Abstract
Over the past decade, genetic lesions that cause ribosome dysfunction have been identified in both congenital and acquired human disorders. These discoveries have established a new category of disorders, known as ribosomopathies, in which the primary pathophysiology is related to impaired ribosome function. The protoptypical disorders are Diamond–Blackfan anemia, a congenital bone marrow failure syndrome, and the 5q- syndrome, a subtype of myelodysplastic syndrome. In both of these disorders, impaired ribosome function causes a severe macrocytic anemia. In this review, we will discuss the evidence that defects in ribosomal biogenesis cause the hematologic phenotype of Diamond–Blackfan anemia and the 5q- syndrome. We will also explore the potential mechanisms by which a ribosomal defect, which would be expected to have widespread consequences, may lead to specific defects in erythropoiesis.
Keywords: Myelodysplastic syndrome, Diamond–Blackfan anemia, p53
1 Introduction
More than a decade ago, researchers discovered that a rare congenital bone marrow failure syndrome, Diamond–Blackfan anemia (DBA), is caused by mutations in a ribosomal protein RPS19 [1]. The finding that mutation of a ribosomal protein gene can cause DBA was truly unexpected given that the majority of clinical symptoms are related to erythropoiesis, and defects in ribosomal function might be expected to have global effects. Nevertheless, since that initial discovery, mutations in nine ribosomal proteins have been identified in patients with DBA, accounting for 50% of cases [2]. Mutations have not been discovered in any other type of gene in DBA patients. These genetic data therefore indicate a causal link between defects in ribosome biology and impaired red blood cell production.
The connection between ribosomal abnormalities and defects in erythropoiesis was reinforced by the identification of RPS14 as a candidate gene for the profound macrocytic anemia in the 5q- syndrome [3], a subtype of myelodysplastic syndrome. Heterozygous deletions of chromosome 5q in MDS are large, and haploinsufficiency for multiple genes likely contributes to the phenotype of the 5q- syndrome [4]. Nevertheless, in vitro and in vivo studies indicate that the severe macrocytic anemia, the aspect of the 5q- syndrome phenotype most analogous to Diamond–Blackfan anemia, is caused by RPS14 haploinsufficiency [3, 5].
In this review, we will summarize the evidence that ribosomal defects cause the hematologic phenotype of Diamond–Blackfan anemia and the 5q- syndrome, and discuss the possible mechanisms of how ribosomal dysfunction leads to disorders of erythropoiesis, including the role of the p53 pathway.
2 Genetics of Diamond–Blackfan anemia
DBA is a congenital bone marrow failure syndrome characterized by anemia, macrocytosis, reticulocytopenia, and a selective decrease of erythroid progenitor cells on bone marrow evaluation [2]. Other notable red blood cell features include an elevated erythrocyte adenosine deaminase activity and increased levels of fetal hemoglobin. The disorder is also commonly associated with a range of clinical abnormalities ranging from limb malformations, craniofacial deformities, and cardiac defects in up to 62% of patients [6]. Our understanding of the pathophysiology of the disease has improved significantly since its original report in 1936 by Josephs [7] and further description in 1938 by Blackfan and Diamond [8]. Draptchinskaia et al. [1] identified the breakpoint of a congenital balanced translocation in a patient with DBA that disrupted the gene encoding ribosomal protein S19 (RPS19). Since that report, mutations or deletions have been reported in the RPS7, RPS10, RPS17, RPS24, RPS26, RPL5, RPL11, and RPL35A genes [9–14]. Patients with DBA have also been found to have polymorphisms in genes encoding RPS15, RPS27A, and RPL30 that may or may not be pathogenic [12, 14].
Both in vitro and in vivo models of ribosomal haploinsufficiency have been developed to study DBA. In vitro, using RNA interference, knockdown of RPS19 in normal human hematopoietic progenitor cells leads to a profound defect in the proliferation and differentiation of erythroid cells [15, 16]. This hematopoietic defect can be rescued by the forced overexpression of RPS19. Similarly, expression of a RPS19 cDNA in bone marrow cells from patients with DBA significantly improved the defects in erythropoiesis [17]. Similar experiments using zebrafish models have also been used to model ribosomal dysfunction. Using morpholinos, two groups have found that zebrafish with RPS19 deficiency develop hematologic and developmental abnormalities that resemble DBA [18, 19]. A limitation to note with RNA interference and morpholinos is that the level of knockdown of ribosomal proteins is unlikely to be exactly 50% and it is unknown whether the phenotype of DBA and del(5q) MDS are affected by the precise level of ribosomal activity.
Murine models have the potential to more accurately model ribosomal haploinsufficiency and several models have now been developed. An initial Rps19 knockout mouse model was homozygous lethal and did not have a DBA phenotype in the heterozygous state; but the heterozygous mice had normalized Rps19 levels [20]. Mice with heterozygous missense mutations in Rps19 and Rps20 have only a mild macrocytic anemia, but these mutations may cause hypomorphic alleles, rather than complete loss of function of the allele [21]. A transgenic mouse model expressing a RPS19 mutation acting via a dominant negative mechanism also demonstrated a mild macrocytic anemia [22]. Finally, chronic RPS19 deficiency caused by RNA interference has also been shown to lead to bone marrow failure in a murine model [23]. A potentially very useful model of ribosomal haploinsufficiency is a murine model with conditional deletion of RPS6. These mice have a significant macrocytic anemia with reticulocytopenia as well as an elevated adenosine deaminase [24].
3 Genetics of del(5q) MDS
The 5q- syndrome, first described in 1974 [25] is an acquired bone marrow failure disorder caused by an interstitial deletion of the long arm of chromosome 5 and is categorized by the World Health Organization [26] as an independent subtype of myelodysplastic syndrome (MDS). The phenotype primarily consists of a profound macrocytic anemia as well as thrombocytosis with hypolobated micromegakaryocytes. There is also a female predominance and patients have a lower rate of progression to acute myeloid leukemia. Finally, patients with del(5q) MDS show an exquisite sensitivity to the immunomodulatory agent lenalidomide [27]. RPS14 was identified as a 5q- syndrome gene using an RNA interference screen of every gene within the commonly deleted area [3]. Patients with the 5q- syndrome have been confirmed to have haploinsufficient expression of RPS14 [3, 28].
Similar to the in vitro work with RPS19, decreased expression of RPS14 causes impaired erythropoiesis whereas overexpression of RPS14 in patient samples rescues erythropoiesis [3]. A murine model has now been developed with coordinate deletion of loci syntenic with the common deleted region of the 5q- syndrome, including haploinsufficiency for Rps14. These mice have a profound macrocytic anemia, comparable to the clinical phenotype in humans [5]. This data suggests the molecular pathogenesis of the 5q- syndrome is similar to Diamond–Blackfan anemia and is due to a functionally similar defect in ribosome biogenesis.
4 Ribosome dysfunction in DBA and del(5q) MDS
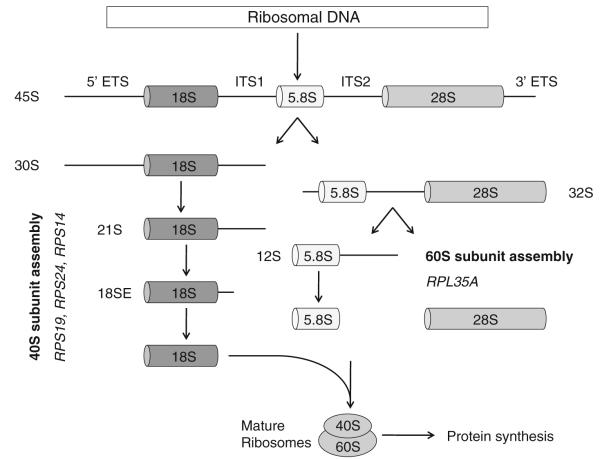
The eukaryotic ribosome is composed of 40S and 60S subunits which associate to form the translationally active 80S ribosome. The 40S subunit contains 18S rRNA and the 60S subunit contains 28S, 5.8S, and 5S rRNAs. In eukaryotes, assembly of ribosomal RNAs and ribosomal proteins, along with associated proteins and small nucleolar RNAs, occurs in the nucleolus, leading to the production of pre-60S and pre-40S pre-ribosomal particles. These particles are exported to the cytoplasm where the final steps in assembly and maturation of ribosomes occur [29]. A simplified overview of this process is outlined in Fig. 1. Ribosomal proteins assemble onto precursor rRNA transcripts and facilitate a series of endonucleolytic and exonucleolytic cleavages required for generation of the mature rRNAs.
Fig. 1.
Simplified overview of eukaryotic ribosome biogenesis. Adapted from Narla and Ebert [2], © the American Society of Hematology
Haploinsufficiencies for distinct ribosomal proteins have been linked to defects at distinct steps in pre-rRNA processing. Mutations in RPS19 and in RPS24, the first two mutations identified in DBA, impair pre-rRNA processing of the 18S rRNA which leads to decreased production of the 40S ribosomal subunit [30–33] RPS14 deficiency causes a block in the processing of the 18S rRNA and the formation of the 40S ribosomal subunit [3]. There is evidence that depletion of any of the RPS proteins causes a reduction in the amount of free 40S subunits and a significant reduction in the amount of mature 80S ribosomes (with the exception of RPS25 which has not been shown to be mutated in DBA) [34]. In contrast, when RPL proteins, including RPL35A which has also been described in DBA, are depleted, the amount of the 60S subunit is reduced, as is the level of mature 80S ribosomes [34].
Mutations in a variety of ribosomal proteins ultimately lead to a decrease in the number of mature ribosomes, which would be expected to have a number of consequences for a cell. This includes defects in the translation of specific proteins which might play a critical role in erythropoiesis as discussed later in this paper. Additionally, several ribosomal proteins have extraribosomal functions, including replication and DNA repair, so mutations in ribosomal proteins may have effects that are independent of the protein translation machinery [35, 36].
5 Activation of p53 by ribosome dysfunction
Mutations in ribosomal protein genes would be expected to have widespread and diverse effects throughout an organism. However, defects in ribosome function appear to cause relatively tissue-specific effects in DBA, with a particularly dramatic erythroid phenotype. The fundamental question of why there are such specific phenotypes remains the source of active investigation. As discussed earlier, multiple in vitro and in vivo models of ribosomal haploinsufficiency have been developed which have helped to confirm the role of ribosomal haploinsufficiency in disorders of erythropoiesis. Ongoing work with these models has increasingly focused on the role of p53 in this process.
Among its myriad of roles, p53 monitors ribosome function [37] and plays a fundamental role in the surveillance of protein translation [38]. The murine double minute 2 protein (MDM2, or HDM2 in humans) functions as a link between ribosome biogenesis and the p53 pathway. MDM2 is a central regulator of p53, acting as a ubiquitin ligase that leads to the degradation of p53 [39]. MDM2 has also been shown to bind specifically to several free ribosomal proteins including RPL5, RPL23, RPL11, RPS7 and RPL26 [40–46]. In an elegant series of experiments, nucleolar disruption experimentally induced by treatment with actinomycin D, was shown to lead to the release of RPL11 and other ribosomal proteins into the nucleoplasm, the binding of RPL11 to MDM2, the inhibition of MDM2 activity, and the consequent accumulation of p53 [43, 47]. A schematic view of this pathway is shown in Fig. 2.
Fig. 2.
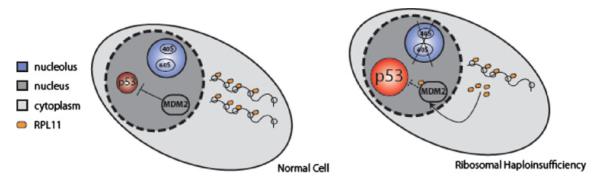
Overview of the potential mechanism of the cellular consequences of ribosomal haploinsufficiency via p53 modulation. On the left, a normal cell is portrayed with unperturbed ribosome biogenesis and steady state levels of p53. On the right, a cell is depicted with ribosomal haploinsufficiency which leads to up-regulation of RPL11, which binds MDM2 resulting in p53 activation. This leads to apoptosis and cell cycle arrest. Adapted from Narla and Ebert [2], © the American Society of Hematology
Morpholinos targeting RPS19 in zebrafish cause an accumulation of p53 and a block in erythropoiesis that is reversed in the absence of p53 [18]. In a murine model with a heterozygous missense mutation in Rps19, induction of p53 and p53 target genes was identified in the hyperpigmented foot pads of the mice [21]. When the RPS19 mutant mouse line had one allele of p53 genetically inactivated, there was an increase in RBC count and decrease in MCV. Homozygous inactivation of p53 in RPS19 mutant mice fully corrected the hematologic phenotype [21]. Another murine model involves the conditional deletion of a set of genes on 5q including RPS14. These mice develop a severe macrocytic anemia [5]. When these mice were crossed with p53 null mice, there was a complete rescue of the erythroid phenotype [5].
A murine model of Treacher-Collins syndrome offers tantalizing clues to the potential management of patients via modulation of the p53 pathway [48]. Treacher-Collins syndrome is an autosomal dominant condition which includes characteristic craniofacial changes [49] that arise from symmetrically and bilaterally diminished growth of the structures derived from the first and second pharyngeal arch, groove, and pouch [50]. TCOF1 was identified as the gene responsible for TCS; TCOF1 encodes a protein known as Treacle [51]. Mice haploinsufficient for Tcof1 exhibit diminished production of ribosomes and this deficiency correlates with decreased proliferation of both neural ectoderm and neural crest cells [52]. Interestingly, a study by Jones et al. [48] showed that chemical and genetic inhibition of p53 activity in these mice can prevent the craniofacial abnormalities.
6 Tissue-specific effects of ribosomal dysfunction
Erythroid progenitor cells are notable for their incredibly rapid rate of proliferation, doubling every 12–24 h [53]. This extraordinary rate of proliferation requires a very high level of ribosome biogenesis and ribosomal activity. Indeed, rapid proliferation may be important for the sensitivity of cells with ribosomal haploinsufficiency to p53 activation as stimulated but not resting T cells activate p53 in response to haploinsufficient RPS6 protein [54].
Recent work in our lab has focused on understanding the molecular basis for the lineage-restricted effects of ribosome dysfunction [55]. We found that decreased expression of the RPS14 or RPS19 genes causes selective activation of the p53 pathway in erythroid progenitor cells compared to cells from other hematopoietic lineages. This activation of the p53 pathway resulted in erythroid-specific cell cycle arrest and apoptosis. Consistent with the work in the RPS6 model, decreased expression of RPS14 increased the levels of RPL11 which binds to HDM2, providing a mechanism for p53 activation. Furthermore, we found accumulation of nuclear p53 in the erythroid progenitor cells in bone marrow specimens from patients with DBA and del(5q) MDS [55].
7 Conclusion
Despite the universal requirement for ribosomes in all human cells, erythropoiesis is particularly sensitive to ribosome dysfunction. Heterozygous loss-of-function mutations for many different ribosomal protein genes causes DBA, and heterozygous loss of RPS14 is linked to the macrocytic anemia of the 5q- syndrome. Activation of p53 activity appears be central to the hematopoietic phenotype of ribosomal dysfunction. Efforts are currently underway to investigate whether pharmacologic modulation of p53 activity or ribosome function may have therapeutic utility in treating anemia in patients with disorders of ribosome biogenesis.
Contributor Information
Anupama Narla, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA; Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Karp Research Building, CHRB 05.211, 1 Blackfan Circle, Boston, MA 02115, USA; Department of Medicine, Children’s Hospital Boston, Boston, MA 02115, USA.
Slater N. Hurst, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Karp Research Building, CHRB 05.211, 1 Blackfan Circle, Boston, MA 02115, USA
Benjamin L. Ebert, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA; Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Karp Research Building, CHRB 05.211, 1 Blackfan Circle, Boston, MA 02115, USA; Harvard Stem Cell Institute, Cambridge, MA 02138, USA
References
- 1.Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond–Blackfan anaemia. Nat Genet. 1999;21(2):169–75. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 2.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebert BL, Pretz J, Bosco J, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–9. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ebert BL. Deletion 5q in myelodysplastic syndrome: a paradigm for the study of hemizygous deletions in cancer. Leukemia. 2009;23(7):1252–6. doi: 10.1038/leu.2009.53. [DOI] [PubMed] [Google Scholar]
- 5.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipton JM, Ellis SR. Diamond–Blackfan anemia: diagnosis, treatment, and molecular pathogenesis. Hematol Oncol Clin North Am. 2009;23(2):261–82. doi: 10.1016/j.hoc.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Josephs HW. Anaemia of infancy and early childhood. Medicine. 1936;15:307. [Google Scholar]
- 8.Diamond LK, Blackfan KD. Hypoplastic anemia. Am J Dis Child. 1938;56:464. [Google Scholar]
- 9.Gazda HT, Grabowska A, Merida-Long LB, et al. Ribosomal protein S24 gene is mutated in Diamond–Blackfan anemia. Am J Hum Genet. 2006;79(6):1110–8. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond– Blackfan anemia. Hum Mutat. 2007;28(12):1178–82. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 11.Farrar JE, Nater M, Caywood E, et al. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond–Blackfan anemia. Blood. 2008;112(5):1582–92. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazda HT, Sheen MR, Vlachos A, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond–Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769–80. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cmejla R, Cmejlova J, Handrkova H, et al. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond–Blackfan anemia. Hum Mutat. 2009;30(3):321–7. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- 14.Doherty L, Sheen MR, Vlachos A, et al. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond–Blackfan anemia. Am J Hum Genet. 2010;86(2):222–8. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flygare J, Kiefer T, Miyake K, et al. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond–Blackfan anemia. Blood. 2005;105(12):4627–34. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 16.Ebert BL, Lee MM, Pretz JL, et al. An RNA interference model of RPS19 deficiency in Diamond–Blackfan anemia recapitulates defective hematopoiesis and rescue by dexamethasone: identification of dexamethasone-responsive genes by microarray. Blood. 2005;105(12):4620–6. doi: 10.1182/blood-2004-08-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamaguchi I, Ooka A, Brun A, Richter J, Dahl N, Karlsson S. Gene transfer improves erythroid development in ribosomal protein S19-deficient Diamond–Blackfan anemia. Blood. 2002;100(8):2724–31. doi: 10.1182/blood.V100.8.2724. [DOI] [PubMed] [Google Scholar]
- 18.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–37. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 19.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond–Blackfan anemia. Hum Mol Genet. 2008;17(20):3204–11. doi: 10.1093/hmg/ddn216. [DOI] [PubMed] [Google Scholar]
- 20.Matsson H, Davey EJ, Frojmark AS, et al. Erythropoiesis in the Rps19 disrupted mouse: analysis of erythropoietin response and biochemical markers for Diamond–Blackfan anemia. Blood Cells Mol Dis. 2006;36(2):259–64. doi: 10.1016/j.bcmd.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.McGowan KA, Li JZ, Park CY, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–70. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devlin EE, Dacosta L, Mohandas N, Elliott G, Bodine DM. A transgenic mouse model demonstrates a dominant negative effect of a point mutation in the RPS19 gene associated with Diamond–Blackfan anemia. Blood. 2010;116(15):2826–35. doi: 10.1182/blood-2010-03-275776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaako PFJ, Olsson K, Quere R, Larsson J, Bryder D, Karlsson S. Chronic RPS19 deficiency leads to bone marrow failure in a mouse model for Diamond–Blackfan anemia. Oral session: bone marrow failure: genetics and pathogenetics; Abstract #193, 52nd ASH Annual Meeting and Exposition; Orlando, FL. 4–7 December, 2010. [Google Scholar]
- 24.Park CMK, Glader B, Barsh G, Weissman I. Haploinsufficiency of ribosomal protein S6 in mice mimics bone marrow failure syndromes in humans. Oral session: bone marrow failure: genetics and pathogenetics; Abstract #194. 52nd ASH Annual Meeting and Exposition; Orlando, FL. 4–7 December, 2010. [Google Scholar]
- 25.Van den Berghe H, Cassiman JJ, David G, Fryns JP, Michaux JL, Sokal G. Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature. 1974;251(5474):437–8. doi: 10.1038/251437a0. [DOI] [PubMed] [Google Scholar]
- 26.Vardiman JW, Harris NL, Brunning RD. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood. 2002;100(7):2292–302. doi: 10.1182/blood-2002-04-1199. [DOI] [PubMed] [Google Scholar]
- 27.List A, Dewald G, Bennett J, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. N Engl J Med. 2006;355(14):1456–65. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 28.Pellagatti A, Hellstrom-Lindberg E, Giagounidis A, et al. Haploinsufficiency of RPS14 in 5q- syndrome is associated with deregulation of ribosomal- and translation-related genes. Br J Haematol. 2008;142(1):57–64. doi: 10.1111/j.1365-2141.2008.07178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henras AK, Soudet J, Gerus M, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65(15):2334–59. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choesmel V, Bacqueville D, Rouquette J, et al. Impaired ribosome biogenesis in Diamond–Blackfan anemia. Blood. 2007;109(3):1275–83. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flygare J, Aspesi A, Bailey JC, et al. Human RPS19, the gene mutated in Diamond–Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109(3):980–6. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Idol RA, Robledo S, Du HY, et al. Cells depleted for RPS19, a protein associated with Diamond–Blackfan anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39(1):35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Choesmel V, Fribourg S, Aguissa-Toure AH, et al. Mutation of ribosomal protein RPS24 in Diamond–Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17(9):1253–63. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 34.Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14(9):1918–29. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21(5):164–5. [PubMed] [Google Scholar]
- 36.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–77. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Constantinou C, Elia A, Clemens MJ. Activation of p53 stimulates proteasome-dependent truncation of eIF4E-binding protein 1 (4E-BP1) Biol Cell. 2008;100(5):279–89. doi: 10.1042/BC20070121. [DOI] [PubMed] [Google Scholar]
- 39.Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM. Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem. 2000;275(12):8945–51. doi: 10.1074/jbc.275.12.8945. [DOI] [PubMed] [Google Scholar]
- 40.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–82. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 41.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–68. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin A, Itahana K, O’Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24(17):7669–80. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–87. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Wolf GW, Bhat K, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–12. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D, Zhang Z, Li M, et al. Ribosomal protein S7 as a novel modulator of p53–MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–37. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 46.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32(2):180–9. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fumagalli S, Di Cara A, Neb-Gulati A, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11(4):501–8. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones NC, Lynn ML, Gaudenz K, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14(2):125–33. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treacher-Collins E. Case with symmetrical congenital notches in the outer part of each lower lid and defective development of the malar bones. Trans Opthalmol Soc UK. 1900;20:90. [Google Scholar]
- 50.Sakai D, Trainor PA. Treacher Collins syndrome: unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41(6):1229–32. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. The Treacher Collins Syndrome Collaborative Group. Nat Genet. 1996 Feb;12(2):130–6. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 52.Dixon J, Jones NC, Sandell LL, et al. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103(36):13403–8. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lajtha LG, Oliver R. A kinetic model of the erythron. Proc R Soc Med. 1961;54:369–71. [PubMed] [Google Scholar]
- 54.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19(24):3070–82. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dutt S, Narla A, Lin K, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2010 doi: 10.1182/blood-2010-07-295238. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]