Abstract
Background
Gastric cancer is the second leading cause of cancer mortality worldwide and projected to rise to tenth in all-cause mortality in the near term. Early detection requires improved sensitivity and specificity of endoscopic imaging with novel methods.
Objective
Evaluate the utility of activatable molecular probes for the detection of gastric cancer both in vivo and ex vivo in a preclinical model.
Methods
Smad4+/− mice, which develop spontaneous gastric neoplasia, were compared with normal wild type controls. Cathepsin-activatable and matrix metalloproteinase (MMP)-activatable molecular probes were injected 24 hours and 6 hours before imaging, respectively. In vivo imaging was performed using quantitative tomographic near infrared fluorescence (NIRF) imaging. For validation, ex vivo imaging and histologic examination were performed.
Results
Molecular imaging in vivo of Smad4+/− gastric cancer murine models revealed intense activation of both cathepsin B and MMP probes. Ex vivo imaging and histology confirmed that the detected neoplasms were adenocarcinomas and hyperplastic lesions.
Conclusions
This study provides proof of principle that the cathepsin and MMP-activatable molecular probes are activated in the Smad4+/− murine model of spontaneous gastric adenocarcinoma, and can be imaged by both in vivo and ex vivo NIRF methods. The cathepsin probe also detects hyperplastic lesions.
Keywords: near-infrared, molecular imaging, gastric neoplasia, probe, mouse model
INTRODUCTION
Gastric cancer is the second leading cause of cancer mortality, with an expected incidence of over one million worldwide in 2010 (1–3). Early sensitive and specific detection of gastric cancer is essential for treatment optimization. Traditional endoscopy for gastric cancer screening has several limitations, including the inherent invasiveness, potential complications, and resource utilization. In addition, standard endoscopy may miss early lesions, as well as yield false positive results. Yalamarthi et al (4) reported significant rates of missed diagnosis and endoscopist or pathologist errors in gastric cancer patients who had undergone endoscopy with 3 years before diagnosis. Magnification endoscopy, chromoendoscopy, and confocal endoscopy have shown promise to improve detection and potentially avoid conventional biopsy with histology (4). The challenge for the next generation of endoscopic imaging is to improve efficiency, and to increase sensitivity and specificity for the detection of gastrointestinal tumors (5–7).
Molecular imaging of biological targets, such as cell surface receptors, enzymes, or proteins, provide functional visualization which may help achieve the clinical goals of early detection and characterization (8–11). Among the optical imaging technologies, near infrared fluorescent (NIRF) imaging has demonstrated promise. Compared with optical imaging at other visible wavelengths, NIRF has the advantages of imaging deeper tissues and minimizing tissue auto-fluorescence, since the major light absorbers (hemoglobin, water, and lipids) have their lowest absorption coefficient in the near infrared spectral region of 600–900 nm (8–10). Activatable probes are substrates of enzymes which are highly expressed in a spectrum of pathological processes, and a variety of activatable agents have been recently developed. Cathepsin-activatable and matrix metalloproteinase (MMP)-activatable probes have been used to detect cancers (12–14) and inflammation (7, 15, 16) in mouse models. In our previous study, we demonstrated the ability of the cathepsin-activatable probe to detect precancerous colonic adenomas in murine models. Importantly, discrimination of colonic adenomas within a background of chronic inflammation in an ulcerative colitis mouse model was exhibited using this probe (17). We also performed NIRF imaging with a capsule endoscope with the goal of combining a non-invasive imaging technique with these activatable probes (7). Our aim in the present study was to examine whether cathepsin- or MMP-activatable molecular probes can successfully detect and image gastric neoplasia in a Smad4+/− model of spontaneous gastric adenocarcinoma.
MATERIALS AND METHODS
Animal models
The study was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of North Carolina at Chapel Hill. Specific pathogen-free (SPF) male heterozygous Smad4m4Mag (18, 19) mice (Samd4 +/−) on C57BL/6 background were provided by Dr. Yijing Chen (Department of Biological Sciences, Kent State University). These mice have targeted disruption of one Smad4 allele. The smad4 protein is a crucial component of signaling by transforming growth factor (TGF-β), which negatively regulates cell growth and promotes apoptosis of epithelial cells. Studies have demonstrated that loss of Smad4 expression promotes gastric carcinoma (20–22). Xu et al reported that disruption of one Smad4 allele initiates progressive gastric polyposis, dysplasia and adenocarcinoma in mice (23). Wild type C57BL/6 mice (n=3) were maintained in our SPF animal facility and used as controls. Water and food were provided ad libitum, until 4 days before imaging when Smad4 +/− and WT controls were fed a Nutren 1.0 fiber liquid diet (Nestle Nutrition, Minnetonka, MN) diluted in 1:1 dH2O. Liquid diet was administered to clear the bowel of solid fecal matter which may auto-fluoresce or interfere with disease detection. Our preliminary studies confirmed that 4 days on this liquid diet effectively reduced background fluorescence.
Molecular imaging methodology
The activatable molecular probes, ProSense® 680 (mainly targeting cathepsin B) and MMPSense® 750 FAST (primarily targeting MMP-9), were obtained from PerkinElmer (Waltham, MA). The probes were administered via retro orbital injection at the recommended doses by the vendor (ProSense® 680, 2nmol/150µL; MMPSense® 750 FAST, 2nmol/100 µL) in phosphate buffered saline. Per guidelines, in vivo imaging was performed 24 hours (ProSense® 680) or 6 hours (MMPSense® 750 FAST) after the probe injection.
In Vivo tissue imaging
The tomographic and planar imaging studies were performed using a fluorescence molecular tomography (FMT) system (FMT 2500™ LX, PerkinElmer, Waltham, MA). Mice were anaesthetized and imaged, as previously described (7). Tomographic imaging with the FMT system use a NIR laser diode to transilluminate, with signal detection via a CCD camera. Optical filters allowed excitation of the probes at their respective excitation wavelengths (ProSense: 680 ± 10nm; MMPSense: 750nm) and imaging of emitted fluorescence at their emission wavelengths (ProSense: 700 ± 10nm; MMPSense: 770nm). The entire imaging acquisition sequence took approximately 3–5 min per animal model. In addition, two dimensional fluorescence reflectance imaging (FRI) was also performed with the FMT system using the built-in LED front illuminators and a collection of single camera images.
Ex Vivo tissue imaging
Mice were euthanized immediately after live imaging in vivo. Each stomach was resected and then imaged ex vivo with FRI.
Histology
Following in vivo and ex vivo imaging, to confirm that the fluorescent signal originated from the neoplasms, the fresh stomach tissue was fixed in 10% zinc formalin and paraffin embedded. Regions of stomach tissue that yielded signals with activated probe in Smad4+/− mice and corresponding regions from WT controls were sectioned for hemotoxylin and eosin (H&E) staining. An experienced pathologist blinded to the sample source, categorized the tissue sections as normal, hyperplasia, dysplasia or adenocarcinoma.
Image and Statistical Analysis
For in vivo imaging data analysis, the fluorochrome quantification of total fluorescence and signal intensity within the gastric region of each mouse was determined using three dimensional (3D) region of interest (ROI) analysis. Briefly, images were displayed as rotatable reconstructed three-dimensional datasets, allowing views in transverse, sagittal and coronal planes. The target 3D region was defined by ROI placement in all three viewing planes to delineate appropriate regions of fluorescence within the imaging dataset. Image calibration was performed using a threshold applied to all animals equal to 30% of the mean fluorescence in the stomach ROI of normal control mice. This threshold setting minimizes the background fluorescence but allows optical detection of fluorescence signals in activated targets. Fluorochrome concentration in the target tissue was then automatically calculated from the reconstructed images (FMT TrueQuant® Imaging Software, ver. 2.0.0.19) and pre-acquired calibrations for the specific probe. Results are expressed in total fluorescence calculated as picomoles (specifically, pmol equivalents of activated probe per ROI based on comparisons with calibration standards).
In order to measure the signal intensity of fluorescence signal, potentially another measure of protease activation in dysplastic lesions, we divided total fluorescence (pmol equivalents of activated probe) by volume of the ROI used for 3D tomographic imaging (fluorescence/volume of ROI). For ex vivo 2D FRI data analysis, identical 2D ROIs were placed on appropriate regions of reflective fluorescence in both WT and Smad4+/− mice stomach tissue, with imaging calibration, as described above.
In vivo FMT data (total fluorescence or fluorescence intensity) and ex vivo FRI data (counts/energy) obtained on individual animals with each of the two probes tested were used to generate the mean of signals (± SE) in observed WT or Smad4+/− mice. Mean signal intensity (SI) was recorded. Data from the Smad4+/− mice and WT controls were compared by students t-test. The neoplastic lesion to background (normal mucosa) target contrast calculation (TBC) was determined according to published methods (24). The TBC, expressed as a percentage, represents the ratio of the difference in signal in a macroscopically evident lesion or region of activated probe and grossly normal mucosa divided by the signal in grossly normal mucosa. Regions of normal mucosa were obtained as size matched gastric mucosa from tissue adjacent to macroscopically evident tumors in Smad4 +/− mice or WT mice gastric tissue.
TBC (%) = ([SI dysplasia- SI mucosa]/ SI mucosa) X 100.
In addition, for in vivo FMT data, we compared signals (total fluorescence and fluorescence intensity) of the ProSense® 680 and MMPSense® 750 probes. For ex vivo data, we compared FRI and FMT results from ProSense® 680 and MMPSense® 750 FAST probe activation.
In the statistical analysis, data are expressed as mean ± standard error and compared for significance using the t-test. All statistical analyses were performed using STATISTICA software version 9 (Statsoft, Inc. Tulsa, OK). A P value of less than 0.05 was considered statistically significant.
RESULTS
Detection of gastric tumors by molecular probes in Smad4+/− mice
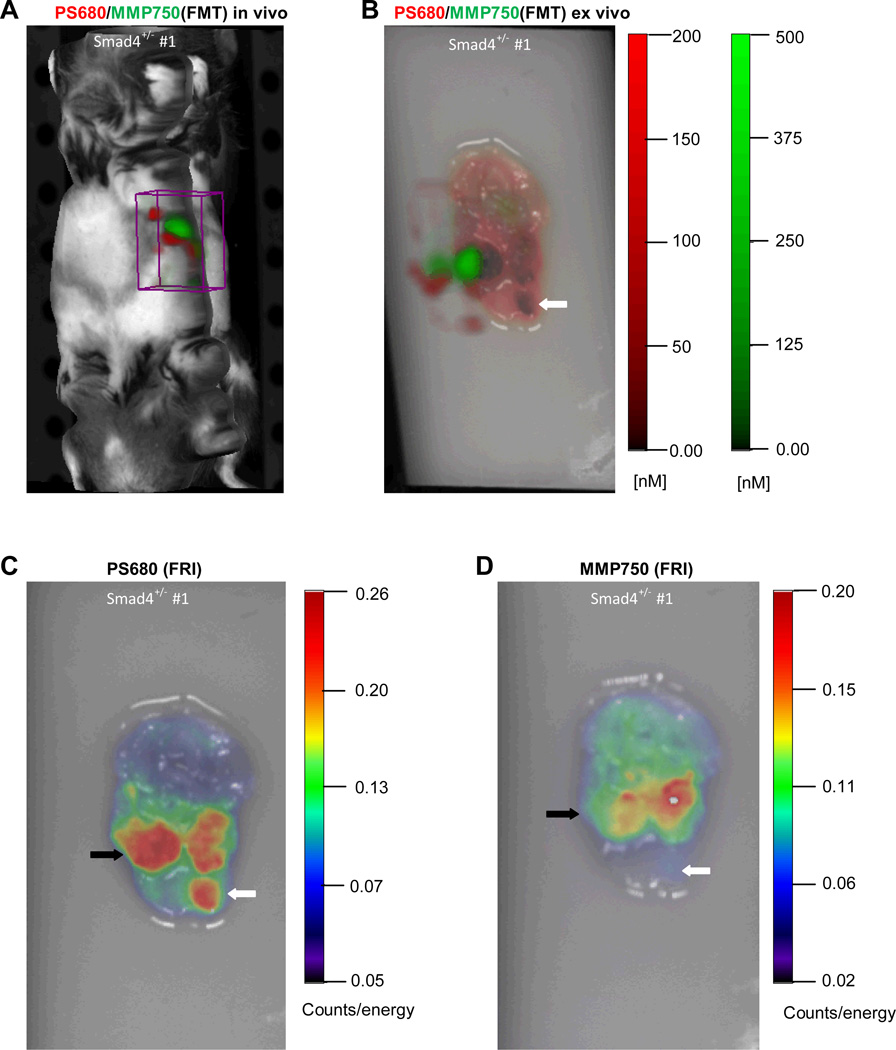
Discrete adenocarcinoma and hyperplastic lesions were successfully detected in the stomachs of each of Smad4+/− animal models by both the cathepsin and MMP molecular probes. Figure 1 shows in vivo imaging of a Smad4+/− mouse which yielded signals in the region of the stomach at each of the distinct emission wavelengths for cathepsin (ProSense® 680) and MMP (MMPSense® 750 FAST) probes. Ex vivo imaging of the stomach using FMT (Figure 1B, with the 3D FMT image is displayed on the top of 2D FRI image) and FRI (Figure 1C and D) confirmed significant activation of both the cathepsin- and MMP-activatable probes in regions corresponding to visible gastric tumors. We observed that cathepsin activation and MMP activation in gastric neoplasia were not identical. As shown in Figure1C and D, activation of the cathepsin probe was detected in three individual lesions, whereas activation of the MMP probe was only observed in two of the three lesions. Importantly, the tumor localized by the cathepsin probe but not by the MMP probe was a hyperplastic lesion. Lastly, relative signal intensity appears to be greater with cathepsin- versus the MMP-activated probe.
Figure 1.
In vivo and ex vivo imaging of gastric tumors in a Smad4+/− mice injected with activatable cathepsin and MMP probes using FMT and FRI. (A) In vivo FMT image of Smad4+/− mouse. ROI (boxed area) reveals NIRF at specific emission wavelengths for each probe in the anatomical region of the stomach. (B) Ex vivo FMT imaging of stomach from Smad4+/− mouse shows signal in visible gastric tumors. The 3D FMT image is superimposed on the 2D FRI image. (C) 2D FRI image of gastric tumor activated by cathepsin. (D) 2D FRI image of gastric tumors activated by MMP. Noted are differences between the two probes: the open arrow demonstrates probe activation with cathepsin but not MMP; and the closed arrow showing a neoplasm with cathepsin versus MMP.
Quantitative comparisons of cathepsin- and MMP-activatable probes in vivo
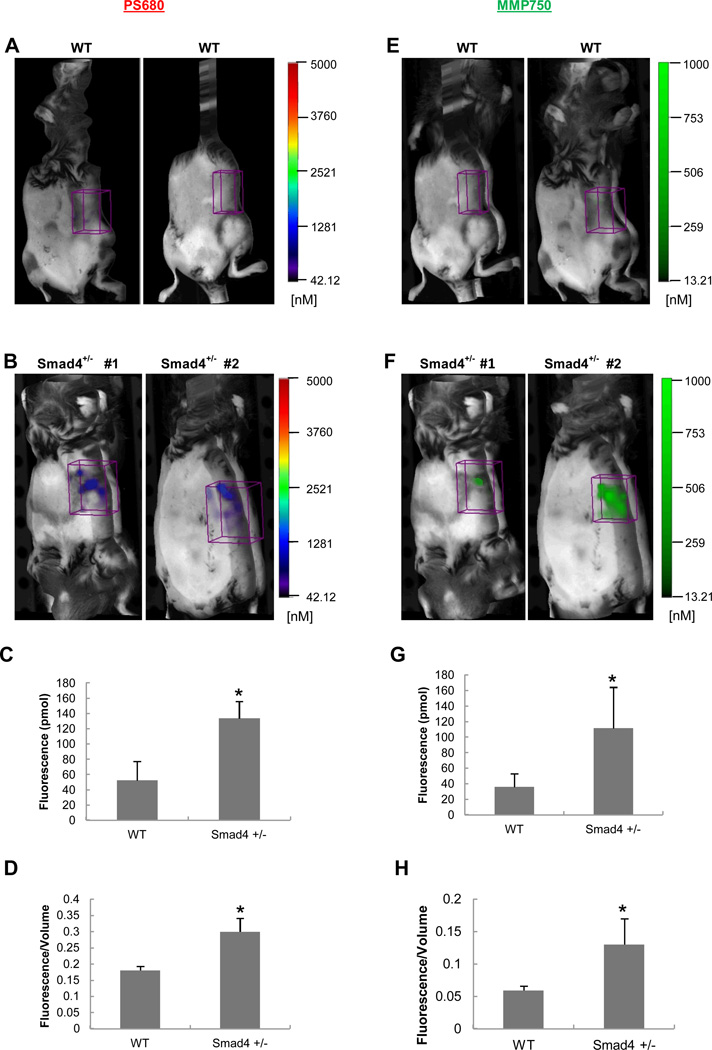
For the cathepsin probe, the 3D FMT quantification measured a 2.5-fold greater total fluorescence signal (Figure 2C) and a significant 1.8-fold greater in fluorescence signal intensity in the stomach ROI of Smad4 +/− mice versus WT mice (Figure 2D). Similarly, for the MMP probe, 3D FMT in vivo imaging yielded a 3.0-fold increase in total fluorescence (Figure 2G) and 2.2-fold increase in fluorescence signal intensity (Figure 2H) in the stomach ROI of Smad4 +/− mice compared with WT controls.
Figure 2.
In vivo imaging of WT and Smad4 +/− mice injected with cathepsin and MMP probes. ROIs were placed in the stomach region from each animal. (A, E) Representative images from WT controls; (B, F) Representative images from Smad4+/− mice; (C, G) 3D quantification of total fluorescence (pmol) and (D, H) fluorescence intensity (total fluorescence divided by volume). Histograms show mean ± SEM; n=4 for Smad4 +/− mice and n=3 for the WT mice; *p=<0.05 compared with WT.
Quantitative comparisons for ex vivo detection of gastric tumors
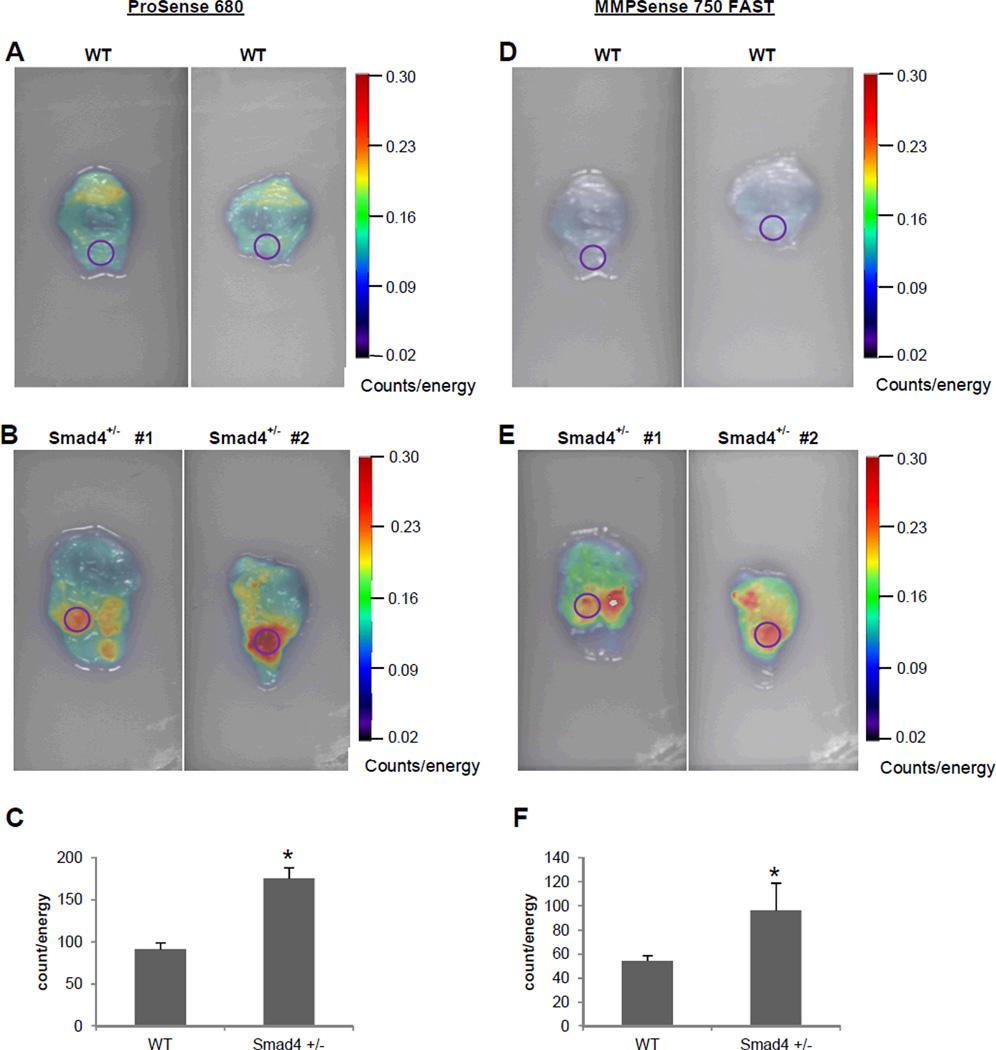
Ex vivo imaging of freshly dissected stomach from WT and Smad4+/− mice confirmed the ability of detecting tumors by using NIRF probes, cathepsin (Figure 3A-C) and MMP (Figure 3D-F). FRI imaging had modest background fluorescence signal in the stomach of WT mice (Figure 3A, D) but significantly stronger signals were observed in focus regions of the stomach of Smad4 +/− mice corresponding to grossly evident tumors (Figure 3B, E). Mean data for WT vs. Smad4+/− mice indicated a significant greater signal in Smad4+/− mouse stomach compared to WT (ProSense® 680: 175.81±12.8 vs. 91.4±8.0 counts/energy; MMPSense® 750 FAST: 96.41±22.9 vs. 54.4±4.3 counts/energy). Furthermore, on average, the TBC value indicated an increased protease activity in the region of the tumor in Smad4+/− mice compared with WT: TBC was 92% increased with cathepsin, and 77% increased with MMP. (Similar values for TBC were calculated in the comparison of tumor and grossly normal areas in Smad4+/− mice).
Figure 3.
Ex vivo FRI imaging of stomach tissue from WT and Smad4 +/− mice injected with cathepsin and MMP molecular probes. Stomach tissues were dissected immediately after in vivo imaging and imaged at 680 and 750 excitation wavelengths. Identical ROI (circles) were placed on area of interest in WT and Smad4+/− mice stomach. (A, D) Representative FRI images from WT. (B, E) Representative FRI images from Smad4+/− mice injected with the cathepsin and MMP probes (D). (C, F) 2D quantification of total counts (counts/energy) in WT and Smad4+/− mice. Histograms show mean ± SEM; n=4 for Smad4 +/− mice and n=3 for the WT mice; *p=<0.05 compared with WT.
Histologic confirmation of gastric neoplasia detected by activatable NIRF probes
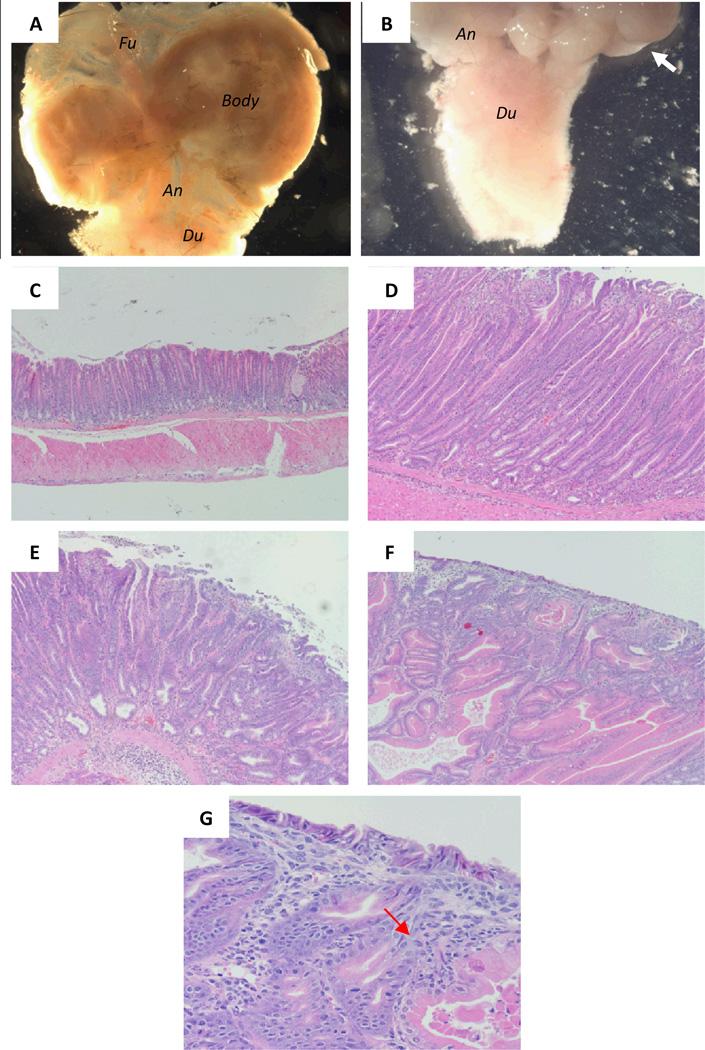
Upon dissection, macroscopically evident tumors were present in the antrum of each of the Smad4+/− animal models (Figure 4A, B), which is consistent with previous findings in these mice (23). To validate that the fluorescence signals observed with activatable NIRF probes are specific, histology was performed on sections of stomach tissue from Smad4+/− and WT mice. As shown in the representative H&E images in Figure 4 and in Table 1, all Smad4 +/− mice showed evidence of tumorigenesis with hyperplastic lesions, dysplastic lesions and adenocarcinoma. The number of gross lesions matched the number detected with activated probes, with the exception of the lack of MMP probe activation in the hyperplastic lesions.
Figure 4.
Histologic analysis of gastric tumors detected by activatable probes of in Smad4+/− mice. (A) Normal stomach. (B) Antral tumor. White arrow points to antral tumors. (C-F) Progression of tumor formation from atypical hyperplasia (D), dysplasia (E) and adenocarcinoma (F, G). Black arrow points to the infiltrating lymphocytes.
Magnifications: 7.11X for A and B, 10X for C-F, 40X for G.
Abbreviations: Fu, fundus; An, antrum; Du, duodenum.
Table 1.
Summary of pathology of individual tumors that detected by probes in Smad4+/− mice.
| Mouse | Tumor | PS680 | MMP750 FAST | Pathology |
|---|---|---|---|---|
| Smad4+/− #1 | A | Yes | Yes | hyperplasia, dysplasia, carcinoma |
| B | Yes | Yes | hyperplasia, dysplasia, carcinoma | |
| C | Yes | No | hyperplasia | |
| Smad4+/− #2 | A | Yes | Yes | hyperplasia, dysplasia, carcinoma |
| Smad4+/− #3 | A | Yes | No | hyperplasia |
| B | Yes | Yes | hyperplasia, dysplasia, carcinoma | |
| Smad4+/− #4 | A | Yes | Yes | hyperplasia, dysplasia, carcinoma |
| B | Yes | Yes | hyperplasia, dysplasia, carcinoma | |
DISCUSSION
In the present study, we provide proof of principle of the utility of NIRF-based molecular imaging for detection of gastric adenocarcinoma. Specifically, we described the efficacy of NIRF imaging by both in vivo and ex vivo methods with the cathepsin-activatable and the MMP-activatable probes to detect gastric neoplasia in the Smad4 +/− gastric cancer mouse model. The MMP probe is activated by gastric dysplasia and adenocarcinoma, while the cathepsin probe is activated by hyperplastic lesions as well as dysplastic and adenocarcinoma.
The cathepsin probe is composed of cathepsin substrates, representing preferred substrates for cleavage by cathepsin B compared with other cathepsins (24). Cathepsin B is a 30 kDa cysteine endopeptidase found mainly in the lysosomes of normal tissues (25, 26). It is involved in proteolytic degradation and turnover of extracellular proteins which is an essential step in cancer invasion and metastasis (27, 28). A number of studies have shown that cathepsin B is over-expressed in gastric cancer tissues compared with noncancerous regions (29–31). The MMP probe comprises substrates of MMPs, representing preferred substrates for cleavage by MMP-2 and MMP-9 (10). Evidence suggests that MMP-9 is highly expressed in human gastric carcinoma and can serve as a prognostic marker (6, 20, 32, 33). Thus, detection of elevated cathepsin or MMP activity in regions of gastric dysplasia or cancer is potentially an effective tool for molecular probe based detection of gastric tumors. Both probes are optically quiescent and undetectable prior to interaction with their respective cysteine proteases. They become activated upon protease cleavage permitting emission of NIRF of sites of protease up-regulation and probe activation. The probe activation is visualized at specific times after injection (ProSense® 680, 24 hours; MMPSense® 750 FAST, 6 hours) which have been shown in studies to permit sufficient concentrations of the agent to be cleaved by cathepsins or MMPs, respectively, in tissues where these enzymes are highly expressed.
The fluorescence imaging technologies FMT and FRI are widely used imaging techniques. FMT permits in vivo detection of specific cellular and sub-cellular targets (34) because of its advantages of deep tissue penetration and high target to background ratio. In the current study, we tested FMT for in vivo imaging and the results demonstrated that FMT for activatable NIRF probes is effective for in vivo detection of gastric tumors. We demonstrate and quantify in vivo, that both ProSense® 680 and MMPSense® 750 FAST were activated in the gastric region of Smad4+/− mice at significantly higher levels than in WT control mice.
Our study suggests that the MMP probe is activated by gastric dysplasia and adenocarcinoma, while the cathepsin probe is activated by hyperplastic lesions as well as dysplastic and adenocarcinoma. Interestingly, in our prior study of molecular imaging in colon cancer animal models, cathepsin B was not significantly activated in colonic hyperplastic polyps. These differences warrant further study and analyses of molecular and histopathologic features of tumors yielding differing signals with the probes. This finding also suggests important and useful differential sensitivity and specificity of two probes for the detection of gastric neoplasms.
In conclusion, this proof of principle study in murine models demonstrates the feasibility of the use of activatable molecular probes and NIRF imaging to improve detection of gastric neoplasia and cancer. Future studies combining endoscopy or capsule endoscopy, with novel molecular probes and imaging techniques, may significantly improve gastric tumor detection for future clinical application.
ACKNOWLEDGEMENTS
We thank Dr. Howard Zhang (Department of Internal Medicine, School of Medicine, Case Western Reserve University) for important input. We acknowledge Kirk McNaughton and the Histology Core Facility in the Department of Cell and Molecular Physiology at UNC-Chapel Hill. The authors also like to thank Dr. Hong Yuan from the Biomedical Research Imaging Center (BRIC) and Dr. Weihua Tang of the Department of Pathology in UNC-Chapel Hill. This work was supported by a grant from the University Cancer Research Foundation at UNC-Chapel Hill (PKL, DM), U01 grant (CA105417-07), NIH DK47769-11(PKL), NIH HD058290 (YC) and CA125588 (DM)
Abbreviations
- FMT
fluorescence molecular tomography
- FRI
fluorescence reflectance imaging
- MMP
matrix metalloproteinase
- NIRF
near infrared fluorescent
REFERENCES
- 1.Lochhead P, El-Omar EM. Gastric cancer. Br Med Bull. 2008;85:87–100. doi: 10.1093/bmb/ldn007. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.den Hoed CM, van Eijck BC, Capelle LG, et al. The prevalence of premalignant gastric lesions in asymptomatic patients: Predicting the future incidence of gastric cancer. Eur J Cancer. 2011;47:1211–1218. doi: 10.1016/j.ejca.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Yalamarthi S, Witherspoon P, McCole D, et al. Missed diagnoses in patients with upper gastrointestinal cancers. Endoscopy. 2004;36:874–879. doi: 10.1055/s-2004-825853. [DOI] [PubMed] [Google Scholar]
- 5.Wong Kee Song LM, Wilson BC. Endoscopic detection of early upper GI cancers. Best Pract Res Clin Gastroenterol. 2005;19:833–856. doi: 10.1016/j.bpg.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 6.Sampieri CL, de la Pena S, Ochoa-Lara M, et al. Expression of matrix metalloproteinases 2 and 9 in human gastric cancer and superficial gastritis. World J Gastroenterol. 2010;16:1500–1505. doi: 10.3748/wjg.v16.i12.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korideck H, Peterson JD. Noninvasive quantitative tomography of the therapeutic response to dexamethasone in ovalbumin-induced murine asthma. J Pharmacol Exp Ther. 2009;329:882–889. doi: 10.1124/jpet.108.147579. [DOI] [PubMed] [Google Scholar]
- 8.Seibel EJ, Brentnall TA, Dominitz JA. New endoscopic and cytologic tools for cancer surveillance in the digestive tract. Gastrointest Endosc Clin N Am. 2009;19:299–307. doi: 10.1016/j.giec.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahmood U. Optical molecular imaging approaches in colorectal cancer. Gastroenterology. 2010;138:419–422. doi: 10.1053/j.gastro.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 11.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes Dev. 2003;17:545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 12.Alencar H, Funovics MA, Figueiredo J, et al. Colonic adenocarcinomas: near-infrared microcatheter imaging of smart probes for early detection--study in mice. Radiology. 2007;244:232–238. doi: 10.1148/radiol.2441052114. [DOI] [PubMed] [Google Scholar]
- 13.Kossodo S, Pickarski M, Lin SA, et al. Dual in vivo quantification of integrin-targeted and protease-activated agents in cancer using fluorescence molecular tomography (FMT) Mol Imaging Biol. 2010;12:488–499. doi: 10.1007/s11307-009-0279-z. [DOI] [PubMed] [Google Scholar]
- 14.Weissleder R, Tung CH, Mahmood U, et al. In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 15.Peterson JD, Labranche TP, Vasquez KO, et al. Optical tomographic imaging discriminates between disease-modifying anti-rheumatic drug (DMARD) and non-DMARD efficacy in collagen antibody-induced arthritis. Arthritis Res Ther. 2010;12:R105. doi: 10.1186/ar3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaffer FA, Vinegoni C, John MC, et al. Real-time catheter molecular sensing of inflammation in proteolytically active atherosclerosis. Circulation. 2008;118:1802–1809. doi: 10.1161/CIRCULATIONAHA.108.785881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aisenberg J. Gastrointestinal endoscopy nears "the molecular era". Gastrointest Endosc. 2008;68:528–530. doi: 10.1016/j.gie.2008.03.1075. [DOI] [PubMed] [Google Scholar]
- 18.Chen, et al. A novel mouse Smad4 mutation reduces protein stability and wild-type protein levels. Mamm Genome. 2006;17:211–219. doi: 10.1007/s00335-005-0074-3. [DOI] [PubMed] [Google Scholar]
- 19.Vivian, et al. An allelic series of mutations in Smad2 and Smad4 identified in a genotype-based screen of N-ethyl-N-nitrosourea-mutagenized mouse embryonic stem cells. PNAS. 2002;99:15542–15547. doi: 10.1073/pnas.242474199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mroczko B, Lukaszewicz-Zajac M, Guzinska-Ustymowicz K, et al. Expression of matrix metalloproteinase-9 in the neoplastic and interstitial inflammatory infiltrate cells in gastric cancer. Folia Histochem Cytobiol. 2009;47:491–496. doi: 10.2478/v10042-009-0070-7. [DOI] [PubMed] [Google Scholar]
- 21.Powell SM, Harper JC, Hamilton SR, et al. Inactivation of Smad4 in gastric carcinomas. Cancer Res. 1997;57:4221–4224. [PubMed] [Google Scholar]
- 22.Goetz M, Wang TD. Molecular imaging in gastrointestinal endoscopy. Gastroenterology. 2010;138:828 e1–833 e1. doi: 10.1053/j.gastro.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Brodie SG, Yang X, et al. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene. 2000;19:1868–1874. doi: 10.1038/sj.onc.1203504. [DOI] [PubMed] [Google Scholar]
- 24.Marten K, Bremer C, Khazaie K, et al. Detection of dysplastic intestinal adenomas using enzyme-sensing molecular beacons in mice. Gastroenterology. 2002;122:406–414. doi: 10.1053/gast.2002.30990. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Morgan D, Cecil G, et al. Biochromoendoscopy: molecular imaging with capsule endoscopy for detection of adenomas of the GI tract. Gastrointest Endosc. 2008;68:520–527. doi: 10.1016/j.gie.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mort JS, Buttle DJ. Cathepsin B. Int J Biochem Cell Biol. 1997;29:715–720. doi: 10.1016/s1357-2725(96)00152-5. [DOI] [PubMed] [Google Scholar]
- 27.Palermo C, Joyce JA. Cysteine cathepsin proteases as pharmacological targets in cancer. Trends Pharmacol Sci. 2008;29:22–28. doi: 10.1016/j.tips.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Nomura T, Katunuma N. Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J Med Invest. 2005;52:1–9. doi: 10.2152/jmi.52.1. [DOI] [PubMed] [Google Scholar]
- 29.Ebert MP, Kruger S, Fogeron ML, et al. Overexpression of cathepsin B in gastric cancer identified by proteome analysis. Proteomics. 2005;5:1693–1704. doi: 10.1002/pmic.200401030. [DOI] [PubMed] [Google Scholar]
- 30.Chung SM, Kawai K. Protease activities in gastric cancer tissues. Clin Chim Acta. 1990;189:205–210. doi: 10.1016/0009-8981(90)90093-8. [DOI] [PubMed] [Google Scholar]
- 31.Herszenyi L, Plebani M, Carraro P, et al. Proteases in gastrointestinal neoplastic diseases. Clin Chim Acta. 2000;291:171–187. doi: 10.1016/s0009-8981(99)00227-2. [DOI] [PubMed] [Google Scholar]
- 32.Gao ZL, Zhang C, Du GY, et al. Clinical significance of changes in tumor markers, extracellular matrix, MMP-9 and VEGF in patients with gastric carcinoma. Hepatogastroenterology. 2007;54:1591–1595. [PubMed] [Google Scholar]
- 33.Choi EH, Kim JT, Kim JH, et al. Upregulation of the cysteine protease inhibitor, cystatin SN, contributes to cell proliferation and cathepsin inhibition in gastric cancer. Clin Chim Acta. 2009;406:45–51. doi: 10.1016/j.cca.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Ntziachristos V, Ripoll J, Wang LV, et al. Looking and listening to light: the evolution of whole-body photonic imaging. Nat Biotechnol. 2005;23:313–320. doi: 10.1038/nbt1074. [DOI] [PubMed] [Google Scholar]