Abstract
Developmental dyslexia is a disorder characterized by a specific deficit in reading despite adequate overall intelligence and educational resources. The neurological substrate underlying these significant behavioral impairments is not known. Studies of post mortem brain tissue from male and female dyslexic individuals revealed focal disruptions of neuronal migration concentrated in the left hemisphere, along with aberrant symmetry of the right and left the planum temporale, and changes in cell size distribution within the medial geniculate nucleus of the thalamus (Galaburda et al., 1985; Humphreys et al., 1990). More recent neuroimaging studies have identified several changes in the brains of dyslexic individuals, including regional changes in gray matter, changes in white matter, and changes in patterns of functional activation. In a further effort to elucidate the etiology of dyslexia, epidemiological and genetic studies have identified several candidate dyslexia susceptibility genes. Some recent work has investigated associations between some of these genetic variants and structural changes in the brain. Variants of one candidate dyslexia susceptibility gene, KIAA0319, have been linked to morphological changes in the cerebellum and functional activational changes in the superior temporal sulcus (Jamadar et al., 2011; Pinel et al., 2012). Animal models have been used to create a knockdown of Kiaa0319 (the rodent homolog of the human gene) via in utero RNA interference in order to study the gene’s effects on brain development and behavior. Studies using this animal model have demonstrated that knocking down the gene leads to focal disruptions of neuronal migration in the form of ectopias and heterotopias, similar to those observed in the brains of human dyslexics. However, further changes to the structure of the brain have not been studied following this genetic disruption. The current study sought to determine the effects of embryonic Kiaa0319 knockdown on volume of the cortex and hippocampus, as well as midsagittal area of the corpus callosum in male rats. Results demonstrate that Kiaa0319 knockdown did not change the volume of the cortex or hippocampus, but did result in a significant reduction in the midsagittal area of the corpus callosum. Taken in the context of previous reports of behavioral deficits following Kiaa0319 knockdown (Szalkowski et al., 2012), and reports that reductions of corpus callosum size are related to processing deficits in humans (Paul et al., 2011), these results suggest that Kiaa0319 has a specific involvement in neural systems important for temporal processing.
Keywords: Dyslexia, KIAA0319, RNA interference, Neuronal migration, Rapid auditory processing1
1. INTRODUCTION
Developmental dyslexia is a common disorder affecting 5–10% of the population (Peterson and Pennington, 2012), and is defined as a significant impairment in reading despite adequate intelligence and educational opportunity. While dyslexia is specifically considered a disorder of reading, it is in fact characterized by a wide array of more basic behavioral impairments, including deficits in phonological processing (Kovelman et al. 2012; Melby-Lervag et al. 2012; Peyrin et al., 2012), short term and/or working memory (Beneventi et al., 2010; Gathercole et al., 2006; Menghini et al., 2010, 2011; Smith-Spark and Fisk 2007), visuospatial attention (Franceschini et al., 2012; Gabrieli and Norton 2012), and rapid auditory processing (Cohen-Mimran and Sapir 2007; Fitch and Szalkowski, 2012; Hamalainen et al., 2012; Vandermosten et al., 2011). Deficits in these core component behaviors, or “endophenotypes” vary greatly across individuals, but are thought to summate in various combinations—possibly as a function of risk factors, including genetics—to produce a high degree of heterogeneity in this population.
Despite years of research, the etiology of dyslexia is still not fully understood. However, studies that examined the post mortem brains of dyslexic individuals were among the first to report potential neurological markers for the disorder. Specifically, Galaburda et al., (1985) and Humphreys et al., (1990) observed microscopic focal disruptions to the cortex and underlying white matter in the form of ectopic collections of cells and aberrant microgyric infolding of the cortex, predominantly in the left hemisphere perisylvian regions of the brains of male and female dyslexic individuals. Additional anomalies were observed subcortically in the lateral (Livingstone et al., 1991) and medial (Galaburda and Eidelberg, 1982; Galaburda et al., 1994) geniculate nucleus of the thalamus of dyslexic subjects’ brains, with a shift in the distribution to more small cells and fewer large cells as compared to the control subjects’ brains. This finding suggested that the mechanism(s) underlying the apparent disruptions to neuronal migration also had widespread effects on the morphology of distal brain structures. However, the relationship between these anomalies regarding causation (i.e., whether cortical anomalies induced sub-cortical changes, or whether both phenomena evidence common underlying causal factors) is not well understood. The nature of these disruptions led researchers to conclude that the disruptive mechanism causing these widespread changes had likely taken place during prenatal life, and specifically during the process of neuronal migration. More recent studies have used in vivo neuroimaging to examine the brains of dyslexic patients in search of neurobiological markers of the disease. Studies of independent populations of dyslexic individuals from several different countries have indicated that there are also significant, region-specific reductions in gray matter in the cortex, subcortical structures, and cerebellum of dyslexic individuals as compared to age-matched controls (Brambati et al., 2004; Kronbichler et al., 2008; Menghini et al., 2008; Siok et al., 2008). Studies of changes in the corpus callosum have yielded conflicting results. In general, there are region specific changes, with some areas exhibiting increases in size and others exhibiting decreases in size in dyslexic individuals as compared to controls (Elnakib et al., 2012; Hasan et al., 2012; Paul et al., 2011). Changes in functional activation, with associated microstructural changes in white matter, have also been reported in some dyslexic populations (Carter et al., 2009; Hoeft et al., 2011; Pugh et al., 2000; Rimrodt et al., 2010; Wolf et al., 2010). Additionally, these gross neurological anomalies have been significantly associated with specific behavioral deficits, such as performance on tasks of phonological processing and short term memory (Fine et al., 2007; Leonard et al., 2001). Moreover, recent work has demonstrated that many of these observed brain changes are present in children at risk for dyslexia, even before learning to read, and so they are present from birth and do not reflect experience-dependent changes (Raschle et al., 2010).
Additional advances in understanding the etiology of dyslexia have been achieved through epidemiological and genetic studies of dyslexic populations. Recent work in this field has led to the identification of several candidate dyslexia susceptibility genes (Anthoni et al., 2012; Francks et al., 2004; Hannula-Jouppi et al., 2005; Matsson et al., 2011; Meng et al., 2005; Poelmans et al., 2008; Scerri et al., 2010; Taipale et al., 2003). Some recent reports have even used in vivo brain imaging to draw correlations between specific variants in these CDSGs and significant changes in gross brain structure and functional activation (Jamadar et al., 2011 Pinel et al., 2012). One of the discovered CDSGs, KIAA0319, has been linked to altered functional activation patterns in superior temporal sulcus and morphological changes in the cerebellum, both known language-related areas, in the brains of both dyslexic and unaffected individuals (Jamadar et al., 2011; Pinel et al., 2012).
In the wake of the discovery of CDSGs for dyslexia, animal models were developed to study neuroanatomical and behavioral effects following manipulation of the homologs of these genes. Studies utilizing RNA interference to embryonically knock down genes in rats have demonstrated that the rodent homologs of a handful of these genes, including KIAA0319, are involved in neuronal migration (Burbridge et al., 2008; Meng et al., 2005; Paracchini et al., 2006; Peschansky et al., 2010; Rosen et al., 2007; Wang et al., 2006). Knocking down these genes in utero leads to the development of ectopic and heterotopic collections of cells in the brains of adult animals; malformations that are strikingly similar to those observed by Galaburda et al., (1985) and Humphreys et al., (1990) in human dyslexic brains. There have not been any reports to date of gross changes in structural size or volume in the brains of animals following knockdown of Kiaa0319 (or any of the other CDSGs). In fact, the specific function(s) of these genes (and their protein products) in brain development and growth in animal and human populations remains elusive. Current data suggests that the Kiaa0319 protein is important for cell adhesion. The Kiaa0319 protein is characterized by a large polycystic kidney domain, which is a structure known to play a role in intracellular adhesion in other proteins (Ibraghimov-Beskrovnaya et al., 2000). Additionally, high magnification observation of Kiaa0319 shRNA transfected neurons revealed loss of association between neurons and radial glial fibers, further suggesting that the protein is necessary for adhesion (Paracchini et al., 2006). A potential role for Kiaa0319 in extracellular signaling has also been suggested based on the observation that some variants of the protein are excreted into extracellular space (Velayos-Baeza et al., 2008, 2009; 2010).
The current study sought to characterize the effects of embryonic Kiaa0319 knockdown on specific structures in the brains of adult male rats. We specifically examined the effects of this genetic disruption on volume of the cortex, volume of the hippocampus, and midsagittal area of the corpus callosum in adulthood. We have previously reported on the behavioral deficits in this sample of animals (including impairments of rapidly changing/short duration acoustic stimuli, but not working memory), and so results from the current study may provide a better understanding of the biological underpinnings of the observed behavioral profile (see Szalkowski et al., 2012 for review).
2. METHODS
Prepared brain sections from a total of 50 male Sprague-Dawley (Shams, n = 11; Kiaa0319 shRNA, n = 14) and Wistar (Shams, n = 9; Kiaa0319 shRNA, n = 16) rats were used for the current study. The histological sections analyzed in the current study were derived from the animals used in our recent report on the behavioral effects of Kiaa0319 knockdown (Szalkowski et al., 2012). Specifically, the Wistar rats in the current study are those that underwent rapid auditory processing testing in the Szalkowski et al., 2012 paper, while the Sprague Dawley rats in the current study underwent spatial working memory testing in the 8 arm radial water maze. For detailed information on the housing and behavioral testing conditions that these rats were exposed to, refer to Szalkowski et al., (2012). All animals were sacrificed at postnatal day 110 (P110), and so the brain sections used in the current study were derived from adult animals. All procedures were in accordance with National Institutes of Health guidelines and were approved by the University of Connecticut Institute for Animal Care and Use Committee.
2.1 In utero electroporation
In utero electroporation surgeries for these subjects were performed in two batches on embryonic day 15 (E15), as described in Szalkowski et al., 2012. Surgeries were performed by C.F. at the University of Connecticut. Briefly, time-mated Sprague Dawley or Wistar dams were anesthetized with an intraperitoneal injection of Ketamine (100 mg/kg) and Xylazine (15 mg/kg). A vertical incision was made through the skin and muscle of the lower abdomen and the uterine horns were exposed. Each pup received a single injection of either the Kiaa0319 short hairpin RNA (shRNA) solution (see below) or the sham solution (see below) into one randomly chosen lateral ventricle. Injections were made with air pressure (General Valve Picospritzer) using pulled glass capillary needles. Fluorescent proteins were used as markers of transfection, with one color used to identify Kiaa0319 shRNA transfection and a second color used to identify sham transfection in each batch of surgeries. The fluorescent proteins were then visualized after histological preparation of brain tissue and used to classify each subject as either shRNA or sham for subsequent data analyses. Kiaa0319 shRNA injections consisted of 1.5 μg/μL of Kiaa0319 shRNA (pU6shRNA-Kiaa0319) and 0.5 μg/μL enhanced green fluorescent protein (eGFP, used in Batch 1) or 0.5 μg/μL monomeric red fluorescent protein (mRFP, used in Batch 2). Note that the specificity and effectiveness of the vector used in the current study at knocking down Kiaa0319 protein translation has previously been reported (Paracchini et al., 2006; Peschansky et al., 2010). Sham injections consisted of 0.5 μg/μL of mRFP (in Batch 1) or eGFP (in Batch 2) alone. Following injection into the lateral ventricle, a pair of copper alloy plates (1 × .5 cm) were placed over the injection site and delivered a 70 mV electrical pulse. The paddle positions were then reversed and a second electrical pulse was delivered, leading to bilateral transfection of cells. This created a favorable electrical environment for the shRNA plasmids and fluorescent proteins to be taken up into the nuclei of transfected cells. Roughly equal numbers of sham and Kiaa0319 shRNA injections were made in each litter. After all pups had received an injection the uterine horns were carefully placed back in the abdominal cavity and sutures were placed. Subcutaneous injections of 0.5% Metacam (2 mg/kg) were administered to alleviate post-operative pain.
2.2 Histology
At the end of behavioral testing, all animals were weighed, deeply anesthetized with Ketamine (100 mg/kg) and Xylazine (15 mg/kg) and were transcardially perfused with 0.4 M phosphate buffered saline, followed by 4% paraformaldehyde. Heads were removed and brains were extracted and shipped to GDR for histological preparation. Brains were cryoprotected in a 30% sucrose solution before being sliced at 40 μm in the coronal plane. A 1-in-10 series was mounted onto glass slides and stained for Thionin. An adjacent series of free-floating sections was analyzed with fluorescence microscopy for the presence of GFP or RFP (Chemicon, 1:200) using ABC protocols. A third adjacent series of sections was immunohistochemically processed for GFP and RFP antibodies. Nissl stained sections were examined with light microscopy for the presence of malformations in the form of ectopias and heterotopias.
2.3 Stereological Assessment: Midsagittal area of the corpus callosum measurement
Midsagittal area of the corpus callosum was measured for each subject using the Axio 2 Zeiss microscope and Stereo Investigator (MBFBioscience, Williston, VT). Measurements were taken at 10X magnification. The corpus callosum was analyzed on systemically sampled section, beginning rostrally where the corpus callosum was first seen to cross the midline and continuing caudally until the last section where the corpus callosum crossed the midline. On each coronal section measured, a line was drawn along the dorsal-ventral axis of the brain at the midline, extending beyond the dorsal and ventral boundaries of the corpus callosum, and we then measured the dorsal-ventral length of the midline of the corpus callosum. These measurements, along with the thickness of the mounted slices, and the distance between each counted section were entered into the Cavalieri equation, which yielded an estimate of the midsagittal area of the corpus callosum for each subject (see Newbury and Rosen, 2012; Rosen and Harry, 1990 for details).
2.4 Stereological Assessment: Cortical and hippocampal volume measurement
Volumes of the cerebral cortex and hippocampus were measured under 2.5X magnification on an Axio 2 Zeiss Microscope using Stereo Investigator software. The Cavalieri Estimator probe was used to overlay a grid of 600 μm × 600 μm on each section, and all points of intersection on the grid within the boundaries of the region of interest are counted.. Volume estimates for right and left cortex and hippocampus were calculated separately using Cavalieri’s rule. An average of 10 sections each was counted for the cortex and hippocampus for each subject using the boundaries of Paxinos and Watson (2007). Specifically, all regions dorsal to the rhinal fissure were included in the cortex measurements, including areas extending up and onto the medial surface. The hippocampal measurements included the subiculum, dentate gyrus, CA1, CA2, and CA3.
2.5 Data analyses
Average midsagittal area of the corpus callosum, volume of the cortex, and volume of the hippocampus were compared for Strains (Wistar and Sprague Dawley) and Treatment (Kiaa0319 shRNA and sham). Left and right cortex and hippocampus volumes (Hemisphere) were also compared within-subjects. All statistical analyses were performed using IBM SPSS Standard Statistics Package v. 19.0. All reported P values are two-tailed unless otherwise stated.
3. RESULTS
3.1 Body weight in Kiaa0319 shRNA and sham Wistar and Sprague Dawley subjects
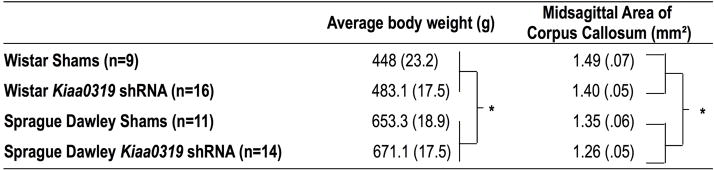
Adult body weight was analyzed as a function of Strain and Treatment to ensure that any structural brain size differences found were not solely, or partly, reflections of overall differences in size or development (Table 1). A univariate ANOVA (Strain (2 levels) × Treatment (2 levels) revealed a significant Strain effect [F(1,44) = 102.5, P < .01]. There was not a significant Treatment effect, nor a Treatment × Strain interaction [F(1,44) < 1, NS for both effects]. The Strain effect reflects the fact that Wistar rats, across Treatment groups, were smaller than Sprague Dawley rats.
Table 1.
Average adult body weight and Midsagittal Area of Corpus Callosum
|
Average weights and midsagittal area of corpus callosum, +/− SEM. For average body weight,
indicates significant Strain effect (P < .01), with Sprague Dawley animals significantly larger than Wistar animals, across Treatment groups. There was an overall main effect of Treatment on Midsagittal area of corpus callosum, with Kiaa0319 shRNA animals having significantly (*, P < .05) smaller callosa than shams.
3.2 Midsagittal area of the corpus callosum in Kiaa0319 shRNA and sham subjects
A univariate analysis of variance (ANOVA) (Strain (2 levels) × Treatment (2 levels)) revealed a significant main effect of Strain [F(1,46) = 10.8, P < .01], and also a significant main effect of Treatment [F(1,46) = 4.3, P <.05 on midsagittal corpus callosum area (see Table 1). There was not a significant Strain × Treatment interaction [F(1,46) < 1, NS]. The main effect of Strain is reflective of the fact that Wistar rats were observed to have a larger midsagittal area of the corpus callosum than Sprague Dawley rats, across Treatment groups. Importantly, the main effect of Treatment indicates that, across Strain, Kiaa0319 shRNA animals had a significantly smaller midsagittal area of the corpus callosum as compared to sham counterparts. The lack of a Strain × Treatment interaction tells us that the observed significant Treatment effects were also independent of the observed body size differences between Wistar and Sprague Dawley rats. (Note that an analysis of covariance with body weight as a covariate revealed that this variable did not significantly influence midsagittal area of corpus callosum [F(1,42) = 1.7, NS], and the Treatment effect remained significant [F(1,42) = 4.927, P < .05]). Because clinical data suggests that reductions in corpus callosum volume are relevant to impairments in reading and phonological processing in dyslexic individuals (Fine et al., 2007; Paul et al., 2011), we performed a bivariate correlation on the midsagittal area of the corpus callosum and performance scores from a rapid auditory processing test that had been used on all subjects in adulthood (mean attenuation scores on FM Sweep 125 ms task, see Szalkowski et al., 2012 for further details). There was not a significant correlation between this measure of auditory performance and mean midsagittal corpus callosum area in sham [r = .30, NS] or Kiaa0319 shRNA treated animals [r = −.16, NS].
3.3 Volume of the cortex in Kiaa0319 shRNA and sham subjects
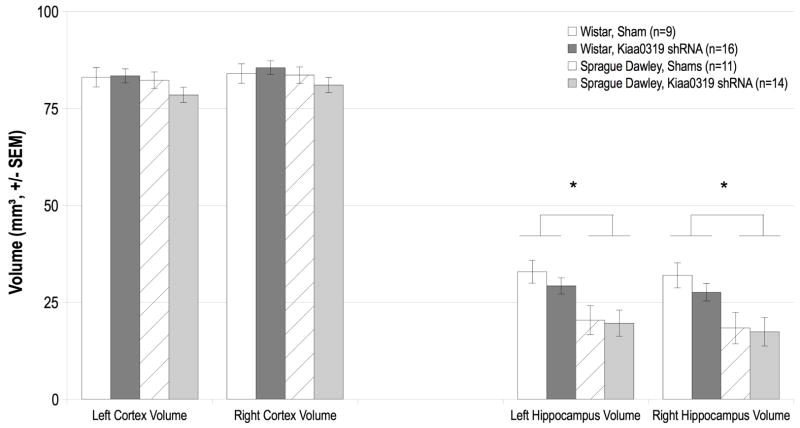
A 2 between (Strain and Treatment) one within (Hemisphere) ANOVA was used to compare right and left cortex volume in Kiaa0319 shRNA and sham treated Wistar and Sprague Dawley rats (see Figure 1). There was not a significant main effect of Hemisphere, Strain, nor Treatment [F(1,44) < 1, NS in all cases], nor were there significant interactions among these independent variables [F(1,31) < 2, NS for all effects]. These results indicate that there were no differences in cortical volume in Kiaa0319 shRNA and sham subjects.
Figure 1.
Volume measurements from right and left cortex and hippocampus of Sprague and Wistar rats. There were no significant effects of Treatment, Strain, or Hemisphere, (nor any significant interactions) on cortex volume. There was a significant main effect of Strain on hippocampal volume, demonstrating that Wistar rats—across Treatment groups—had larger hippocampi than Sprague counterparts. However, the Kiaa0319 shRNA treatment was not found to have an impact on hippocampal volume.
3.4 Volume of the hippocampus in Kiaa0319 shRNA and sham subjects
A 2 between (Strain and Treatment) one within (Hemisphere) was used to compare right and left hippocampus volume in Kiaa0319 shRNA and sham treated Wistar and Sprague Dawley rats (see Figure 1). There was a significant main effect of Strain [F(1,31) = 36.2, P < .001]— Wistar animals have significantly larger hippocampal volumes than Sprague Dawley animals, across Treatment groups.. There were no significant main effects of Hemisphere or Treatment [F(1,31) < 2, NS for both effects], nor were there significant interactions among the independent variables [F(1,31) < 2, NS for all effects]. This indicates that there were no significant differences in hippocampal volume between Kiaa0319 shRNA and sham subjects.
4. DISCUSSION
We find a significant reduction in midsagittal area of the corpus callosum in Kiaa0319 shRNA transfected rats as compared to shams. General reductions in the midsagittal area of the corpus callosum are known to be associated with phonological processing deficits in human dyslexics. However, we did not find a significant correlation between this anatomical measure and rapid auditory processing abilities in Kiaa0319 shRNA or sham animals. Finally, Kiaa0319 knockdown did not affect volume of the cortex and hippocampus in this study.
4.1 Cortical dysgenesis and morphological changes in the brain: animal models
Studies have examined the impact of cortical dysgenesis on widespread changes to brain structure using other animal models of developmental brain disruption. Our lab has previously reported that whole brain volume, and specifically neocortical volume, are reduced in rats following induction of a microgyric lesion via a focal freezing probe on P1 (Peiffer et al., 2003). An additional study from our lab demonstrated significant reductions in the volume of neocortex, corpus callosum, and hippocampus in a teratogenic model of nodular heterotopia in rats (Threlkeld et al., 2009). A study that looked at differences in the midsagittal area of the corpus callosum and neocortical volume in strains of mice with spontaneously-occurring ectopias did not find any differences between mice with and without ectopias in these morphological measures (Rosen et al., 1990). Taken together with the current results, these data suggest that the observed changes in the corpus callosum are not simply part of a generic cascade of reorganization that occurs following any disruption to neuronal migration. Instead it seems likely that the observed pattern of pathological brain changes in the Kiaa0319 knockdown animals is due to specific effects of the genetic manipulation and not just to the resulting cortical dysgenesis.
4.2 Structural brain changes in developmental dyslexia
Several structural neural anomalies have been reported in dyslexic individuals. On a more general level, region-specific gray matter reductions are a consistent finding in language-impaired populations, with unilateral and bilateral volume reductions often reported in language-related cortical regions such as the superior temporal gyrus, fusiform gyrus, cerebellum, and planum temporale (Brambati et al., 2004; Kronbichler et al., 2008; Menghini et al., 2008; Siok et al., 2008; also see Sun et al., 2010 for a complete review). Changes in white matter have also been reported in dyslexic individuals (Carter et al., 2009; Elnakib et al., 2012; Hasan et al., 2012; Hoeft et al., 2011; Paul et al., 2011; Pugh et al., 2000; Rimrodt et al., 2010; Wolf et al., 2010; see Vandermosten et al., 2012 for a complete review). Region specific differences in the corpus callosum of dyslexics have been reported, but there is a great deal of variability in the findings across different studies. For example, increases in size have been reported in posterior aspects of the callosum, while decreases have been reported in anterior and middle aspects (Duara et al., 1991; Hasan et al., 2012; Kilian et al., 2008; Rumsey et al., 1996). Studies have also reported decreases in the area of the midbody of the corpus callosum in dyslexic individuals (Fine et al., 2007). It has been suggested that this reduction in callosal midbody size has been linked to deficits in temporal processing (Aboitiz et al., 1992; Fine et al., 2007; Paul et al., 2011). This is interesting given our current findings suggesting that Kiaa0319 knockdown led to a reduction in callosal size in rats, and moreover in rats that had demonstrated specific deficits in rapid auditory processing (although we did not find these measures to be correlated).
The relatively small reduction in callosal size observed following Kiaa0319 knockdown in the current study parallels the subtle changes to the callosum in clinical language impaired populations. In fact, as discussed above, changes in callosal size are typically apparent in specific subregions and not in measures of the total rostral-caudal area of the corpus callosum. Despite their subtlety, these small changes in callosal size are associated with impaired interhemispheric conduction and a range of sensory and cognitive impairments (see Paul et al., 2011 for review). Thus, in spite of the lack of a significant correlation between midsagittal area of the corpus callosum and rapid auditory processing abilities in the current study, it is likely that the observed changes in callosal size are functionally significant. The lack of a correlation between the midsagittal area of the corpus callosum and the rapid auditory processing data from the animals in the current study may be explained several ways. First, it is possible that the overall reduction in callosal size observed here is the result of a large reduction in one particular area of the callosum that may correlate more closely with sensory processing measures. It would be interesting to repeat this analysis using brain sections cut in the sagittal plane, which would enable us to analyze changes in callosal size by subregion. It is also worth noting that the correlation only compared RAP scores and callosal area in 16 animals that had experienced the Kiaa0319 knockdown. It is possible that a correlation may become apparent with a larger sample size. Similarly, a correlation between auditory processing abilities and midsagittal area of the corpus callosum may become apparent following a greater reduction in callosal size. The degree of Kiaa0319 knockdown achieved following electroporation may have influenced the degree of reduction of the midsagittal area of the corpus callosum, and it would be interesting to repeat this analysis in animals following a “larger” knockdown (perhaps via bilateral instead of unilateral injections of shRNA plasmids), or in a mouse model using a total genetic knockout. Moreover, we cannot verify whether or not genetic compensation may have dampened the effects of the Kiaa0319 knockdown on callosal size; this would be a more likely scenario in a knockout model in which the gene/protein is absent from all cells throughout all of development, as opposed to the RNAi model that knocks down levels of the protein by affecting translation in a restricted subset of cells at a specific developmental time point. Finally, it is possible that this reduction in callosal size would correlate better with different tasks of auditory processing than with the task we tested our animals on (short frequency modulated (FM) sweep detection). Further studies are needed to explore the relationship between this genetically-mediated callosal size reduction and a broader range of auditory processing abilities, as well as abilities in other sensory and cognitive domains. Given human behavioral data showing that KIAA0319 variants are specifically linked to deficits in phonological processing, along with animal work showing that Kiaa0319 knockdown leads to temporal auditory processing deficits, and anatomical data suggesting that Kiaa0319 knockdown results in reduced callosal size—which is related to temporal processing deficits in humans—it seems possible that KIAA0319 may impact behavior at least in part through anatomical changes to the corpus callosum.
It is worth noting that anatomical findings are not consistent across all studies of dyslexic individuals, and in fact some studies have reported the opposite (that dyslexic individuals have region-specific increases in cortical volume as compared to controls) (Vinckenbosch et al., 2005). The majority of papers reporting size differences in brain regions in dyslexic and control individuals use heterogeneous populations of dyslexics that are often recruited based on categorical diagnosis alone. Thus, it is difficult to say whether or not these anatomical findings would be present in every sub-population of dyslexic individuals. It seems possible that, like behavioral expression of the disorder, the neuroanatomical substrate of dyslexia may vary from one individual or sub-population to the next. It also seems plausible that different candidate dyslexia susceptibility genes may contribute uniquely to the patterns of disruptions of neuroanatomy in dyslexic individuals. For example, it is possible that reductions in cortical volume would not be detected in a population of dyslexics selected based on having a particular KIAA0319 variant. It is even possible that different variants of the same gene may influence brain morphology differently, depending on the functional implication of the mutation. Further clinical neuroimaging studies using samples selected based on genotype will be necessary to address these issues.
4.3 The impact of KIAA0319 variants on human brain morphology and function
In the clinical literature, there are few reports detailing putative associations between variants in specific candidate dyslexia susceptibility genes and morphological changes in the brain. However, one recent study of a population of undiagnosed (i.e., not dyslexic) French adults demonstrated that a genomic region containing a variant of KIAA0319—a region found to be associated with dyslexia—significantly correlated with reduced functional symmetry in the superior temporal sulcus during a speech listening task (Pinel et al., 2012). This led the authors to hypothesize that KIAA0319 may play a role in cortical lateralization of phonological processing abilities in both typical and impaired populations. Given that functional asymmetry in the superior temporal sulcus is present at birth in humans, it also seems possible that any effects of KIAA0319 on the functional neuroanatomy of this region would be established early in development. Moreover, based on the superior temporal sulcus’ known role in language, these neuroanatomic findings are complementary to other reports detailing the influence of KIAA0319 variants on language-related behavioral traits in both dyslexic and control populations (Paracchini et al., 2008; Scerri et al., 2011).
Jamadar and colleagues reported on another clinical study that has investigated the relationship between specific KIAA0319 variants and brain morphology. In their sample of control and schizophrenic patients, they found significant associations between dyslexia-related KIAA0319 variants and gray matter volume in the inferior cerebellum, but not in the cortex, in both controls and schizophrenic individuals (2011). Specifically, a negative correlation was detected between the KIAA0319 variant and cerebellar networks, suggesting that the genetic variant was related to reductions in cerebellar size. This is an interesting finding given the cerebellum’s putative role in dyslexia, and reports of cerebellar structural abnormalities in dyslexic individuals (Stoodley and Stein, 2011). The latter supports the view that KIAA0319 variants may differentially influence the structure of language-related regions in the brain.
5. CONCLUSIONS
We report novel findings of reduced midsagittal area of the corpus callosum in adult male rats following embryonic knockdown of Kiaa0319. The specific effects on the corpus callosum in the absence of volumetric anomalies the neocortex or hippocampus—which is in contrast to observations from other animal models of developmental brain disruption— suggest that this change may be due to specific genetic effects rather than a “side effect” or epiphenomena of disruption to neuronal migration. Kiaa0319 knockdown has also been shown to specifically influence rapid auditory processing abilities in rats, which is interesting given clinical literature that suggests that reduced corpus callosum size is related to deficits in acoustic temporal processing. The neurological phenotype associated with KIAA0319 variants in humans remains unclear, although preliminary evidence suggests that they are associated with changes in language-related brain structures. Further studies examining associations between brain morphology and CDSG variants are needed, and future work in our lab will continue to characterize the effects of knocking down other rodent homologs of CDSGs on rodent brain structure.
Highlights.
We replicate reports that interference with Kiaa0319 disrupts neuronal migration in rodents.
Kiaa0319 interference leads specific reductions in midsagittal area of the corpus callosum.
Kiaa0319 interference did not lead to changes in the size of the cortex of hippocampus.
Footnotes
Abbreviations: CDSG: candidate dyslexia susceptibility gene; RAP: rapid auditory processing; RNAi: RNA interference; shRNA: short hairpin RNA; RFP: red fluorescent protein; GFP: green fluorescent protein
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aboitiz F, Scheibel AB, Fisher RS, Zaidel E. Individual differences in brain asymmetries and fiber composition in the human corpus callosum. Brain Res. 1992;598:154–161. doi: 10.1016/0006-8993(92)90179-d. [DOI] [PubMed] [Google Scholar]
- Anthoni H, Sucheston LE, Lewis BA, Tapia-Paez I, Fan X, Zucchelli M, Taipale M, Stein CM, Hokkanen ME, Castren E, Pennington BF, Smith SD, Olson RK, Tomblin JB, Schulte-Korne G, Nothen M, Schumacher J, Muller-Myhsok B, Hoffmann P, Gilger JW, Hynd GW, Nopola-Hemmi J, Leppanen PH, Lyytinen H, Schoumans J, Nordenskjold M, Spencer J, Stanic D, Boon WC, Simpson E, Makela S, Gustafsson JA, Peyrard-Janvid M, Iyengar S, Kere J. The aromatase gene CYP19A1: Several genetic and functional lines of evidence supporting a role in reading, speech and language. Behav Genet. 2012 doi: 10.1007/s10519-012-9532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beneventi H, Tonnessen FE, Ersland L, Hugdahl K. Executive working memory processes in dyslexia: Behavioral and fMRI evidence. Scand J Psychol. 2010;51:192–202. doi: 10.1111/j.1467-9450.2010.00808.x. [DOI] [PubMed] [Google Scholar]
- Brambati SM, Termine C, Ruffino M, Stella G, Fazio F, Cappa SF, Perani D. Regional reductions of gray matter volume in familial dyslexia. Neurology. 2004;63:742–745. doi: 10.1212/01.wnl.0000134673.95020.ee. [DOI] [PubMed] [Google Scholar]
- Burbridge TJ, Wang Y, Volz AJ, Peschansky VJ, Lisann L, Galaburda AM, LoTurco JJ, Rosen GD. Postnatal analysis of the effect of embryonic knockdown and overexpression of candidate dyslexia susceptibility gene homolog Dcdc2 in the rat. Neuroscience. 2008;152:723–733. doi: 10.1016/j.neuroscience.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JC, Lanham DC, Cutting LE, Clements-Stephens AM, Chen X, Hadzipasic M, Kim J, Denckla MB, Kaufmann WE. A dual DTI approach to analyzing white matter in children with dyslexia. Psychiatry Res. 2009;172:215–219. doi: 10.1016/j.pscychresns.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Mimran R, Sapir S. Auditory temporal processing deficits in children with reading disabilities. Dyslexia. 2007;13:175–192. doi: 10.1002/dys.323. [DOI] [PubMed] [Google Scholar]
- Cope N, Harold D, Hill G, Moskvina V, Stevenson J, Holmans P, Owen MJ, O’Donovan MC, Williams J. Strong evidence that KIAA0319 on chromosome 6p is a susceptibility gene for developmental dyslexia. Am J Hum Genet. 2005;76:581–591. doi: 10.1086/429131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deffenbacher KE, Kenyon JB, Hoover DM, Olson RK, Pennington BF, DeFries JC, Smith SD. Refinement of the 6p21.3 quantitative trait locus influencing dyslexia: Linkage and association analyses. Hum Genet. 2004;115:128–138. doi: 10.1007/s00439-004-1126-6. [DOI] [PubMed] [Google Scholar]
- Duara R, Kushch A, Gross-Glenn K, Barker WW, Jallad B, Pascal S, Loewenstein DA, Sheldon J, Rabin M, Levin B. Neuroanatomic differences between dyslexic and normal readers on magnetic resonance imaging scans. Arch Neurol. 1991;48:410–416. doi: 10.1001/archneur.1991.00530160078018. [DOI] [PubMed] [Google Scholar]
- Elbert A, Lovett MW, Cate-Carter T, Pitch A, Kerr EN, Barr CL. Genetic variation in the KIAA0319 5′ region as a possible contributor to dyslexia. Behav Genet. 2011;41:77–89. doi: 10.1007/s10519-010-9434-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elnakib A, Casanova M, Gimelfarb G, Switala A, El-Baz A. Dyslexia diagnostics by 3D shape analysis of the corpus callosum. Trans Inf Technol Biomed. 2012 doi: 10.1109/TITB.2012.2187302. [DOI] [PubMed] [Google Scholar]
- Fine JG, Semrud-Clikeman M, Keith TZ, Stapleton LM, Hynd GW. Reading and the corpus callosum: An MRI family study of volume and area. Neuropsychology. 2007;21:235–241. doi: 10.1037/0894-4105.21.2.235. [DOI] [PubMed] [Google Scholar]
- Fitch RH, Szalkowski CE. Using animal models to dissociate genetic, neural, and behavioral contributors to language disability. In: Benasich AA, Fitch RH, editors. Developmental dyslexia: Early precursors, neurobehavioral markers, and biological substrates. Baltimore: Brookes Publishing Company; 2012. [Google Scholar]
- Franceschini S, Gori S, Ruffino M, Pedrolli K, Facoetti A. A causal link between visual spatial attention and reading acquisition. 2012;22:814–819. doi: 10.1016/j.cub.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Francks C, Paracchini S, Smith SD, Richardson AJ, Scerri TS, Cardon LR, Marlow AJ, MacPhie IL, Walter J, Pennington BF, Fisher SE, Olson RK, DeFries JC, Stein JF, Monaco AP. A 77-kilobase region of chromosome 6p22.2 is associated with dyslexia in families from the united kingdom and from the united states. Am J Hum Genet. 2004;75:1046–1058. doi: 10.1086/426404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrieli JE, Norton E. Reading abilities: Importance of visual-spatial attention. 2012;22:R298–R299. doi: 10.1016/j.cub.2012.03.041. [DOI] [PubMed] [Google Scholar]
- Galaburda AM, Eidelberg D. Symmetry and asymmetry in the human posterior thalamus. II Thalamic lesions in a case of developmental dyslexia. Arch Neurol. 1982;39:333–336. doi: 10.1001/archneur.1982.00510180011002. [DOI] [PubMed] [Google Scholar]
- Galaburda AM, Menard MT, Rosen GD. Evidence for aberrant auditory anatomy in developmental dyslexia. Proc Natl Acad Sci USA. 1994;91:8010–8013. doi: 10.1073/pnas.91.17.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda AM, Sherman GF, Rosen GD, Aboitiz F, Geschwind N. Developmental dyslexia: Four consecutive patients with cortical anomalies. Ann Neurol. 1985;18:222–233. doi: 10.1002/ana.410180210. [DOI] [PubMed] [Google Scholar]
- Gathercole SE, Alloway TP, Willis C, Adams A. Working memory in children with reading disabilities. J Exp Child Psychol. 2006;93:265–281. doi: 10.1016/j.jecp.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Hamalainen JA, Salminen HK, Leppanen PH. Basic auditory processing deficits in dyslexia: Systematic review of the behavioral and event-related Potential/Field evidence. J Learn Disabil. 2012 doi: 10.1177/0022219411436213. [DOI] [PubMed] [Google Scholar]
- Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kaariainen H, Kere J. The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet. 2005;1:e50. doi: 10.1371/journal.pgen.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan KM, Molfese DL, Walimuni IS, Stuebing KK, Papanicolaou AC, Narayana PA, Fletcher JM. Diffusion tensor quantification and cognitive correlates of the macrostructure and microstructure of the corpus callosum in typically developing and dyslexic children. NMR Biomed. 2012 doi: 10.1002/nbm.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeft F, McCandliss BD, Black JM, Gantman A, Zakerani N, Hulme C, Lyytinen H, Whitfield-Gabrieli S, Glover GH, Reiss AL, Gabrieli JD. Neural systems predicting long-term outcome in dyslexia. Proc Natl Acad Sci USA. 2011;108:361–366. doi: 10.1073/pnas.1008950108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys P, Kaufmann WE, Galaburda AM. Developmental dyslexia in women: Neuropathological findings in three patients. Ann Neurol. 1990;28:727–738. doi: 10.1002/ana.410280602. [DOI] [PubMed] [Google Scholar]
- Ibraghimov-Beskrovnaya O, Bukanov NO, Donohue LC, Dackowski WR, Klinger KW, Landes GM. Strong homophilic interactions of the Ig-like domains of polycystin-1, the protein product of an autosomal dominant polycystic kidney disease gene, PKD1. Hum Mol Genet. 2000;9:1641–1649. doi: 10.1093/hmg/9.11.1641. [DOI] [PubMed] [Google Scholar]
- Jamadar S, Powers NR, Meda SA, Gelernter J, Gruen JR, Pearlson GD. Genetic influences of cortical gray matter in language-related regions in healthy controls and schizophrenia. Schizophr Res. 2011;129:141–148. doi: 10.1016/j.schres.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian S, Brown WS, Hallam BJ, McMahon W, Lu J, Johnson M, Bigler ED, Lainhart J. Regional callosal morphology in autism and macrocephaly. Dev Neuropsychol. 2008;33:74–99. doi: 10.1080/87565640701729821. [DOI] [PubMed] [Google Scholar]
- Kovelman I, Norton ES, Christodoulou JA, Gaab N, Lieberman DA, Triantafyllou C, Wolf M, Whitfield-Gabrieli S, Gabrieli JD. Brain basis of phonological awareness for spoken language in children and its disruption in dyslexia. Cereb Cortex. 2012;22:754–764. doi: 10.1093/cercor/bhr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronbichler M, Wimmer H, Staffen W, Hutzler F, Mair A, Ladurner G. Developmental dyslexia: Gray matter abnormalities in the occipitotemporal cortex. Hum Brain Mapp. 2008;29:613–625. doi: 10.1002/hbm.20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard CM, Eckert MA, Lombardino LJ, Oakland T, Kranzler J, Mohr CM, King WM, Freeman A. Anatomical risk factors for phonological dyslexia. Cereb Cortex. 2001;11:148–157. doi: 10.1093/cercor/11.2.148. [DOI] [PubMed] [Google Scholar]
- Livingstone M, Rosen G, Drislane F, Galaburda A. Physiological and anatomical evidence for a magnocellular defect in developmental dyslexia. Proceedings of the National Academy of Sciences (USA) 1991;88:7943–7947. doi: 10.1073/pnas.88.18.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson H, Tammimies K, Zucchelli M, Anthoni H, Onkamo P, Nopola-Hemmi J, Lyytinen H, Leppanen PH, Neuhoff N, Warnke A, Schulte-Korne G, Schumacher J, Nothen MM, Kere J, Peyrard-Janvid M. SNP variations in the 7q33 region containing DGKI are associated with dyslexia in the finnish and german populations. Behav Genet. 2011;41:134–140. doi: 10.1007/s10519-010-9431-4. [DOI] [PubMed] [Google Scholar]
- Melby-Lervag M, Lyster SA, Hulme C. Phonological skills and their role in learning to read: A meta-analytic review. Psychol Bull. 2012;138:322–352. doi: 10.1037/a0026744. [DOI] [PubMed] [Google Scholar]
- Meng H, Smith SD, Hager K, Held M, Liu J, Olson RK, Pennington BF, DeFries JC, Gelernter J, O’Reilly-Pol T, Somlo S, Skudlarski P, Shaywitz SE, Shaywitz BA, Marchione K, Wang Y, Paramasivam M, LoTurco JJ, Page GP, Gruen JR. DCDC2 is associated with reading disability and modulates neuronal development in the brain. Proc Natl Acad Sci USA. 2005;102:17053–17058. doi: 10.1073/pnas.0508591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menghini D, Carelsimo GA, Marotta L, Finzi A, Vicari S. Developmental dyslexia and explicit long-term memory. Dyslexia. 2010;16:213–225. doi: 10.1002/dys.410. [DOI] [PubMed] [Google Scholar]
- Menghini D, Finzi A, Carelsimo GA, Vicari S. Working memory impairment in children with developmental dyslexia: Is it just a phonological deficity? Dev Neuropsychol. 2011;36:199–213. doi: 10.1080/87565641.2010.549868. [DOI] [PubMed] [Google Scholar]
- Menghini D, Hagberg GE, Petrosini L, Bozzali M, Macaluso E, Caltagirone C, Vicari S. Structural correlates of implicit learning deficits in subjects with developmental dyslexia. Ann NY Acad Sci. 2008;1145:212–221. doi: 10.1196/annals.1416.010. [DOI] [PubMed] [Google Scholar]
- Newbury AJ, Rosen GD. Genetic, morphometric, and behavioral factors linked to the midsagittal area of the corpus callosum. Front Genet. 2012;3:91. doi: 10.3389/fgene.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Paracchini S, Scerri TS, Winchester L, Addis L, Richardson AJ, Walter J, Stein JF, Talcott JB, Monaco AP. Investigation of dyslexia and SLI risk variants in reading- and language-impaired subjects. Behav Genet. 2011;41:90–104. doi: 10.1007/s10519-010-9424-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paracchini S, Steer CD, Buckingham LL, Morris AP, Ring S, Scerri T, Stein J, Pembrey ME, Ragoussis J, Golding J, Monaco AP. Association of the KIAA0319 dyslexia susceptibility gene with reading skills in the general population. Am J Psychiatry. 2008;165:1576–1584. doi: 10.1176/appi.ajp.2008.07121872. [DOI] [PubMed] [Google Scholar]
- Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, Francks C, Richardson AJ, Wade-Martins R, Stein JF, Knight JC, Copp AJ, Loturco J, Monaco AP. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–1666. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- Paul LK. Developmental malformation of the corpus callosum: A review of typical callosal development and examples of developmental disorders with callosal involvement. J Neurodev Disord. 2011;3:3–27. doi: 10.1007/s11689-010-9059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 6. Amsterdam: Elsevier; 2007. [DOI] [PubMed] [Google Scholar]
- Peiffer AM, Fitch RH, Thomas JJ, Yurkovic AN, Rosen GD. Brain weight differences associated with induced focal microgyria. BMC Neurosci. 2003;4:12. doi: 10.1186/1471-2202-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschansky VJ, Burbridge TJ, Volz AJ, Fiondella C, Wissner-Gross Z, Galaburda AM, LoTurco JJ, Rosen GD. The effect of variation in expression of the candidate dyslexia susceptibility gene homolog Kiaa0319 on neuronal migration and dendritic morphology in the rat. Cereb Cortex. 2010;20:884–897. doi: 10.1093/cercor/bhp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RL, Pennington BF. Developmental dyslexia [Google Scholar]
- Peyrin C, Lallier M, Demonet JF, Pernet C, Baciu M, Le Bas JF, Valdois S. Neural dissociation of phonological and visual attention span disorders in developmental dyslexia: FMRI evidence from two case reports. Brain Lang. 2012;120:381–394. doi: 10.1016/j.bandl.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Pinel P, Fauchereau F, Moreno A, Barbot A, Lathrop M, Zelenika D, Le Bihan D, Poline JB, Bourgeron T, Dehaene S. Genetic variants of FOXP2 and KIAA0319/TTRAP/THEM2 locus are associated with altered brain activation in distinct language-related regions. J Neurosci. 2012;32:817–825. doi: 10.1523/JNEUROSCI.5996-10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelmans G, Engelen JJ, Van Lent-Albrechts J, Smeets HJ, Schoenmakers E, Franke B, Buitelaar JK, Wuisman-Frerker M, Erens W, Steyaert J, Schrander-Stumpel C. Identification of novel dyslexia candidate genes through the analysis of a chromosomal deletion. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:140–147. doi: 10.1002/ajmg.b.30787. [DOI] [PubMed] [Google Scholar]
- Pugh KR, Mencl WE, Jenner AR, Katz L, Frost SJ, Lee JR, Shaywitz SE, Shaywitz BA. Functional neuroimaging studies of reading and reading disability (developmental dyslexia) Ment Retard Dev Disabil Res Rev. 2000;6:207–213. doi: 10.1002/1098-2779(2000)6:3<207::AID-MRDD8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Raschle NM, Chang M, Gaab N. Structural brain alterations associated with dyslexia predate reading onset. Neuroimage. 2011;57:742–749. doi: 10.1016/j.neuroimage.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice ML, Smith SD, Gayan J. Convergent genetic linkage and associations to language, speech and reading measures in families of probands with specific language impairment. J Neurodev Disord. 2009;1:264–282. doi: 10.1007/s11689-009-9031-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimrodt SL, Peterson DJ, Denckla MB, Kaufmann WE, Cutting LE. White matter microstructural differences linked to left perisylvian language network in children with dyslexia. Cortex. 2010;46:739–749. doi: 10.1016/j.cortex.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen GD, Harry JD. Brain volume estimation from serial section measurements: A comparison of methodologies. J Neurosci. 1990;35:115–124. doi: 10.1016/0165-0270(90)90101-k. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Sherman GF, Emsbo K, Mehler C, Galaburda AM. The midsagittal area of the corpus callosum and total neocortical volume differ in three inbred strains of mice. Exp Neurol. 1990;107:271–276. doi: 10.1016/0014-4886(90)90145-i. [DOI] [PubMed] [Google Scholar]
- Rosen GD, Bai J, Wang Y, Fiondella CG, Threlkeld SW, LoTurco JJ, Galaburda AM. Disruption of neuronal migration by RNAi of Dyx1c1 results in neocortical and hippocampal malformations. Cerebral Cortex. 2007;17:2562–2572. doi: 10.1093/cercor/bhl162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumsey JM, Casanova M, Mannheim GB, Patronas N, De Vaughn N, Hamburger SD, Aquino T. Corpus callosum morphology, as measured with MRI, in dyslexic men. Biol Psychiatry. 1996;39:769–775. doi: 10.1016/0006-3223(95)00225-1. [DOI] [PubMed] [Google Scholar]
- Scerri TS, Morris AP, Buckingham LL, Newbury DF, Miller LL, Monaco AP, Bishop DV, Paracchini S. DCDC2, KIAA0319 and CMIP are associated with reading-related traits. Biol Psychiatry. 2011;70:237–245. doi: 10.1016/j.biopsych.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scerri TS, Paracchini S, Morris A, MacPhie IL, Talcott J, Stein J, Smith SD, Pennington BF, Olson RK, DeFries JC, Monaco AP, Richardson AJ. Identification of candidate genes for dyslexia susceptibility on chromosome 18. PLoS One. 2010;5:e13712. doi: 10.1371/journal.pone.0013712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siok WT, Niu Z, Jin Z, Perfetti CA, Tan LH. A structural-functional basis for dyslexia in the cortex of chinese readers. Proc Natl Acad Sci USA. 2008;105:5561–5566. doi: 10.1073/pnas.0801750105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Spark JH, Fisk JE. Working memory functioning in developmental dyslexia. Memory. 2007;15:34–56. doi: 10.1080/09658210601043384. [DOI] [PubMed] [Google Scholar]
- Stoodley CJ, Stein JF. The cerebellum and dyslexia. Cortex. 2011;47:101–116. doi: 10.1016/j.cortex.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Sun YF, Lee JS, Kirby R. Brain imaging findings in dyslexia. Pediatr Neonatol. 2010;51:89–96. doi: 10.1016/S1875-9572(10)60017-4. [DOI] [PubMed] [Google Scholar]
- Szalkowski CE, Fiondella CG, Galaburda AM, Rosen GD, Loturco JJ, Fitch RH. Neocortical disruption and behavioral impairments in rats following in utero RNAi of candidate dyslexia risk gene Kiaa0319. Int J Dev Neurosci. 2012;30:293–302. doi: 10.1016/j.ijdevneu.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalkowski CE, Hinman JR, Threlkeld SW, Wang Y, LePack A, Rosen GD, Chrobak JJ, LoTurco JJ, Fitch RH. Persistent spatial working memory deficits in rats following in utero RNAi of Dyx1c110. 2011:244–252. doi: 10.1111/j.1601-183X.2010.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Kaminen N, Nopola-Hemmi J, Haltia T, Myllyluoma B, Lyytinen H, Muller K, Kaaranen M, Lindsberg PJ, Hannula-Jouppi K, Kere J. A candidate gene for developmental dyslexia encodes a nuclear tetratricopeptide repeat domain protein dynamically regulated in brain. Proc Natl Acad Sci USA. 2003;100:11553–11558. doi: 10.1073/pnas.1833911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlkeld SW, Hill CA, Cleary CE, Truong DT, Rosen GD, Fitch RH. Developmental learning impairments in a rodent model of nodular heterotopia. J Neurodev Disord. 2009;1:237–250. doi: 10.1007/s11689-009-9026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlkeld SW, McClure MM, Bai J, Wang Y, LoTurco JJ, Rosen GD, Fitch RH. Developmental disruptions and behavioral impairments in rats following in utero RNAi of Dyx1c1. Brain Res Bull. 2007;71:508–514. doi: 10.1016/j.brainresbull.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermosten M, Boets B, Luts H, Poelmans H, Wouters J, Ghesquiere P. Impairments in speech and nonspeech sound categorization in children with dyslexia are driven by temporal processing difficulties. Res Dev Disabil. 2011;32:593–603. doi: 10.1016/j.ridd.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Vandermosten M, Boets B, Wouters J, Ghesquiere P. A qualitative and quantitative review of diffusion tensor imaging studies in reading and dyslexia. Neurosci Biobehav Rev. 2012;36:1532–1552. doi: 10.1016/j.neubiorev.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Velayos-Baeza A, Levecque C, Kobayashi K, Holloway ZG, Monaco AP. The dyslexia-associated KIAA0319 protein undergoes proteolytic processing with {gamma}-secretase-independent intramembrane cleavage. J Biol Chem. 2010;285:40148–40162. doi: 10.1074/jbc.M110.145961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayos-Baeza A, Toma C, da Roza S, Paracchini S, Monaco AP. Alternative splicing in the dyslexia-associated gene KIAA0319. Mamm Genome. 2007;18:627–634. doi: 10.1007/s00335-007-9051-3. [DOI] [PubMed] [Google Scholar]
- Velayos-Baeza A, Toma C, Paracchini S, Monaco AP. The dyslexia-associated gene KIAA0319 encodes highly N- and O-glycosylated plasma membrane and secreted isoforms. Hum Mol Genet. 2008;17:859–871. doi: 10.1093/hmg/ddm358. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch E, Robichon F, Eliez S. Gray matter alteration in dyslexia: Converging evidence from volumetric and voxel-by-voxel MRI analyses. Neuropsychologia. 2005;43:324–331. doi: 10.1016/j.neuropsychologia.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Wang Y, Paramasivam M, Thomas A, Bai J, Kaminen-Ahola N, Kere J, Voskuil J, Rosen GD, Galaburda AM, Loturco JJ. DYX1C1 functions in neuronal migration in developing neocortex. Neuroscience. 2006;143:515–522. doi: 10.1016/j.neuroscience.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Wolf RC, Sambataro F, Lohr C, Steinbrink C, Martin C, Vasic N. Functional brain network abnormalities during verbal working memory performance in adolescents and young adults with dyslexia. Neuropsychologia. 2010;48:309–318. doi: 10.1016/j.neuropsychologia.2009.09.020. [DOI] [PubMed] [Google Scholar]