cells (AsPC-1, PANC1, PaCa-2, PANC02, and AH367) grown in 10-cm dishes using 1x RIPA buffer (Cell Signaling Technology, Danvers, MA) containing protease inhibitors (Roche, Indianapolis, IN) and homogenized using a 23-gauge syringe needle. Protein concentrations were determined using the micro BCAassay (Pierce, Rockford, IL). Tris-glycine 4% to 12% gradient gels (Invitrogen/Life Technologies) were loaded with equal protein per well and run at 175 V using the mini-cell apparatus (Invitrogen/Life Technologies). Proteins were transferred onto polyvinylidene difluoride (PVDF) membrane using the iBlot apparatus (Invitrogen/Life Technologies). The blots were blocked in SuperBlock buffer (Invitrogen/Life Technologies) for 1 hour at room temperature, incubated in primary antibodies against CTSE (C7 clone; Santa Cruz Biotechnology) or glyceraldehyde 3-phosphate dehydrogenase (GAPDH; R&D Systems) in a 5% milk-TBS-Tween 20 solution overnight at 4°C, then washed in TBS-Tween 20. Blots were then incubated in secondary antibodies conjugated to HRP in 5% milk-TBS-Tween 20 solution at room temperature for 1 hour and then washed in TBS-Tween 20. West Pico chemiluminescent substrate (Pierce) was applied to the membranes for 5 minutes, at which point blots were exposed to film (Eastman Kodak) and developed.
Quantitative Reverse Transcription-Polymerase Chain Reaction
Lysate was collected from cells grown in 10-cm dishes using RLT buffer from the RNeasy extraction kit (Qiagen) and homogenized using a 23-gauge syringe needle. RNA was extracted using the manufacturer's recommended protocol, including the optional DNAse step. Quantitative reverse transcription-polymerase chain reaction was carried out using the TaqMan RNA-to-CT One-Step Kit and TaqMan Gene Expression Assays for human CTSE and GAPDH (Applied Biosystems/Life Technologies), with the ABI7500 Fast Thermal Cycler. Relative expression was determined using the 2-ΔΔCt method, with PaCa-2 parent as the calibrator.
Dose-Response Assays
CTSE-mCherry cells (1 x 105 cells/well) were seeded into 48-well black-wall, clear-bottom plates (Corning) and incubated for 24 hours before adding different concentrations of RIT-TMB. After incubating for 15 minutes at 37°C, cells were washed three times with phosphate-buffered saline (PBS) and the fluorescence levels were measured with a SynergyMx plate reader (Biotek, Winooski, VT) with excitation at 503 and 587 nm and emission at 510 and 610 nm for TMB and mCherry, respectively. Following fluorescence measurement, proteins were extracted using RIPA lysis buffers and quantified using BCA assay (Pierce).
Blocking Experiments
CTSE-mCherry cells (1 x 105 cells/well) were seeded into 48-well black-wall, clear-bottom plates (Corning) and incubated for 24 hours before incubation either with RIT (50 µM) for 30 minutes at 37°C and subsequently RIT-TMB (10 nM) for 15 minutes at 37°C or with just RIT-TMB (10 nM) for 15 minutes at 37°C. Following the incubation, cells were washed three times with PBS and the fluorescence levels were measured with a SynergyMx plate reader (Biotek) with excitation at 503 and 587 nm and emission at 510 and 610 nm for TMB and mCherry, respectively. Following fluorescence measurement, proteins were extracted using RIPA lysis buffers and quantified using BCA assay (Pierce).
Immunohistochemistry and Co-staining
To detect CTSE expression in PDAC tumor tissue, paraffin-embedded human tissue sections (tissue microarray) were deparaffinized and rehydrated before antigen retrieval procedure. Heat-induced epitope retrieval was performed using retrieval solution (pH 6.0; BD Biosciences) according to the manufacturer's protocol. The tissue sections were incubated in 1% hydrogen peroxide solution to block endogenous peroxidase activity and blocked with 4% normal rabbit serum in PBS. The sections were stained with goat anti-human CTSE antibody (R&D Systems), followed by biotinylated anti-goat IgG (Vector Laboratories, Inc, Burlingame, CA). For paraffin-embedded mouse tissue, goat anti-mouse CTSE antibody (R&D Systems) was used. Vectastain ABC Kit (Vector Laboratories, Inc) and a 3-amino-9-ethylcarbazole substrate (Dako, Carpinteria, CA) were used for color development, and all sections were counterstained with Harris hematoxylin solution (Sigma-Aldrich, St Louis, MO). The images of slides were captured and digitized automatically at a magnification of x40 (NanoZoomer 2.0RS; Hamamatsu, Hamamatsu City, Japan).
Human biopsy samples, obtained through an Institutional Review Board (IRB)-approved protocol, were used to section 5-µm-thick frozen sections. The frozen sections were briefly fixed and CTSE staining was performed by applying goat anti-human CTSE antibody (R&D Systems) and biotinylated anti-goat IgG (Vector Laboratories, Inc). The sections were incubated with streptavidin Alexa Fluor 594 (Invitrogen) to detect CTSE expression in tissue. After washing the slides with PBS, 20 µM RIT-TMB was incubated at room temperature for 1 hour and the images were captured using BX63 (Olympus, Center Valley, PA) equipped with Neo sCMOS (ANDOR Technology, South Windsor, CT)—TXRED: EX562/40, EM624/40, DM593 for CTSE and green fluorescent protein (GFP): EX472/30, EM520/35, DM495 for RIT-TMB.
Immunofluorescence Staining and Microscopy
PaCa-2 CTSE-mCherry cells were seeded in eight-well chamber slides coated with gelatin and grown overnight at 37°C and 5% CO2. Fixed with 4% paraformaldehyde in PBS, the cells were permeabilized with 0.1% Triton X-100/1% BSA in PBS for 30 minutes. Cells were incubated with anti-CTSE monoclonal antibody (1:50; Santa Cruz Biotechnology) for 1 hour at room temperature, then washed three times with PBS/0.1% Triton X-100. The cells were then stained with an IgG-Cy5 secondary antibody (1:100; Millipore, Billerica, MA) for 30 minutes at room temperature and washed three times with 0.1% Triton X-100/PBS. Finally, cells were stained with RIT-TMB in DMSO for 30 minutes, washed three times with PBS, stained with Hoechst dye for 10 minutes, and washed three times with PBS. Prolong gold reagent was added to each of the wells, and a coverslip was mounted over the slide. Images were acquired using an Olympus 80i microscope.
In Vitro Live Cell Imaging
Both parent and CTSE-mCherry cell lines (PaCa-2, PANC-1, AsPC1, Panc02, AH367) were seeded in ibidi coat 96-well plates (ibidi, Verona, WI) and allowed to attach overnight. The cells were incubated at 37°C with varying concentrations of RIT-TMB or TMB alone in DMSO for 20 minutes, at which point the cells were washed with PBS and put into phenol red-free growth medium. Live cell microscopy was carried out using a DeltaVision imaging system (Applied Precision, Issaquah, WA), consisting of an environmental chamber heated to 37°C, with CO2 bubbled through a water bath, an automated Olympus IX70 inverted microscope, and a CoolSNAP HQ2 CCD camera.
Pharmacokinetics Experiments
Blood half-life of RIT-TMB was quantified by intravital microscopy imaging of vessels in the ear of Nu/Nu control mice. AngioSenseR 680 (PerkinElmer, Waltham, MA) was injected intravenously into the tail vein with a catheter to allow focusing on the vessels in the ear. A 40-minute time-lapse imaging series with 10-second interval was initiated in the fluorescein isothiocyanate and 680 channel before injection of RIT-TMB [7.5 µl of 10 mM RIT-TMB in DMSO (75 nmol) diluted with 17.5 µl of dimethylacetamide/Solutol (1:1), then diluted with 10 x 17.5 µl of 0.9% saline] into the tail vein. Average fluorescence intensity of three ROIs within vessels minus average background of three regions of interest (ROIs) of images collected before injection of probe was plotted against the time after probe injection using Prism 5.0a (GraphPad Software).
Intravital Imaging
Titanium dorsal skin fold chambers (APJ Trading Co, Inc, Ventura, CA) were surgically implanted in the dorsal skin fold of Nu/Nu mice. PaCa-2 CTSE-mCherry tumor cells (5 x 106/injection) mixed 1:1 with Matrigel (Becton Dickinson) were injected into the window chamber subcutaneously and grown for approximately 1 week, at which point the skin within the window was removed and replaced with a 10-mm diameter glass coverslip that was secured with a C-clip. On the day of imaging, the window chambers of anesthetized mice were immobilized using a custom-made holder seated on a 37°C heating pad. RIT-TMB [7.5 µl of 10 mMRIT-TMB in DMSO (75 nmol) diluted with 17.5 µl of dimethylacetamide/Solutol (1:1), then diluted with 10 x 17.5 µl 0.9% saline] was injected intravenously into the tail vein. The window chambers were imaged for at least 2 hours following injection to monitor accumulation of the probe in tumor cells. Static and time-lapse images were collected using a customized Olympus FV1000 based on a BX61-WI confocal microscope. The objectives used were 2/340 XLFluor (NA 1/4 0.14, air), 20 UPlanFL (NA 1/4 0.50, water), and 60 LUMFL N (NA 1/4 1.10, water). TMB and mCherry were excited using the 488-nm line of an argon ion laser and a 559-nm diode laser, respectively. Emitted light was separated using a 405/488/559/635-nm dichroic beam splitter and refined using the band-pass filters BA505-540 and BA575-605.
Orthotopic Tumor In Vivo Imaging
Orthotopic tumors were prepared by first locating and sterilizing the skin over the spleen (∼5 mm below the rib cage). An incision (1 cm) starting at the apex of the spleen was made through the abdominal skin and muscle allowing visualization of the pancreas. The pancreas was externalized using a damp cotton swab and PaCa-2 CTSE-mCherry tumor cells (5 x 105/injection) mixed 1:1 with Matrigel (Becton Dickinson) were injected into the exposed pancreatic lobe. The pancreas was positioned back into the abdominal cavity and the incision closed with 5-0 suture. Following surgery, mice were administered a nonfluorescent diet until day of imaging. Images were collected with an intravital laser scanning microscope as described above. A custom-made tissue stabilizer [22] was used to reduce motion artifacts induced by breathing and cardiac activity and to stabilize the imaging focal plane. Samples were excited at 488 nm with an air-cooled argon laser (Melles Griot, Carlsbad, CA) for visualization of the RIT-TMB.
Results
Design of RIT-TMB was guided by binding assays of two truncated derivatives of the HIV protease inhibitor RIT, RIT-NH-Ac, 5, and Ac-NH-RIT, 6, as well as docking simulations of RIT (Autodock Tools 1.5.6rc3 [21] and Autodock Vina [20]) with a previously described homology model (SWISS-MODEL Workspace, http://swissmodel.expasy.org) [19] of the mature CTSE structure. Competitive binding assay of RIT-NH-Ac and Ac-NH-RIT against recombinant CTSE (Figure W1C) demonstrated that RIT-NH-Ac has a 30-fold higher affinity over Ac-NH-RIT. This was supported by SPR binding assays (Figure W1D). RIT-NH-Ac was found to have a low nanomolar affinity for CTSE (41 nM), while no SPR binding was observed for Ac-NH-RIT. Docking simulations of the parent RIT compound demonstrated the P2′ region of the molecule extended out from the binding pocket of the CTSE homology model (Figure W2). We therefore designed imaging agents based on modification at this position.
Synthesis of RIT-TMB(Figure 1A) was achieved from the commercial reagents, desthiazolylmethyloxycarbonyl ((S)-N-((2S,4S,5S)-5-amino-4-hydroxy-1,6-diphenylhexan-2-yl)-2-(3-((2-isopropylthiazol-4-yl) methyl)-3-methylureido)-3-methylbutanamide), 1, and amine reactive tetramethyl BODIPY NHS ester, 2. After stirring for 14 hours in the presence of base and subsequent HPLC purification, RIT-TMB, 3, was isolated in 63% yield and 99% purity (Figure 1C). The identity of RIT-TMB was confirmed by HPLC-LC/MS (Figure 1C), 1H-NMR, and 13C-NMR.
The inhibitory strength of RIT-TMB was tested against human recombinant CTSE (results shown in Figure 1D) and compared to that of the parent molecule RIT. RIT-TMB was found to have an IC50 of 39.9 ± 1.2 nM, while RIT had an IC50 of 5.5 ± 1.3 nM (similar to previous reports [17]), demonstrating that conjugation of RIT and tetramethyl BODIPY fluorochrome was tolerated by the enzyme.
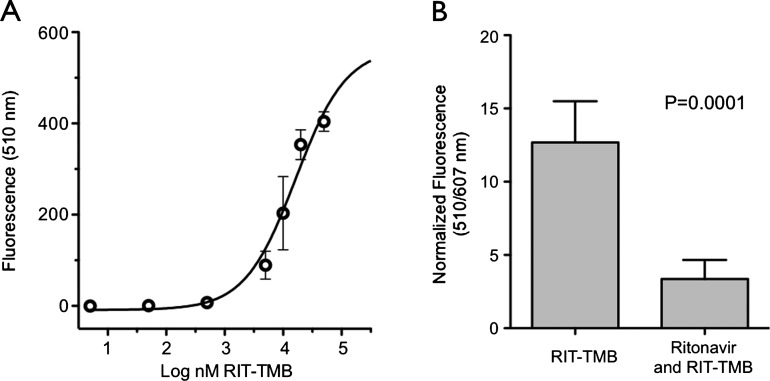
We initially examined the native protein and mRNA expression levels of a series of mouse and human pancreatic cancer cell lines that showed MIA PaCa-2 (PaCa-2) cells as having moderate amounts of protein (Figure W5). To visualize CTSE in co-localization studies, we generated PaCa-2 cells that express a CTSE/mCherry fusion protein. Experiments were then conducted to determine cell permeability of the probe and the minimal concentration required for imaging (Figure 2A). PaCa-2 CTSE-mCherry cells were plated and treated with serial dilutions of RIT-TMB. Following 15-minute incubation at 37°C and washes, we found that probe uptake was rapid (completion within 5–10 minutes). Preincubation of PaCa-2 CTSE-mCherry cells with RIT resulted in a substantial reduction in cellular uptake of RIT-TMB (73.5% reduction in signal; Figure 2B), while incubation with TMB not conjugated to the truncated RIT resulted in no cellular uptake (Figure W4). Probe uptake in treated and untreated wells was normalized against the fluorescence signal from the PaCa-2 CTSE-mCherry signal.
Figure 2.
PDAC cell line characterization. (A) Dose-response curve of RIT-TMB in PaCa-2 CTSE-mCherry cells. (B) Competitive inhibition of RIT-TMB with RIT in PaCa-2 CTSE-mCherry cells.
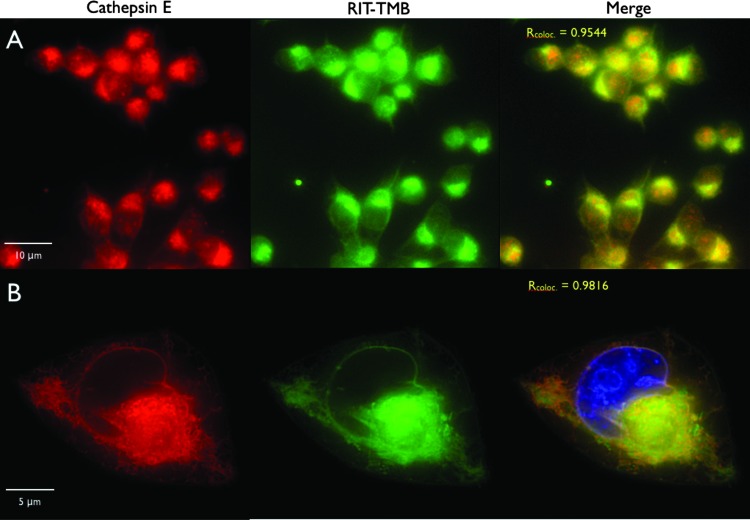
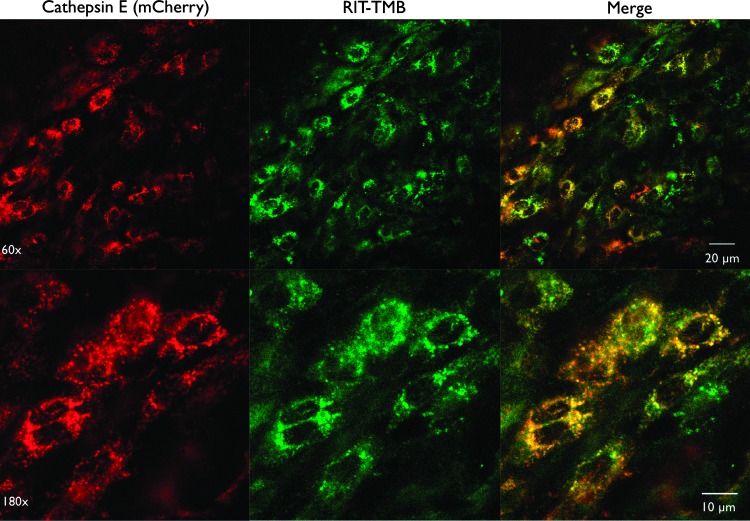
Co-localization of RIT-TMB with CTSE was tested in various PDAC cell lines using an anti-CTSE Cy5 monoclonal antibody. RIT-TMB demonstrated exceptional co-localization with anti-CTSE mAb and mCherry fluorescence in native and transfected PaCa2 lines (Pearson coefficients of 0.9544 and 0.9816, respectively; Figure 3).
Figure 3.
In vitro live cell microscopy imaging of RIT-TMB in PaCa-2 and PaCa-2 CTSE-mCherry. (A) PaCa-2 cell line: anti-CTSE Cy5 mAb (red), RIT-TMB (green). (B) Single PaCa-2 CTSE-mCherry cell: CTSE-mCherry (red), RIT-TMB (green), Hoechst nuclear stain (blue).
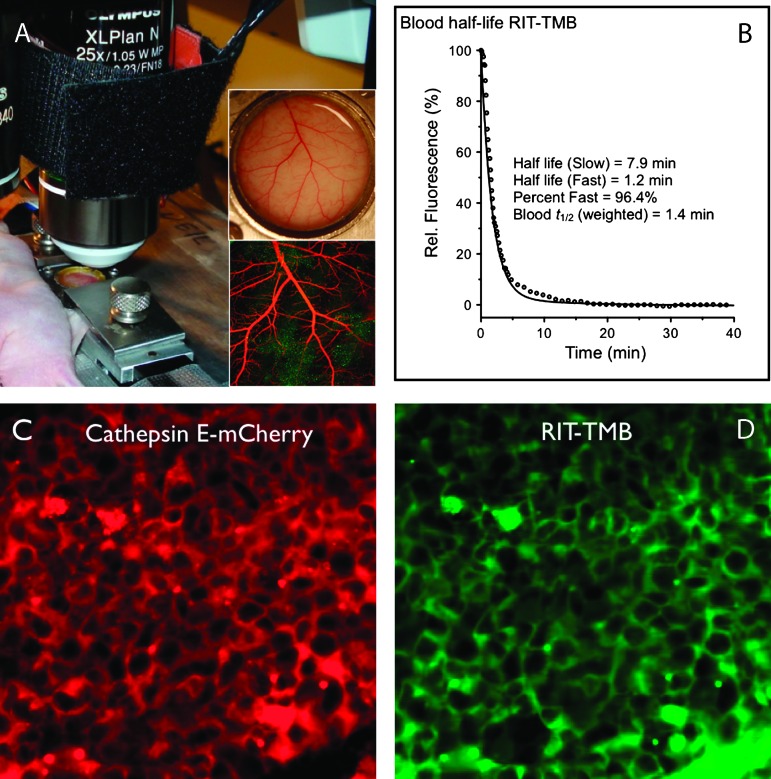
The blood half-life was determined by serial imaging and fitted to a two-compartment decay model where the weighted half-life was found to be 1.4 minutes (Figure 5B). We tested RIT-TMB in two in vivo models of PDAC to evaluate its performance. The first in vivo PDAC model, a dorsal window chamber model [23], was prepared with the PaCa-2 CTSE-mCherry cell line. After allowing a 7-day cell expansion period, time-lapse imaging experiments of RIT-TMB were performed for 2 hours post-injection to monitor accumulation of the probe in tumor cells. Figure 4 shows representative examples that confirm in vivo cellular uptake of RIT-TMB with good co-localization.
Figure 5.
In vivo PDAC orthotopic intravital microscopy. (A) Setup of intravital window chamber imaging. (B) Blood half-life as measured by serial fluorescence imaging. (C) Representative section showing CTSE-mCherry expressing PDAC cells. (D) Identical section as shown in C, following intravenous administration of RIT-TMB. Some of the agent can still be seen in large vessels (bottom right). Note the good co-localization with CTSE-mCherry.
Figure 4.
In vivo dorsal window chamber intravital microscopy. In vivo imaging, 1 hour post injection, of RIT-TMB, PaCa-2 CTSE-mCherry cells in dorsal window chamber.
RIT-TMB was also tested in a second in vivo model, an orthotopic injection of PaCa-2 CTSE-mCherry cells into the pancreas. PaCa-2 CTSE-mCherry cells were injected into the pancreas and expanded for 7 days. On the day of imaging, mice were anesthetized and the pancreas subsequently exposed. Before injection, no signal was observed at 488-nm excitation and 522-nm emission. After intravenous RITTMB injection, accumulation of RIT-TMB within the tumor was observed by monitoring 488/522 nm (Figure 5, C and D) with good co-localization with CTSE-mCherry signal of the PaCa-2-transfected cancer cells.
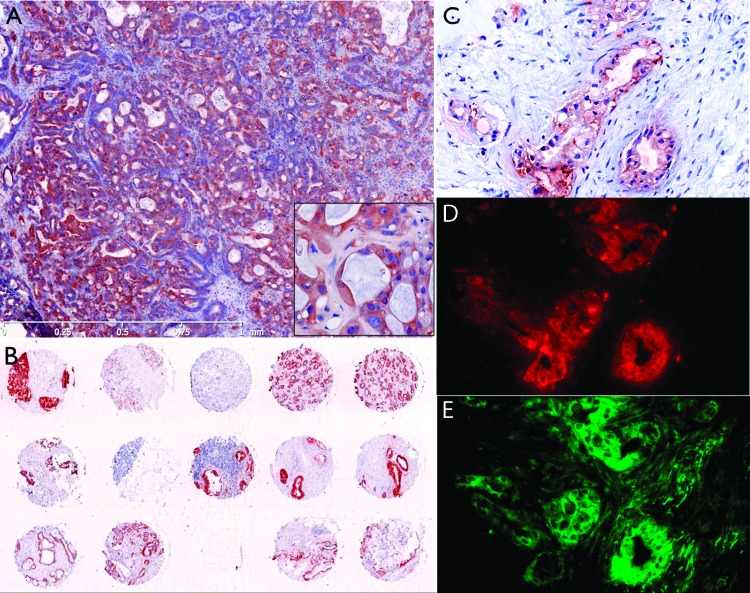
We finally tested RIT-TMB by immunofluorescence in murine and human biopsy specimens. Tumor tissue sections excised from 10-week-old KRAS p53-/- PDAC mice demonstrated good co-localization between double staining with anti-CTSE immunofluorescence and RIT-TMB (Figure W2) with particularly high accumulation within the pancreatic ducts. Human immunohistologic staining demonstrated consistent positive CTSE staining in 13 of 14 microarray samples analyzed (Figure 6B). Finally, RIT-TMB demonstrated good colocalization with the anti-human CTSE antibody (Figure 6, C–E) when tested in fresh human biopsy samples.
Figure 6.
Histology of human biopsy samples. (A) Human tissue tumor microarray. (B) Human PDAC metathesis in the liver (fine needle aspirate). (C) Anti-CTSE immunohistochemistry (adjacent slice as D and E). (D) Anti-CTSE immunofluorescence. (E) RIT-TMB staining, same slice as D (20 µM RIT-TMB for 1 hour).
Discussion
There are currently no clinically approved real-time imaging methods that can effectively detect invasive pancreatic cancer cells or discriminate cancer cells from adjacent noncancerous cells during surgery. While surgical removal of localized PDAC remains the clinical gold standard, teasing out what is cancer and what is surrounding inflammation remains highly challenging due to diffuse tumor infiltration, microscopic disease, and complex anatomy. The availability of an intraoperative imaging agent with high affinity for PDAC at the whole body and cellular levels would be an invaluable addition to our current surgical and research armamentarium.
Here, we show the synthesis and characterization of a non-peptide small molecule dual function inhibitor and imaging agent of CTSE based on an FDA-approved protease inhibitor. This was achieved in a single synthetic step with high chemical purity from commercial starting materials. Chemical modification of the parent compound, RIT, was shown to result in a CTSE affinity in the tens of nanomolar range. We demonstrated excellent in vitro imaging of RIT-TMB with a transfected mCherry-CTSE protein within a PDAC cell line (PaCa-2). No signal was observed for the free fluorochrome in the same cell line thus supporting the targeted nature of the probe. More significantly, rapid co-localization was observed in two in vivo models of PDAC.
In addition to PDAC, CTSE overexpression has been described in gastric carcinomas [11], cervical adenocarcinoma [24], and adenocarcinoma of the lung [25]. Molecularly targeted imaging agents such as RIT-TMB could be used to explore the biology of CTSE in these cancers as well as monitor response to targeted therapies. Similar to other targeted low-affinity reagents with pancreatic cell specificity [26], it is expected that these approaches could be useful not only during surgery but also in endoscopy, interventional procedures, and whole-body imaging.
Supplementary Material
Acknowledgments
The authors thank Joshua Dunham and Rainer Kohler for intravital microscopy, Peter Waterman, Alex Zaltsman, and Claudio Vinegoni for fluorescence imaging, Yoshiko Iwamoto for histology, Melina Pectasides for obtaining clinical samples, Matthew Sebas for animal surgery, and Jessica Lacy for assisting in cell biology.
Abbreviations
- PDAC
pancreatic ductal adenocarcinoma
- CTSE
cathepsin E
- SPR
surface plasmon resonance
- RIT
ritonavir
- RIT-TMB
ritonavir tetramethyl-BODIPY
Footnotes
This work was supported in part by a grant from the Lustgarten Foundation for Pancreatic Cancer Research and by grants from the National Institute of Health (2P01CA117969, 2P50CA086355, and PO1-AI54904).
This article refers to supplementary materials, which are designated by Figures W1 to W5 and are available online at www.neoplasia.com.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qu W, Oya S, Lieberman BP, Ploessl K, Wang L, Wise DR, Divgi CR, Chodosh LP, Thompson CB, Kung HF. Preparation and characterization of L-[5-11C]-glutamine for metabolic imaging of tumors. J Nucl Med. 2012;53:98–105. doi: 10.2967/jnumed.111.093831. [DOI] [PubMed] [Google Scholar]
- 3.Cruz-Monserrate Z, Abd-Elgaliel WR, Grote T, Deng D, Ji B, Arumugam T, Wang H, Tung CH, Logsdon CD. Detection of pancreatic cancer tumours and precursor lesions by cathepsin E activity in mouse models. Gut. 2012;61:1315–1322. doi: 10.1136/gutjnl-2011-300544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abd-Elgaliel WR, Cruz-Monserrate Z, Logsdon CD, Tung CH. Molecular imaging of cathepsin E-positive tumors in mice using a novel protease-activatable fluorescent probe. Mol Biosyst. 2011;7:3207–3213. doi: 10.1039/c1mb05215b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abd-Elgaliel WR, Tung CH. Selective detection of cathepsin E proteolytic activity. Biochim Biophys Acta. 2010;1800:1002–1008. doi: 10.1016/j.bbagen.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eser S, Messer M, Eser P, von Werder A, Seidler B, Bajbouj M, Vogelmann R, Meining A, von Burstin J, Algul H, et al. In vivo diagnosis of murine pancreatic intraepithelial neoplasia and early-stage pancreatic cancer by molecular imaging. Proc Natl Acad Sci USA. 2011;108:9945–9950. doi: 10.1073/pnas.1100890108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sessa F, Bonato M, Frigerio B, Capella C, Solcia E, Prat M, Bara J, Samloff IM. Ductal cancers of the pancreas frequently express markers of gastrointestinal epithelial cells. Gastroenterology. 1990;98:1655–1665. doi: 10.1016/0016-5085(90)91104-e. [DOI] [PubMed] [Google Scholar]
- 8.Uno K, Azuma T, Nakajima M, Yasuda K, Hayakumo T, Mukai H, Sakai T, Kawai K. Clinical significance of cathepsin E in pancreatic juice in the diagnosis of pancreatic ductal adenocarcinoma. J Gastroenterol Hepatol. 2000;15:1333–1338. [PubMed] [Google Scholar]
- 9.Azuma T, Yamada M, Murakita H, Nishikawa Y, Kohli Y, Yamamoto K, Hori H. Cathepsin E expressed in pancreatic cancer. Adv Exp Med Biol. 1995;362:363–366. doi: 10.1007/978-1-4615-1871-6_47. [DOI] [PubMed] [Google Scholar]
- 10.Azuma T, Hirai M, Ito S, Yamamoto K, Taggart RT, Matsuba T, Yasukawa K, Uno K, Hayakumo T, Nakajima M. Expression of cathepsin E in pancreas: a possible tumor marker for pancreas, a preliminary report. Int J Cancer. 1996;67:492–497. doi: 10.1002/(SICI)1097-0215(19960807)67:4<492::AID-IJC5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 11.Matsuo K, Kobayashi I, Tsukuba T, Kiyoshima T, Ishibashi Y, Miyoshi A, Yamamoto K, Sakai H. Immunohistochemical localization of cathepsins D and E in human gastric cancer: a possible correlation with local invasive and metastatic activities of carcinoma cells. Hum Pathol. 1996;27:184–190. doi: 10.1016/s0046-8177(96)90373-1. [DOI] [PubMed] [Google Scholar]
- 12.Weissleder R, Tung CH, Mahmood U, Bogdanov AJ. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 13.Tung CH, Mahmood U, Bredow S, Weissleder R. In vivo imaging of proteolytic enzyme activity using a novel molecular reporter. Cancer Res. 2000;60:4953–4958. [PubMed] [Google Scholar]
- 14.Kirsch DG, Dinulescu DM, Miller JB, Grimm J, Santiago PM, Young NP, Nielsen GP, Quade BJ, Chaber CJ, Schultz CP, et al. A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat Med. 2007;13:992–997. doi: 10.1038/nm1602. [DOI] [PubMed] [Google Scholar]
- 15.Aikawa E, Aikawa M, Libby P, Figueiredo JL, Rusanescu G, Iwamoto Y, Fukuda D, Kohler RH, Shi GP, Jaffer FA, et al. Arterial and aortic valve calcification abolished by elastolytic cathepsin S deficiency in chronic renal disease. Circulation. 2009;119:1785–1794. doi: 10.1161/CIRCULATIONAHA.108.827972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffer FA, Kim DE, Quinti L, Tung CH, Aikawa E, Pande AN, Kohler RH, Shi GP, Libby P, Weissleder R. Optical visualization of cathepsin K activity in atherosclerosis with a novel, protease-activatable fluorescence sensor. Circulation. 2007;115:2292–2298. doi: 10.1161/CIRCULATIONAHA.106.660340. [DOI] [PubMed] [Google Scholar]
- 17.Kempf DJ, Marsh KC, Denissen JF, McDonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP, et al. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwan JC, Eksioglu EA, Liu C, Paul VJ, Luesch H. Grassystatins A-C from marine cyanobacteria, potent cathepsin E inhibitors that reduce antigen presentation. J Med Chem. 2009;52:5732–5747. doi: 10.1021/jm9009394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 20.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee S, Vinegoni C, Feruglio PF, Weissleder R. Improved intravital microscopy via synchronization of respiration and holder stabilization. J Biomed Opt. 2012;17:96018–1. doi: 10.1117/1.JBO.17.9.096018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orth JD, Kohler RH, Foijer F, Sorger PK, Weissleder R, Mitchison TJ. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011;71:4608–4616. doi: 10.1158/0008-5472.CAN-11-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tenti P, Romagnoli S, Silini E, Zappatore R, Giunta P, Stella G, Carnevali L. Cervical adenocarcinomas express markers common to gastric, intestinal, and pancreatobiliary epithelial cells. Pathol Res Pract. 1994;190:342–349. doi: 10.1016/S0344-0338(11)80406-1. [DOI] [PubMed] [Google Scholar]
- 25.Ullmann R, Morbini P, Halbwedl I, Bongiovanni M, Gogg-Kammerer M, Papotti M, Gabor S, Renner H, Popper HH. Protein expression profiles in adenocarcinomas and squamous cell carcinomas of the lung generated using tissue microarrays. J Pathol. 2004;203:798–807. doi: 10.1002/path.1584. [DOI] [PubMed] [Google Scholar]
- 26.Reiner T, Thurber G, Gaglia J, Vinegoni C, Liew CW, Upadhyay R, Kohler RH, Li L, Kulkarni RN, Benoist C, et al. Accurate measurement of pancreatic islet β-cell mass using a second-generation fluorescent exendin-4 analog. Proc Natl Acad Sci USA. 2011;108:12815–12820. doi: 10.1073/pnas.1109859108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.